Organoids, Biocybersecurity, and Cyberbiosecurity—A Light Exploration
Abstract
1. Introduction
1.1. Enter Organoids
1.2. On Biocomputing and Organoids
1.3. On Cybersecurity Interfacing with Organoids
1.4. Into Biocybersecurity and Cyberbiosecurity
1.5. On Integration of These Areas
2. Methods
3. Some Organoid Advances
3.1. Diversification of Organoid Types
3.2. More Applications in Research and Medicine
3.3. Breakthroughs, Important Methods Integrated, and Design
4. Organoids and Biocomputing
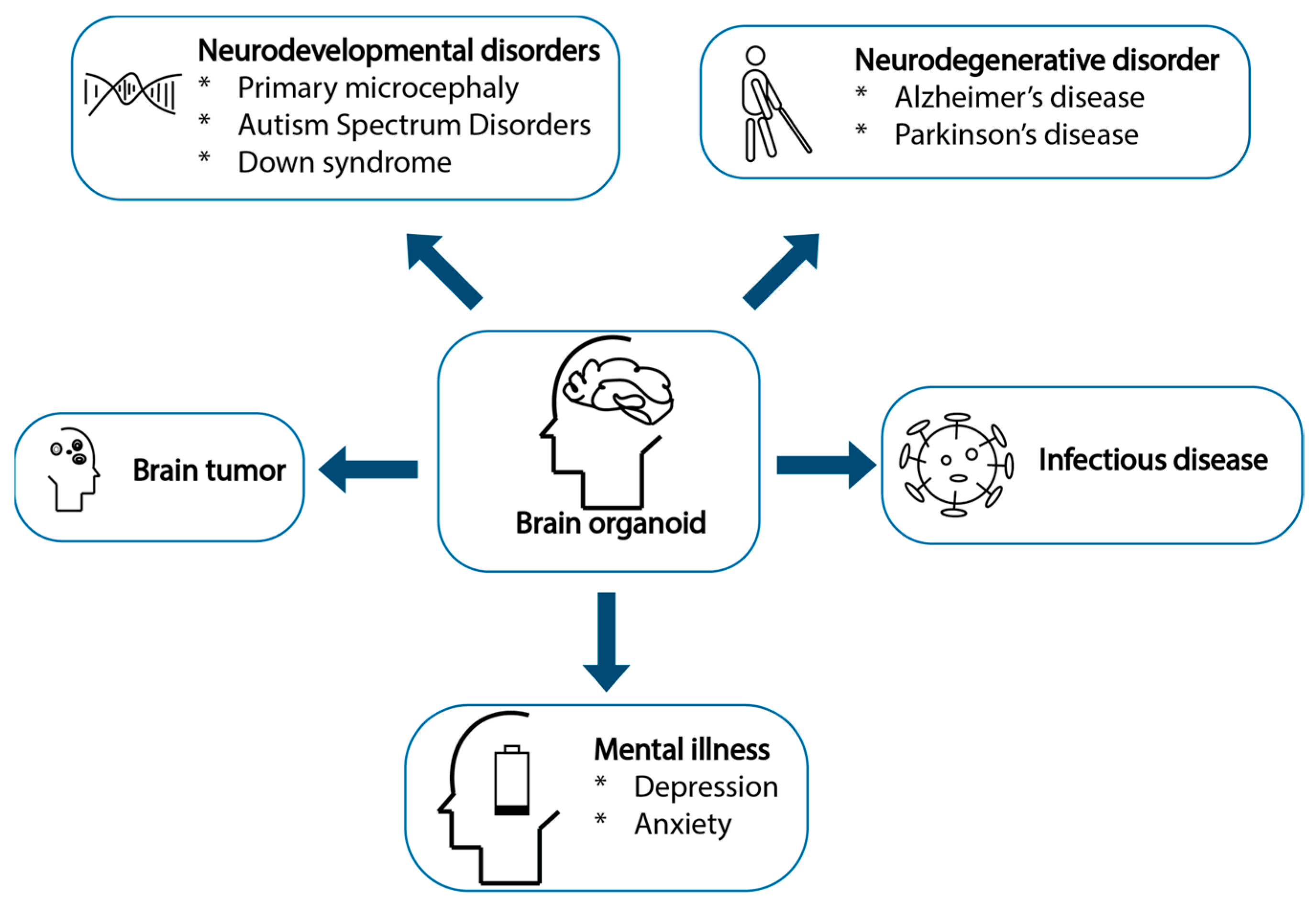
4.1. Brain Organoids: Uses from Now and Potentially into the Future
4.2. On Some Approaches Integrating Organoids in Biocomputing
4.3. Transformative Impacts of Organoids in Biocomputing
4.4. Biocomputing and Organoid Intelligence
4.4.1. Towards Developments in Biocomputing and Organoid Intelligence
4.4.2. Case Studies on Biocomputing and Organoid Intelligence Applications
4.4.3. A Hypothetical Case: Organoids through the Lens of Both Biocybersecurity and Cyberbiosecurity
4.4.4. Organoid Intelligence and Machine Learning Applications
5. Ethical Considerations and Future Prospects
5.1. On Ethical Concerns
5.2. Consent Models for Organoid Research
5.3. Monetization of Organoid Research
5.4. Possible Connections between AI-Driven Cyberattacks and Organoids in Timelines
5.4.1. Organoid Development or Process Manipulation (Research and Development)
5.4.2. Theft of Sensitive and or Otherwise Personal Data
5.4.3. Cases of AI Cyberattacks Focused on Organoids
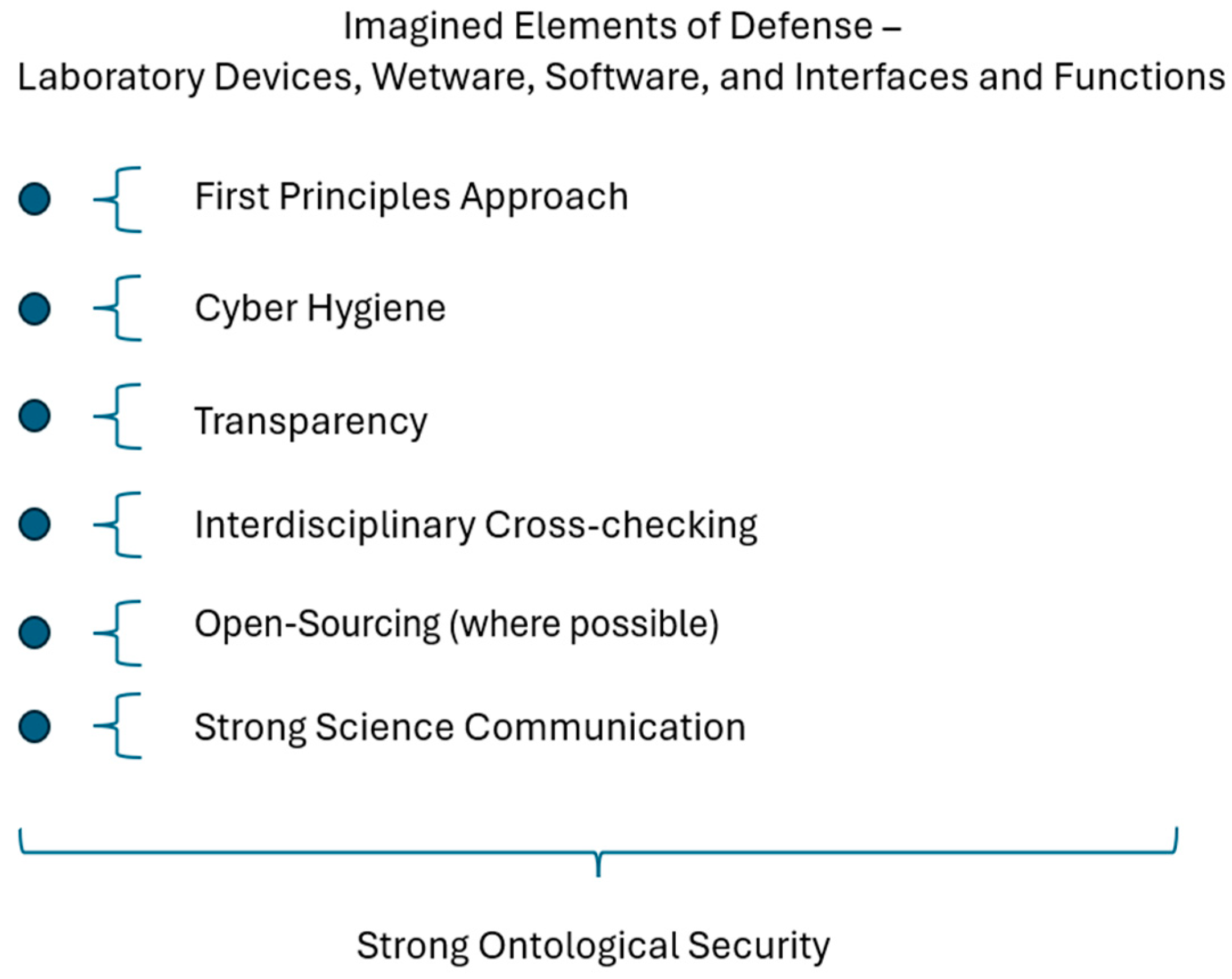
5.4.4. Imagined Defenses
Imagined Defenses for All of the Above: First Principles Approach and Cyber Hygiene
Transparency, Open Sourcing Where Possible, and Interdisciplinary Cross-Checking
- More precise and secure equipment
- ○
- Less reliance on less-maintained hardware and software prone to leak information
- More teams are able to investigate the same core phenomena related to the development, function, and or use of organoids
- ○
- Greater redundancy gives greater portals to available variances in research outcomes and makes it harder for malicious actors to bottleneck and poison avenues of research
- More teams are willing to publish negative results, as they can view the endeavor as productive
- Create a workload that is more feasible and allows for flexibility; this alone has multiple benefits
- ○
- More time on fewer tasks, allowing for more quality work
- ⯀
- More replications and deeper insights
- ⯀
- Fewer mistakes generated through exhaustion
- ⯀
- A greater degree of cyber hygiene
- ○
- A greater degree of collaboration across the globe and between institutions
- ⯀
- Less fear of “scooping” taking research not originally created)
- ⯀
- More shared resources
- Along with greater eyes on resource misuse
- ⯀
- More perspectives shared on pivotal areas of this research
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Glossary
| Term APT | Definition Advanced persistent threat [195]. |
| Amyloid beta | Amyloid beta is produced via the proteolytic cleavage of a transmembrane protein, an amyloid precursor protein (APP), by enzymes called β- and γ-secretases. It is proposed to be an early detrimental occurrence in the development of Alzheimer’s disease [196]. |
| Biocomputing | Biocomputing is an innovative technological domain that operates where biology, engineering, and computer science intersect and aims to utilize cells or their molecular components (such as DNA or RNA) to perform tasks traditionally performed by an electronic computer [197]. |
| Biocybersecurity (BCS) | Biocybersecurity is the intersection of biotechnology, biosecurity, and cybersecurity issues and is part of an initiative aimed at protecting the bioeconomy [198]. |
| Bioinformatics | Bioinformatics is a scientific subdiscipline that involves using computer technology to collect, store, analyze, and distribute biological data and information [199]. |
| CRISPR/Cas9 | CRISPR/Cas9 is a gene editing technology that involves two components: a guide RNA to match a desired target gene and Cas9- an endonuclease that causes a double-stranded DNA break, allowing modifications to the genome [200]. |
| Cyberbiosecurity (CBS) | Cyberbiosecurity refers to attacks originating in hardware and computer systems with a focus on biological or biology-centric systems. A useful primer is presented in past literature [45]. |
| Human Genome Project | The Human Genome Project is an international research project whose main objective is to decode the chemical sequence of the entire human genetic material (genome), identify the 50,000 to 100,000 genes encompassed within it, and provide research tools for the analysis of genetic information [201]. |
| Induced pluripotent stem cells (iPSCs) | Induced pluripotent stem cells are derived from adult somatic cells that have been reprogrammed back into an embryonic-like pluripotent state that enables the development of an unlimited source of any type of human cell needed for therapeutic purposes [8,9,92,100,110,202,203]. |
| In silico | In silico is experimentation performed by computational models that explore pharmacological hypotheses through various methods, including databases, data analysis tools, data mining, homology models, machine learning, pharmacophores, quantitative structure-activity relationships, and network analysis tools [17,204]. |
| Organoid | An organoid is a self-organized 3D tissue that is derived from stem cells (pluripotent, fetal, or adult), which imitate the functional, structural, and biological complexity of an organ [205]. |
| Organoid intelligence (OI) | OI is an emerging interdisciplinary scientific field aiming to establish a new type of biological computing system using 3D cultures of human brain cells (brain organoids) and brain-machine interface technologies [18]. |
| Pathology of tau | The Pathology of Tau are neurodegenerative disorder characterized by the accumulation of abnormal tau protein in the brain [185]. |
| Pluripotent stem cells | Pluripotent stem cells are cells that have the capacity to self-renew by dividing indefinitely, producing unaltered cell daughters maintaining the same properties as the parent cell [206], and are capable of differentiation into any cell type found in the human body |
| Primary microcephaly | Primary microcephaly is a disorder of brain development that results in a head circumference more than three standard deviations below the mean for age and gender [207]. |
| Raspberry Pi | Raspberry Pi is the name of a series of single-board computers made by the Raspberry Pi Foundation, a UK charity dedicated to enhancing computing education and accessibility [208,209]. |
References
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Primers 2022, 2, 94. [Google Scholar] [CrossRef]
- Corrò, C.; Novellasdemunt, L.; Li, V.S. A brief history of organoids. Am. J. Physiol.-Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Zimmermann, B. Lung organoid culture. Differentiation 1987, 36, 86–109. [Google Scholar] [CrossRef] [PubMed]
- Wörsdörfer, P.; I, T.; Asahina, I.; Sumita, Y.; Ergün, S. Do not keep it simple: Recent advances in the generation of complex organoids. J. Neural Transm. 2020, 127, 1569–1577. [Google Scholar] [CrossRef]
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Clevers, H. Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med. 2017, 95, 729–738. [Google Scholar] [CrossRef]
- Menche, C.; Farin, H.F. Strategies for genetic manipulation of adult stem cell-derived organoids. Exp. Mol. Med. 2021, 53, 1483–1494. [Google Scholar] [CrossRef]
- Kollmann, C.; Buerkert, H.; Meir, M.; Richter, K.; Kretzschmar, K.; Flemming, S.; Kelm, M.; Germer, C.-T.; Otto, C.; Burkard, N.; et al. Human organoids are superior to cell culture models for intestinal barrier research. Front. Cell Dev. Biol. 2023, 11, 1223032. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Patrinos, A.; Jordan, E.; Chakravarti, A.; Gesteland, R.; Walters, L.; members of the DOE and NIH planning groups. New goals for the US human genome project: 1998–2003. Science 1998, 282, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Morgan, M.; Patrinos, A. The Human Genome Project: Lessons from large-scale biology. Science 2003, 300, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Sherman, R.M.; Salzberg, S.L. Pan-genomics in the human genome era. Nat. Rev. Genet. 2020, 21, 243–254. [Google Scholar] [CrossRef]
- Goñi-Moreno, A.; Nikel, P.I. High-performance biocomputing in synthetic biology–integrated transcriptional and metabolic circuits. Front. Bioeng. Biotechnol. 2019, 7, 40. [Google Scholar] [CrossRef]
- Thalheim, T.; Aust, G.; Galle, J. Organoid Cultures in Silico: Tools or Toys? Bioengineering 2022, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Caffo, B.S.; Gracias, D.H.; Huang, Q.; Morales Pantoja, I.E.; Tang, B.; Zack, D.J.; Berlinicke, C.A.; Boyd, J.L.; Harris, T.D.; et al. Organoid intelligence (OI): The new frontier in biocomputing and intelligence-in-a-dish. Front. Sci. 2023, 1, 1017235. [Google Scholar] [CrossRef]
- Liu, S.; Kumari, S.; He, H.; Mishra, P.; Singh, B.N.; Singh, D.; Liu, S.; Srivastava, P.; Li, C. Biosensors integrated 3D organoid/organ-on-a-chip system: A real-time biomechanical, biophysical, and biochemical monitoring and characterization. Biosens. Bioelectron. 2023, 231, 115285. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Katz, E. Digital biosensors with built-in logic for biomedical applications—Biosensors based on a biocomputing concept. Anal. Bioanal. Chem. 2010, 398, 1591–1603. [Google Scholar] [CrossRef]
- Yousafzai, M.S.; Hammer, J.A. Using Biosensors to Study Organoids, Spheroids and Organs-on-a-Chip: A Mechanobiology Perspective. Biosensors 2023, 13, 905. [Google Scholar] [CrossRef]
- Öztatlı, H.; Altintas, Z.; Garipcan, B. Biosensors for organs-on-a-chip and organoids. In Advanced Sensor Technology; Elsevier: Amsterdam, The Netherlands, 2023; pp. 471–514. [Google Scholar]
- Susa, K.; Kobayashi, K.; Galichon, P.; Matsumoto, T.; Tamura, A.; Hiratsuka, K.; Gupta, N.R.; Yazdi, I.K.; Bonventre, J.V.; Morizane, R. ATP/ADP biosensor organoids for drug nephrotoxicity assessment. Front. Cell Dev. Biol. 2023, 11, 1138504. [Google Scholar] [CrossRef] [PubMed]
- de Barros, N.R.; Wang, C.; Maity, S.; Peirsman, A.; Nasiri, R.; Herland, A.; Ermis, M.; Kawakita, S.; Carvalho, B.G.; Kouchehbaghi, N.H.; et al. Engineered organoids for biomedical applications. Adv. Drug Deliv. Rev. 2023, 203, 115142. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Feidler, J.C. Biocompution: An emerging discipline at the intersection of the computational and biological sciences. In Proceedings of the 2002 IEEE International Conference on Acoustics, Speech, and Signal Processing, Orlando, FL, USA, 13–17 May 2002; IEEE: New York, NY, USA, 2022; Volume 4, p. IV-4028. [Google Scholar]
- Katz, E. Biocomputing—Tools, aims, perspectives. Curr. Opin. Biotechnol. 2015, 34, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Pitt, W.R.; Williams, M.A.; Steven, M.; Sweeney, B.; Bleasby, A.J.; Moss, D.S. The Bioinformatics Template Library—Generic components for biocomputing. Bioinformatics 2001, 17, 729–737. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bondiau, P.Y.; Konukoglu, E.; Clatz, O.; Delingette, H.; Frenay, M.; Paquis, P. Biocomputing: Numerical simulation of glioblastoma growth and comparison with conventional irradiation margins. Phys. Medica 2011, 27, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Smalley, E. Microsoft makes splash in AI-enabled lab solutions. Nat. Biotechnol. 2019, 37, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Belluati, A.; Jimaja, S.; Chadwick, R.J.; Glynn, C.; Chami, M.; Happel, D.; Guo, C.; Kolmar, H.; Bruns, N. Artificial cell synthesis using biocatalytic polymerization-induced self-assembly. Nat. Chem. 2023, 16, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Gotovtsev, P.M.; Kirillova, D.A.; Vasilov, R.G. Biocomputers: Problems They Solve, State of the Art, and Prospects. Nanotechnol. Russ. 2020, 15, 3–12. [Google Scholar] [CrossRef]
- Grozinger, L.; Amos, M.; Gorochowski, T.E.; Carbonell, P.; Oyarzún, D.A.; Stoof, R.; Fellermann, H.; Zuliani, P.; Tas, H.; Goñi-Moreno, A. Pathways to cellular supremacy in biocomputing. Nat. Commun. 2019, 10, 5250. [Google Scholar] [CrossRef]
- Mathur, M. Bioinformatics challenges: A review. Bioinformatics 2018, 3, 29–33. [Google Scholar]
- Pal, S.K.; Bandyopadhyay, S.; Ray, S.S. Evolutionary computation in bioinformatics: A review. IEEE Trans. Syst. Man Cybern. Part C Appl. Rev. 2006, 36, 601–615. [Google Scholar] [CrossRef]
- Perkhofer, L.; Frappart, P.O.; Müller, M.; Kleger, A. Importance of organoids for personalized medicine. Pers. Med. 2018, 15, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Magré, L.; Verstegen, M.M.; Buschow, S.; van der Laan, L.J.; Peppelenbosch, M.; Desai, J. Emerging organoid-immune co-culture models for cancer research: From oncoimmunology to personalized immunotherapies. J. ImmunoTherapy Cancer 2023, 11, e006290. [Google Scholar] [CrossRef]
- Gong, J.; Gong, Y.; Zou, T.; Zeng, Y.; Yang, C.; Mo, L.; Kang, J.; Fan, X.; Xu, H.; Yang, J. A controllable perfusion microfluidic chip for facilitating the development of retinal ganglion cells in human retinal organoids. Lab A Chip 2023, 23, 3820–3836. [Google Scholar] [CrossRef] [PubMed]
- Männik, J.; Teshima, T.F.; Wolfrum, B.; Yang, D. Lab-on-a-chip based mechanical actuators and sensors for single-cell and organoid culture studies. J. Appl. Phys. 2021, 129, 210905. [Google Scholar] [CrossRef]
- Angotzi, G.N.; Giantomasi, L.; Ribeiro, J.F.; Crepaldi, M.; Vincenzi, M.; Zito, D.; Berdondini, L. Integrated Micro-Devices for a Lab-in-Organoid Technology Platform: Current Status and Future Perspectives. Front. Neurosci. 2022, 16, 842265. [Google Scholar] [CrossRef]
- Sachs, D.M.; Costa, K.D. Robotic System for Organoid Assembly in a Multi-Well Microfluidic Chip. bioRxiv 2023. [Google Scholar] [CrossRef]
- Unagolla, J.M.; Jayasuriya, A.C. Recent advances in organoid engineering: A comprehensive review. Appl. Mater. Today 2022, 29, 101582. [Google Scholar] [CrossRef]
- Wu, J.; Hirai, Y.; Kamei, K.I.; Tsuchiya, T.; Tabata, O. Novel microfluidic device integrated with a fluidic-capacitor to mimic heart beating for generation of functional liver organoids. Electron. Commun. Jpn. 2019, 102, 41–49. [Google Scholar] [CrossRef]
- Zheng, B.; Ko, K.P.; Fang, X.; Wang, X.; Zhang, J.; Jun, S.; Kim, B.-J.; Luo, W.; Kim, M.J.; Jung, Y.-S.; et al. A new murine esophageal organoid culture method and organoid-based model of esophageal squamous cell neoplasia. iScience 2021, 24, 103440. [Google Scholar] [CrossRef]
- Mueller, S. On DNA signatures, their dual-use potential for GMO counterfeiting, and a cyber-based security solution. Front. Bioeng. Biotechnol. 2019, 7, 189. [Google Scholar] [CrossRef]
- Murch, R.S.; So, W.K.; Buchholz, W.G.; Raman, S.; Peccoud, J. Cyberbiosecurity: An emerging new discipline to help safeguard the bioeconomy. Front. Bioeng. Biotechnol. 2018, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Potter, L.; Palmer, X.L. Mission-Aware Differences in Cyberbiosecurity and Biocybersecurity Policies: Prevention, Detection, and Elimination. In Cyberbiosecurity; Springer: Cham, Switzerland, 2023; pp. 37–69. [Google Scholar]
- Potter, L.; Palmer, X.L. Human factors in biocybersecurity wargames. In Advances in Information and Communication: Proceedings of the 2021 Future of Information and Communication Conference (FICC); Springer International Publishing: Cham, Switzerland, 2021; Volume 1, pp. 666–673. [Google Scholar]
- DiEuliis, D.; Lutes, C.D.; Giordano, J. Biodata risks and synthetic biology: A critical juncture. J. Bioterror. Biodef. 2018, 9, 159. [Google Scholar] [CrossRef]
- Titus, A.J.; Hamilton, K.E.; Holko, M. Cyber and Information Security in the Bioeconomy. In Cyberbiosecurity; Springer: Cham, Switzerland, 2023; pp. 17–36. [Google Scholar]
- Pauwels, E. How to Protect Biotechnology and Biosecurity from Adversarial AI Attacks? A Global Governance Perspective. In Cyberbiosecurity; Springer: Cham, Switzerland, 2023; pp. 173–184. [Google Scholar]
- Bai, L.; Wu, Y.; Li, G.; Zhang, W.; Zhang, H.; Su, J. AI-enabled organoids: Construction, analysis, and application. Bioact. Mater. 2024, 31, 525–548. [Google Scholar] [CrossRef]
- Ceze, L.; Nivala, J.; Strauss, K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019, 20, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Ney, P.; Organick, L.; Nivala, J.; Ceze, L.; Kohno, T. DNA Sequencing Flow Cells and the Security of the Molecular-Digital Interface. Proc. Priv. Enhancing Technol. 2021, 2021, 413–432. [Google Scholar] [CrossRef]
- John, S.N.; Noma-Osaghae, E.; Oajide, F.; Okokpujie, K. Cybersecurity Education: The Skills Gap, Hurdle! Innov. Cybersecur. Educ. 2020, 361–376. [Google Scholar]
- Millett, K.; Dos Santos, E.; Millett, P.D. Cyber-biosecurity risk perceptions in the biotech sector. Front. Bioeng. Biotechnol. 2019, 7, 136. [Google Scholar] [CrossRef]
- Reed, J.C.; Dunaway, N. Cyberbiosecurity Implications for the Laboratory of the Future. Front. Bioeng. Biotechnol. 2019, 7, 182. [Google Scholar] [CrossRef]
- Trentesaux, C.; Yamada, T.; Klein, O.D.; Lim, W.A. Harnessing synthetic biology to engineer organoids and tissues. Cell Stem Cell 2023, 30, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Mead, B.E.; Safaee, H.; Langer, R.; Karp, J.M.; Levy, O. Engineering stem cell organoids. Cell Stem Cell 2016, 18, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Boutros, M.; Camp, J.G.; Clarke, L.; Clevers, H.; Knoblich, J.A.; Liberali, P.; Regev, A.; Rios, A.C.; Stegle, O.; et al. The organoid cell atlas. Nat. Biotechnol. 2021, 39, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Kratochvil, M.J.; Seymour, A.J.; Li, T.L.; Paşca, S.P.; Kuo, C.J.; Heilshorn, S.C. Engineered materials for organoid systems. Nat. Rev. Mater. 2019, 4, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G. Organoids as complex (bio) systems. Front. Cell Dev. Biol. 2023, 11, 1268540. [Google Scholar] [CrossRef] [PubMed]
- Nantasanti, S.; de Bruin, A.; Rothuizen, J.; Penning, L.C.; Schotanus, B.A. Concise review: Organoids are a powerful tool for the study of liver disease and personalized treatment design in humans and animals. Stem Cells Transl. Med. 2016, 5, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Kim, H.K.; Jee, J.; Hahn, S.; Jeong, S.; Yoo, J. Development of organoid-based drug metabolism model. Toxicol. Appl. Pharmacol. 2019, 385, 114790. [Google Scholar] [CrossRef] [PubMed]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170217. [Google Scholar] [CrossRef]
- Allam-Ndoul, B.; Castonguay-Paradis, S.; Veilleux, A. Gut microbiota and intestinal trans-epithelial permeability. Int. J. Mol. Sci. 2020, 21, 6402. [Google Scholar] [CrossRef]
- Steinway, S.N.; Saleh, J.; Koo, B.K.; Delacour, D.; Kim, D.H. Human microphysiological models of intestinal tissue and gut microbiome. Front. Bioeng. Biotechnol. 2020, 8, 725. [Google Scholar] [CrossRef]
- Günther, C.; Winner, B.; Neurath, M.F.; Stappenbeck, T.S. Organoids in gastrointestinal diseases: From experimental models to clinical translation. Gut 2022, 71, 1892–1908. [Google Scholar] [CrossRef]
- Richards, D.J.; Coyle, R.C.; Tan, Y.; Jia, J.; Wong, K.; Toomer, K.; Menick, D.R.; Mei, Y. Inspiration from heart development: Biomimetic development of functional human cardiac organoids. Biomaterials 2017, 142, 112–123. [Google Scholar] [CrossRef]
- Miyamoto, M.; Nam, L.; Kannan, S.; Kwon, C. Heart organoids and tissue models for modeling development and disease. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2021; Volume 118, pp. 119–128. [Google Scholar]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Aguirre, A. Heart organoids and engineered heart tissues: Novel tools for modeling human cardiac biology and disease. Biomolecules 2021, 11, 1277. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; van Blitterswijk, C.A.; LaPointe, V.L. Overcoming kidney organoid challenges for regenerative medicine. NPJ Regen. Med. 2020, 5, 8. [Google Scholar] [CrossRef]
- Stein, M.C.; Braun, F.; Krebs, C.F.; Bunders, M.J. Kidney organoid systems for studies of immune-mediated kidney diseases: Challenges and opportunities. Cell Tissue Res. 2021, 385, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Khoshdel Rad, N.; Aghdami, N.; Moghadasali, R. Cellular and molecular mechanisms of kidney development: From the embryo to the kidney organoid. Front. Cell Dev. Biol. 2020, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Astashkina, A.I.; Mann, B.K.; Prestwich, G.D.; Grainger, D.W. A 3-D organoid kidney culture model engineered for high-throughput nephrotoxicity assays. Biomaterials 2012, 33, 4700–4711. [Google Scholar] [CrossRef]
- Astashkina, A.I.; Jones, C.F.; Thiagarajan, G.; Kurtzeborn, K.; Ghandehari, H.; Brooks, B.D.; Grainger, D.W. Nanoparticle toxicity assessment using an in vitro 3-D kidney organoid culture model. Biomaterials 2014, 35, 6323–6331. [Google Scholar] [CrossRef]
- Kim, J.W.; Nam, S.A.; Seo, E.; Lee, J.Y.; Kim, D.; Ju, J.H.; Lin, S.W.; Kim, H.L.; Kim, H.W.; Yang, C.W.; et al. Human kidney organoids model the tacrolimus nephrotoxicity and elucidate the role of autophagy. Korean J. Intern. Med. 2021, 36, 1420. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Cao, Y.; Zhao, P.; Shen, S.; Xi, Y. Organoid: A powerful tool to study lung regeneration and disease. Cell Regen. 2021, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Choi, K.M.; Sicard, D.; Tschumperlin, D.J. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 2017, 113, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Green, R.; Howell, M.; Martinez, T.; Dutta, R.; Mohapatra, S.; Mohapatra, S.S. The design and characterization of a gravitational microfluidic platform for drug sensitivity assay in colorectal perfused tumoroid cultures. Nanomed. Nanotechnol. Biol. Med. 2020, 30, 102294. [Google Scholar] [CrossRef]
- Wang, T.; Wang, M.; Yang, L.; Li, Z.; Loh, X.J.; Chen, X. Cyber–physiochemical interfaces. Adv. Mater. 2020, 32, 1905522. [Google Scholar] [CrossRef] [PubMed]
- Tindle, C.; Fuller, M.; Fonseca, A.; Taheri, S.; Ibeawuchi, S.R.; Beutler, N.; Katkar, G.D.; Claire, A.; Castillo, V.; Hernandez, M.; et al. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. eLife 2021, 10, e66417. [Google Scholar] [CrossRef]
- Liberti, D.C.; Morrisey, E.E. Organoid models: Assessing lung cell fate decisions and disease responses. Trends Mol. Med. 2021, 27, 1159–1174. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Liu, J.; Wang, S.; Chang, M.; Wang, X.; Guo, B.; Yu, Q.; Yan, F.; Su, Y.; Wang, Y. Sweat gland organoids contribute to cutaneous wound healing and sweat gland regeneration. Cell Death Dis. 2019, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Peking, P.; Krisch, L.; Wolf, M.; Hoog, A.; Vári, B.; Muigg, K.; Poupardin, R.; Scharler, C.; Russe, E.; Stachelscheid, H.; et al. Self-assembly of progenitor cells under the aegis of platelet factors facilitates human skin organoid formation and vascularized wound healing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lee, J.; Koehler, K.R. Skin organoids: A new human model for developmental and translational research. Exp. Dermatol. 2021, 30, 613–620. [Google Scholar] [CrossRef]
- Sandoval, A.G.W.; Gim, K.Y.; Huang, J.T.; Koehler, K.R. Applications of Human Pluripotent Stem Cell–Derived Skin Organoids in Dermatology. J. Investig. Dermatol. 2023, 143, 1872–1876. [Google Scholar] [CrossRef] [PubMed]
- Shou, Y.; Liang, F.; Xu, S.; Li, X. The Application of Brain Organoids: From Neuronal Development to Neurological Diseases. Front. Cell Dev. Biol. 2020, 8, 579659. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e5. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Meng, X.; Liu, Y.; Song, D.; Jiang, C.; Cai, J. Applications of brain organoids in neurodevelopment and neurological diseases. J. Biomed. Sci. 2021, 28, 30. [Google Scholar] [CrossRef] [PubMed]
- Costamagna, G.; Comi, G.P.; Corti, S. Advancing drug discovery for neurological disorders using iPSC-derived neural organoids. Int. J. Mol. Sci. 2021, 22, 2659. [Google Scholar] [CrossRef]
- Spiller, E.R.; Ung, N.; Kim, S.; Patsch, K.; Lau, R.; Strelez, C.; Doshi, C.; Choung, S.; Choi, B.; Rosales, E.F.J.; et al. Imaging-based machine learning analysis of patient-derived tumor organoid drug response. Front. Oncol. 2021, 11, 771173. [Google Scholar] [CrossRef]
- Ho, C.; Morsut, L. Novel synthetic biology approaches for developmental systems. Stem Cell Rep. 2021, 16, 1051–1064. [Google Scholar] [CrossRef] [PubMed]
- Katcher, A.; Yueh, B.; Ozler, K.; Nizam, A.; Kredentser, A.; Chung, C.; Frimer, M.; Goldberg, G.L.; Beyaz, S. Establishing Patient-Derived Organoids from Human Endometrial Cancer and Normal Endometrium. Front. Endocrinol. 2023, 14, 1059228. [Google Scholar] [CrossRef]
- He, X.; Jiang, Y.; Zhang, L.; Li, Y.; Hu, X.; Hua, G.; Cai, S.; Mo, S.; Peng, J. Patient-Derived Organoids as a Platform for Drug Screening in Metastatic Colorectal Cancer. Front. Bioeng. Biotechnol. 2023, 11, 1190637. [Google Scholar] [CrossRef]
- Prasad, M.; Kumar, R.; Buragohain, L.; Kumari, A.; Ghosh, M. Organoid technology: A reliable developmental biology tool for organ-specific nanotoxicity evaluation. Front. Cell Dev. Biol. 2021, 9, 696668. [Google Scholar] [CrossRef]
- Takebe, T.; Wells, J.M. Organoids by design. Science 2019, 364, 956–959. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Chen, P.; Chang, R.; Liu, X.; Jhang, J.-W.; Enkhbat, M.; Chen, S.; Wang, H.; Deng, C.; Wang, P.-Y. Artificial tumor matrices and bioengineered tools for tumoroids. Biofabrication 2024, 16, 022004. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.S.; McInnis, M.G. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol. Cell. Neurosci. 2016, 73, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Lavazza, A.; Zilio, F. The Future of Human Cerebral Organoids: A Reply to Commentaries. AJOB Neurosci. 2023, 14, W1–W3. [Google Scholar] [CrossRef] [PubMed]
- News-Medical. Researchers Produce Artificial Cells with Potential for Drug Delivery and Tissue Engineering. [Online]. 2023. Available online: https://www.news-medical.net/news/20231211/Researchers-produce-artificial-cells-with-potential-for-drug-delivery-and-tissue-engineering.aspx (accessed on 16 March 2024).
- Lachance, J.-C.; Rodrigue, S.; Palsson, B.O. Minimal cells, maximal knowledge. eLife 2019, 8, e45379. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kowalczewski, A.; Vu, D.; Liu, X.; Salekin, A.; Yang, H.; Ma, Z. Organoid intelligence: Integration of organoid technology and artificial intelligence in the new era of in vitro models. Med. Nov. Technol. Devices 2024, 21, 100276. [Google Scholar] [CrossRef]
- Morales Pantoja, I.E.; Smirnova, L.; Muotri, A.R.; Wahlin, K.J.; Kahn, J.; Boyd, J.L.; Gracias, D.H.; Harris, T.D.; Cohen-Karni, T.; Caffo, B.S.; et al. First Organoid Intelligence (OI) workshop to form an OI community. Front. Artif. Intell. 2023, 6, 1116870. [Google Scholar] [CrossRef] [PubMed]
- de Hoyos-Vega, J.M.; Yu, X.; Gonzalez-Suarez, A.M.; Chen, S.; Mercado-Perez, A.; Krueger, E.; Hernandez, J.; Fedyshyn, Y.; Druliner, B.R.; Linden, D.R.; et al. Modeling gut neuro-epithelial connections in a novel microfluidic device. Microsyst. Nanoeng. 2023, 9, 144. [Google Scholar] [CrossRef]
- Gamboa, C.M.; Wang, Y.; Xu, H.; Kalemba, K.; Wondisford, F.E.; Sabaawy, H.E. Optimized 3D culture of hepatic cells for liver organoid metabolic assays. Cells 2021, 10, 3280. [Google Scholar] [CrossRef]
- Hu, W.; Lazar, M.A. Modeling metabolic diseases and drug response using stem cells and organoids. Nat. Rev. Endocrinol. 2022, 18, 744–759. [Google Scholar] [CrossRef]
- Bustos, J.F.; Gonzalez, J.C.A.; Angelo, D.; de Abreu, R.; Liebisch-Rey, H.; Silva, A.; Ortiz, D.; Ramírez, L.B.; Ortega, J.; Celis Regalado, L.G. Modeling Metabolic Diseases with Organoids: A Review. J. ISSN 2021, 2766, 2276. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Deng, P.; Chen, W.; Guo, Y.; Tao, T.; Qin, J. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab A Chip 2018, 18, 3606–3616. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.H.; Kolahi, K.S.; Du, Y.; Chang, C.Y.; Krokhotin, A.; Nair, A.; Sobba, W.D.; Karlsson, K.; Jones, S.J.; Longacre, T.A.; et al. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 2021, 11, 1562–1581. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.L.; Larsen, H.L.; Pikkupeura, L.M.; Maciag, G.; Guiu, J.; Müller, I.; Clement, D.L.; Mueller, C.; Johansen, J.V.; Helin, K.; et al. An organoid-based CRISPR-Cas9 screen for regulators of intestinal epithelial maturation and cell fate. Sci. Adv. 2023, 9, eadg4055. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T.; Bendriem, R.M.; Wu, W.W.; Shen, R.F. 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017, 24, 59. [Google Scholar] [CrossRef]
- Aggarwal, D.; Russo, S.; Naik, P.; Bhatia, S.; Spector, D.L. Establishment and Culture of Patient-Derived Breast Organoids. J. Vis. Exp. 2023, 192, e64889. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.; Bhumiratana, S.; Geris, L.; Papantoniou, I.; Grayson, W.L. Bioreactors for engineering patient-specific tissue grafts. Nat. Rev. Bioeng. 2023, 1, 361–377. [Google Scholar] [CrossRef]
- Millen, R.; De Kort, W.W.; Koomen, M.; van Son, G.J.; Gobits, R.; de Vries, B.P.; Begthel, H.; Zandvliet, M.; Doornaert, P.; Raaijmakers, C.P.J.; et al. Patient-derived head and neck cancer organoids allow treatment stratification and serve as a tool for biomarker validation and identification. Med 2023, 4, 290–310. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Park, S.; Song, H.; Ming, G.L. Development and application of brain region–specific organoids for investigating psychiatric disorders. Biol. Psychiatry 2023, 93, 594–605. [Google Scholar] [CrossRef]
- Saorin, G.; Caligiuri, I.; Rizzolio, F. Microfluidic organoids-on-a-chip: The future of human models. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2023; Volume 144, pp. 41–54. [Google Scholar]
- Lou, Y.R.; Leung, A.W. Next generation organoids for biomedical research and applications. Biotechnol. Adv. 2018, 36, 132–149. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Held, M.; Santeramo, I.; Wilm, B.; Murray, P.; Lévy, R. Ex vivo live cell tracking in kidney organoids using light sheet fluorescence microscopy. PLoS ONE 2018, 13, e0199918. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef] [PubMed]
- de Medeiros, G.; Ortiz, R.; Strnad, P.; Boni, A.; Moos, F.; Repina, N.; Meylan, L.C.; Maurer, F.; Liberali, P. Multiscale light-sheet organoid imaging framework. Nat. Commun. 2022, 13, 4864. [Google Scholar] [CrossRef] [PubMed]
- Rios, A.C.; Clevers, H. Imaging organoids: A bright future ahead. Nat. Methods 2018, 15, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Ansari, N.; Stelzer, E.H. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013, 352, 161–177. [Google Scholar] [CrossRef]
- Pham, V.M.; Ha, H.T.; Thakor, N. Microfluidic Culture Platforms in Neuroscience Research. In Handbook of Neuroengineering; Springer: Singapore, 2023; pp. 39–77. [Google Scholar] [CrossRef]
- Miller, C.P.; Shin, W.; Ahn, E.H.; Kim, H.J.; Kim, D.H. Engineering microphysiological immune system responses on chips. Trends Biotechnol. 2020, 38, 857–872. [Google Scholar] [CrossRef]
- Linville, R.M.; DeStefano, J.G.; Sklar, M.B.; Chu, C.; Walczak, P.; Searson, P.C. Modeling hyperosmotic blood–brain barrier opening within human tissue-engineered in vitro brain microvessels. J. Cereb. Blood Flow. Metab. 2020, 40, 1517–1532. [Google Scholar] [CrossRef]
- Blatchley, M.R.; Gerecht, S. Reconstructing the vascular developmental milieu in vitro. Trends Cell Biol. 2020, 30, 15–31. [Google Scholar] [CrossRef]
- Dorsey, P.J. Towards Spatial Computing and Chemical Information Storage in Soft Materials Using DNA Programming. Ph.D. Thesis, Johns Hopkins University, Baltimore, MD, USA, 2020. [Google Scholar]
- Bush, J.; Singh, S.; Vargas, M.; Oktay, E.; Hu, C.H.; Veneziano, R. Synthesis of DNA origami scaffolds: Current and emerging strategies. Molecules 2020, 25, 3386. [Google Scholar] [CrossRef]
- Mishra, S.; Feng, Y.; Endo, M.; Sugiyama, H. Advances in DNA origami–cell interfaces. ChemBioChem 2020, 21, 33–44. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, L.; Wang, B. Customizing cellular signal processing by synthetic multi-level regulatory circuits. Nat. Commun. 2023, 14, 8415. [Google Scholar] [CrossRef]
- Miranda, J. Theology and Ontological Minimalism. Int. eJournal 2019, 9, 171–192. [Google Scholar]
- Norfleet, D.A.; Park, E.; Kemp, M.L. Computational modeling of organoid development. Curr. Opin. Biomed. Eng. 2020, 13, 113–118. [Google Scholar] [CrossRef]
- Cortesi, M.; Liu, D.; Yee, C.; Marsh, D.J.; Ford, C.E. A comparative analysis of 2D and 3D experimental data for the identification of the parameters of computational models. Sci. Rep. 2023, 13, 15769. [Google Scholar] [CrossRef]
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Pain, B.; Baquerre, C.; Coulpier, M. Cerebral organoids and their potential for studies of brain diseases in domestic animals. Vet. Res. 2021, 52, 65. [Google Scholar] [CrossRef]
- Krenn, V.; Bosone, C.; Burkard, T.R.; Spanier, J.; Kalinke, U.; Calistri, A.; Salata, C.; Christoff, R.R.; Garcez, P.P.; Mirazimi, A.; et al. Organoid modeling of Zika and herpes simplex virus 1 infections reveals virus-specific responses leading to microcephaly. Cell Stem Cell 2021, 28, 1362–1379. [Google Scholar] [CrossRef]
- Gabriel, E.; Ramani, A.; Karow, U.; Gottardo, M.; Natarajan, K.; Gooi, L.M.; Goranci-Buzhala, G.; Krut, O.; Peters, F.; Nikolic, M.; et al. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 2017, 20, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Sutarjono, B. Can we better understand how Zika leads to microcephaly? A systematic review of the effects of the Zika virus on human brain organoids. J. Infect. Dis. 2019, 219, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Marotta, N.; Kim, S.; Krainc, D. Organoid and pluripotent stem cells in Parkinson’s disease modeling: An expert view on their value to drug discovery. Expert. Opin. Drug Discov. 2020, 15, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Gao, L.; Wu, X.; Fan, Y.; Liu, M.; Chen, J.; Song, J.; Kong, J.; Dong, Y.; Li, B.; et al. Lung organoids as model to study SARS-CoV-2 infection. Cells 2022, 11, 2758. [Google Scholar] [CrossRef] [PubMed]
- Salahudeen, A.A.; Choi, S.S.; Rustagi, A.; Zhu, J.; van Unen, V.; de la O, S.M.; Flynn, R.A.; Margalef-Català, M.; Santos, A.J.M.; Ju, J.; et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 2020, 588, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, L.; Lacko, L.A.; Chen, S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods 2022, 19, 418–428. [Google Scholar] [CrossRef]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-generation cancer organoids. Nat. Mater. 2022, 21, 143–159. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Fang, Z.; Li, P.; Du, F.; Shang, L.; Li, L. The role of organoids in cancer research. Exp. Hematol. Oncol. 2023, 12, 69. [Google Scholar] [CrossRef]
- Hartung, T.; Smirnova, L.; Morales Pantoja, I.E.; Akwaboah, A.; Alam El Din, D.M.; Berlinicke, C.A.; Boyd, J.L.; Caffo, B.S.; Cappiello, B.; Cohen-Karni, T.; et al. The Baltimore declaration toward the exploration of organoid intelligence. Front. Sci. 2023, 1, 1017235. [Google Scholar] [CrossRef]
- Heaton, A. Phd Student Tries to Get Doom Running on Cells. Destructoid. 12 December 2023. Available online: https://www.destructoid.com/phd-student-doom-running-on-cells-video/ (accessed on 8 March 2024).
- Ledford, H. Neurons in a dish learn to play Pong—what’s next? Nature 2022, 610, 433. [Google Scholar] [CrossRef] [PubMed]
- Kagan, B.J.; Kitchen, A.C.; Tran, N.T.; Habibollahi, F.; Khajehnejad, M.; Parker, B.J.; Bhat, A.; Rollo, B.; Razi, A.; Friston, K.J. In vitro neurons learn and exhibit sentience when embodied in a simulated game-world. Neuron 2022, 110, 3952–3969. [Google Scholar] [CrossRef]
- Webb, A. The Big Nine: How the Tech Titans and Their Thinking Machines Could Warp Humanity; Hachette: London, UK, 2019. [Google Scholar]
- Webb, A.; Hessel, A. The Genesis Machine: Our Quest to Rewrite Life in the Age of Synthetic Biology; Hachette: London, UK, 2022. [Google Scholar]
- Lavazza, A.; Reichlin, M. Human Brain Organoids: Why There Can Be Moral Concerns If They Grow Up in the Lab and Are Transplanted or Destroyed. Camb. Q. Health Ethic 2023, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Ao, Z.; Tian, C.; Wu, Z.; Liu, H.; Tchieu, J.; Gu, M.; Mackie, K.; Guo, F. Brain Organoid Computing for Artificial Intelligence. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Wheeldon, I.; Farhadi, A.; Bick, A.G.; Jabbari, E.; Khademhosseini, A. Nanoscale tissue engineering: Spatial control over cell-materials interactions. Nanotechnology 2011, 22, 212001. [Google Scholar] [CrossRef]
- Le Floch, P.; Li, Q.; Lin, Z.; Zhao, S.; Liu, R.; Tasnim, K.; Jiang, H.; Liu, J. Stretchable Mesh Nanoelectronics for 3D Single-Cell Chronic Electrophysiology from Developing Brain Organoids. Adv. Mater. 2022, 34, 2106829. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; Jung, W.H.; Kim, K.Y.; Koh, B. Toxicity Assessment of SiO2 and TiO2 in Normal Colon Cells, In Vivo and in Human Colon Organoids. Molecules 2020, 25, 3594. [Google Scholar] [CrossRef] [PubMed]
- Isichei, J.C.; Khorsandroo, S.; Desai, S. Cybersecurity and privacy in smart bioprinting. Bioprinting 2023, 36, e00321. [Google Scholar] [CrossRef]
- Parupelli, S.K.; Desai, S. The 3D Printing of Nanocomposites for Wearable Biosensors: Recent Advances, Challenges, and Prospects. Bioengineering 2023, 11, 32. [Google Scholar] [CrossRef]
- Zheng, H.; Feng, Y.; Tang, J.; Ma, S. Interfacing brain organoids with precision medicine and machine learning. Cell Rep. Phys. Sci. 2022, 3, 100974. [Google Scholar] [CrossRef]
- Danelyan, A.A.; Gulyaeva, E.E. International legal aspects of cybersecurity. Mosc. J. Int. Law 2020, 44–53. [Google Scholar] [CrossRef]
- Dittrich, S.; Richardson, L.; Burnette, R.N. Applied Biosecurity in the Face of Epidemics and Pandemics: The COVID-19 Pandemic. In Applied Biosecurity: Global Health, Biodefense, and Developing Technologies; Springer International Publishing: Cham, Switzerland, 2021; pp. 73–88. [Google Scholar]
- Amiri, A.; Shekarchizadeh, M.; Esfahani, A.R.S.; Masoud, G.H. Bio-Cyber Threats and Crimes, the Challenges of the Fourth Industrial Revolution. Bioethics 2021, 81, 97. [Google Scholar]
- Elgabry, M. Towards cyber-biosecurity by design: An experimental approach to Internet-of-Medical-Things design and development. Crime. Sci. 2023, 12, 3. [Google Scholar] [CrossRef]
- Koplin, J.J.; Savulescu, J. Moral Limits of Brain Organoid Research. J. Law Med. Ethics 2019, 47, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, A.J.; Dierickx, K. The Many Moral Matters of Organoid Models: A Systematic Review of Reasons. Medicine, Health Care and Philosophy 2022. Med. Health Care Philos. 2022, 25, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Capatina, L.; Cernian, A.; Moisescu, M.A. Efficient training models of Spiking Neural Networks deployed on a neuromorphic computing architectures. In Proceedings of the 2023 24th International Conference on Control Systems and Computer Science (CSCS), Bucharest, Romania, 24–26 May 2023; IEEE: New York, NY, USA, 2023; pp. 383–390. [Google Scholar]
- Gardner, S.D. Inkjet-Printed Reservoir Computing Network for In Situ Sensor Predictions. Ph.D. Thesis, The University of Alabama at Birmingham, Birmingham, UK, 2023. [Google Scholar]
- Johns Hopkins Medicine. (n.d.). Henrietta Lacks—Her Impact and Legacy. Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/henrietta-lacks (accessed on 8 March 2024).
- Nijhawan, L.P.; Janodia, M.D.; Muddukrishna, B.S.; Bhat, K.M.; Bairy, K.L.; Udupa, N.; Musmade, P.B. Informed consent: Issues and challenges. J. Adv. Pharm. Technol. Res. 2013, 4, 134. [Google Scholar] [PubMed]
- Skloot, R. The immortal life of Henrietta Lacks: The ethical issues behind cells from her tumor. Am. J. Bioeth. 2016, 16, 3–4. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5072843/ (accessed on 8 March 2024).
- Barnhart, A.J.; Dierickx, K. Too-Many-Oids: The paradox in constructing an organoid ethics framework. Mol. Psychol. Brain Behav. Soc. 2023, 2, 10. [Google Scholar] [CrossRef]
- de Jongh, D.; Massey, E.K.; Berishvili, E.; Fonseca, L.M.; Lebreton, F.; Bellofatto, K.; Bignard, J.; Seissler, J.; Buerck, L.W.-V.; Honarpisheh, M.; et al. Organoids: A systematic review of ethical issues. Stem Cell Res. Ther. 2022, 13, 337. [Google Scholar] [CrossRef]
- Boers, S.N.; van Delden, J.J.; Clevers, H.; Bredenoord, A.L. Organoid biobanking: Identifying the ethics. EMBO Rep. 2016, 17, 938–941. [Google Scholar] [CrossRef]
- Potter, L.; Ayala, O.; Palmer, X.L. Biocybersecurity: A converging threat as an auxiliary to war. In Proceedings of the ICCWS 2021 16th International Conference on Cyber Warfare and Security, Cookeville, TN, USA, 25–26 February 2021; p. 291. [Google Scholar]
- Mollaki, V. Ethical Challenges in Organoid Use. BioTech 2021, 10, 12. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobien, D.; Yardimci, M.O.; Nguyen, M.B.; Mao, W.Y.; Fordham, V.; Rahman, A.; Duncan, S.; Batarseh, F.A. AI for Cyberbiosecurity in Water Systems—A Survey. In Cyberbiosecurity; Springer: Cham, Switzerland, 2023; pp. 217–263. [Google Scholar]
- Feng, H.; Conneely, K.N.; Wu, H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Stat. Med. 2018, 37, 1833–1848. Available online: https://pubmed.ncbi.nlm.nih.gov/29611768/ (accessed on 8 March 2024). [CrossRef] [PubMed]
- Johns Hopkins Medicine. (n.d.). Importance of HeLa Cells. Johns Hopkins Medicine. Available online: https://www.hopkinsmedicine.org/henrietta-lacks/importance-of-hela-cells (accessed on 8 March 2024).
- Burrell, D.N.; Mcandrew, I. Addressing Bio-Cybersecurity Workforce Employee Shortages in Biotechnology and Health Science Sectors in the US. Sci. Bull. 2023, 28, 127–141. [Google Scholar]
- Mantas, V.; Pehlivanidis, A.; Kotoula, V.; Papanikolaou, K.; Vassiliou, G.; Papaiakovou, A.; Papageorgiou, C. Factors of influence in prisoner’s dilemma task: A review of medical literature. PeerJ 2022, 10, e12829. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G. Tauopathies. Handb. Clin. Neurol. 2017, 145, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Osborne, M.J. An Introduction to Game Theory; Oxford university press: New York, NY, USA, 2004; Volume 3. [Google Scholar]
- World Intellectual Property Organization. (n.d.). About IP. Available online: https://www.wipo.int/about-ip/en/ (accessed on 8 March 2024).
- Lesaja, S.; Palmer, X.L. Brain-computer interfaces and the dangers of neurocapitalism. arXiv 2020, arXiv:2009.07951. [Google Scholar] [CrossRef]
- Farahany, N.A. The Battle for Your Brain: Defending the Right to Think Freely in the Age of Neurotechnology; St. Martin’s Press: New York, NY, USA, 2023. [Google Scholar]
- Powell, E.; Akogo, D.; Potter, L.; Palmer, X.L. Co-leadership and Cross-pollination of University and DIY Bio Spaces: An Exploration in Consideration of Biocybersecurity. In Proceedings of the Future Technologies Conference (FTC) 2021, Vancouver, BC, Canada, 28–29 November 2021; Springer International Publishing: Cham, Switzerland, 2022; Volume 3, pp. 610–621. [Google Scholar]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Adeoye, S.O.; Batarseh, F.; Brown, A.M.; Kaufman, E.K. Understanding the Landscape of Cyberbiosecurity for Integrative Educational Programming. J. ASABE 2023, 67, 207–217. [Google Scholar] [CrossRef]
- Adeoye, S.; Lindberg, H.; Bagby, B.; Brown, A.; Batarseh, F.; Kaufman, E. Cyberbiosecurity Workforce Preparation. NACTA 2024, 67. [Google Scholar] [CrossRef]
- Schabacker, D.S.; Levy, L.A.; Evans, N.J.; Fowler, J.M.; Dickey, E.A. Assessing cyberbiosecurity vulnerabilities and infrastructure resilience. Front. Bioeng. Biotechnol. 2019, 7, 61. [Google Scholar] [CrossRef]
- Palmer, X.-L.; Potter, L.; Karahan, S. Exploration on APTs in Biocybersecurity and Cyberbiosecurity. International Conference on Cyber Warfare and Security 2022, 17, 532–535. [Google Scholar] [CrossRef]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Ray, S. What Is Biocomputing? Medium. 16 January 2021. Available online: https://medium.com/lansaar/what-is-biocomputing-82671bb381bd (accessed on 8 March 2024).
- Bio-Cybersecurity at CSU. Colorado State University. 25 October 2021. Available online: https://www.research.colostate.edu/bio-cybersecurity/#:~:text=Traditionally%2C%20biosecurity%20focuses%20on%20reducing,information%20in%20technology%2Dbased%20systems (accessed on 8 March 2024).
- Adams, D. Bioinformatics. National Human Genome Research Institute. 15 February 2024. Available online: https://www.genome.gov/genetics-glossary/Bioinformatics (accessed on 8 March 2024).
- Redman, M.; King, A.; Watson, C.; King, D. What Is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Fink, L. The human genome project. Alcohol. Health Res. World 1995, 19, 190. [Google Scholar] [CrossRef] [PubMed]
- UCLA Broad Stem Cell Research Center. Induced Pluripotent Stem Cells (iPS) | UCLA Broad Stem Cell Center. Ucla.edu. 2007. Available online: https://stemcell.ucla.edu/induced-pluripotent-stem-cells (accessed on 8 March 2024).
- Ye, L.; Swingen, C.; Zhang, J. Induced pluripotent stem cells and their potential for basic and clinical sciences. Curr. Cardiol. Rev. 2013, 9, 63–72. [Google Scholar]
- Moore, S. What Is In Silico? News Medical Life Sciences. 27 October 2021. Available online: https://www.news-medical.net/life-sciences/What-is-in-Silico.aspx (accessed on 8 March 2024).
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Romito, A.; Cobellis, G. Pluripotent stem cells: Current understanding and future directions. Stem Cells Int. 2016, 2016, 9451492. [Google Scholar] [CrossRef]
- Jayaraman, D.; Bae, B.I.; Walsh, C.A. The Genetics of Primary Microcephaly. Annu. Rev. Genom. Hum. Genet. 2018, 19, 177–200. [Google Scholar] [CrossRef]
- opensource.com. What Is a Raspberry Pi? Opensource.com. 2012. Available online: https://opensource.com/resources/raspberry-pi (accessed on 12 December 2022).
- What Is Raspberry Pi? Here’s the Best Guide to Get Started. Simplilearn.com. Available online: https://www.simplilearn.com/tutorials/programming-tutorial/what-is-raspberry-pi#:~:text=The%20Raspberry%20Pi%20is%20a,a%20modified%20version%20of%20Linux (accessed on 8 March 2024).
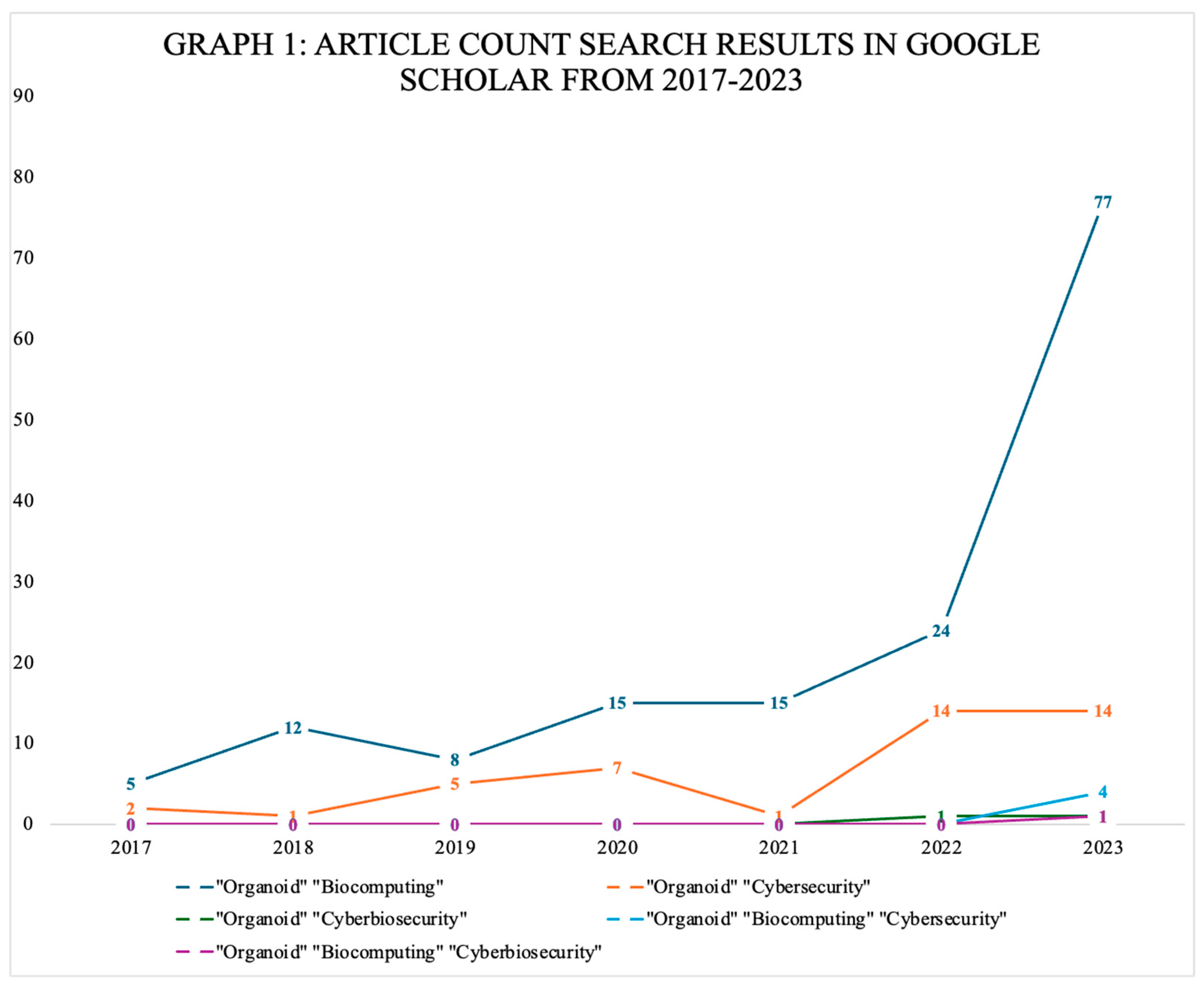


| Terms Entered | Filter | Article Count |
|---|---|---|
| “Organoid” “cyberbiosecurity” | 2018–2023 | 2 |
| “Organoid” “biocybersecurity” | 2018–2023 | 0 |
| “Organoid” “biocomputing” | 2018–2023 | 136 |
| “Organoid” “cybersecurity” | 2018–2023 | 40 |
| “Organoid” “biocomputing” “cybersecurity” | 2018–2023 | 4 |
| “Organoid” “biocomputing” “cyberbiosecurity” | 2018–2023 | 1 |
| “Organoid” “biocomputing” “biocybersecurity” | 2018–2023 | 0 |
| “Organoid” “biocomputing” “cyberbiosafety” | 2018–2023 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palmer, X.; Akafia, C.; Woodson, E.; Woodson, A.; Potter, L. Organoids, Biocybersecurity, and Cyberbiosecurity—A Light Exploration. Organoids 2024, 3, 83-112. https://doi.org/10.3390/organoids3020007
Palmer X, Akafia C, Woodson E, Woodson A, Potter L. Organoids, Biocybersecurity, and Cyberbiosecurity—A Light Exploration. Organoids. 2024; 3(2):83-112. https://doi.org/10.3390/organoids3020007
Chicago/Turabian StylePalmer, Xavier, Cyril Akafia, Eleasa Woodson, Amanda Woodson, and Lucas Potter. 2024. "Organoids, Biocybersecurity, and Cyberbiosecurity—A Light Exploration" Organoids 3, no. 2: 83-112. https://doi.org/10.3390/organoids3020007
APA StylePalmer, X., Akafia, C., Woodson, E., Woodson, A., & Potter, L. (2024). Organoids, Biocybersecurity, and Cyberbiosecurity—A Light Exploration. Organoids, 3(2), 83-112. https://doi.org/10.3390/organoids3020007







