Abstract
A three-dimensional cell culture in hydrogel beads can support cell growth and differentiation into multi-cellular structures, and these gel beads could be used as building blocks for more complex three-dimensional assemblies. This requires hydrogel beads that are robust enough to sort via FACS yet can be degraded by cell-secreted enzymes. Collagen polymers form hydrogels that are excellent cell growth substrates; however, collagen-containing hydrogel beads typically include additional polymers that limit their degradation. Here, we introduce a simple microfluidic method to generate robust, sortable, cell-laden collagen hydrogel beads. We use on-device pH control to trigger collagen gelation without exposing cells to low pH, ensuring high cell viability. We fabricate microfluidic devices to generate droplets with a wide size range, as demonstrated by production of both small (~55 µm diameter) and large (~300 µm diameter) collagen gels. All hydrogels are sufficiently robust to allow for sorting using FACS. Moreover, high cell viability is maintained throughout the process.
1. Introduction
Tissue engineering and organoid research require complex structures comprising numerous cell types at specific stages of differentiation [1,2]. Often, these structures are generated by seeding cells onto scaffolds designed to elicit the desired cell growth and differentiation. Bioprinting, an increasingly used alternative, generates precise, complex structures [3,4], but requires sophisticated instrumentation and is relatively low-throughput. A “building block” system [5], in which cell clusters are cultured to induce the desired cell phenotypes, then phenotype-selected for further assembly into structures with precisely positioned cell clusters, is an attractive alternative. Microfluidics-based cell encapsulation into enzyme-degradable hydrogel beads is an ideal means to produce cell-laden building blocks.
Hydrogels, hydrophilic three-dimensional polymer networks that absorb large quantities of aqueous fluid, are often used as scaffolds in three-dimensional cultures. Cells deposited upon or within appropriate hydrogels can divide, differentiate, and interact with other cells via direct contact and through paracrine signaling.
A variety of cell-compatible polymers, loosely classified as derived from “synthetic” or “natural” sources, are used to generate cell-laden hydrogel spheres (“beads”). Synthetic polymers are commonly based on polyethylene glycol, PEG. While many of these polymers are cell-compatible, cross-linking into hydrogels may impact cell viability. For example, the photopolymerization of PEG-diacrylate (PEG-DA) and other photocurable polymers is usually more efficient in the absence of oxygen. Cells encapsulated into PEG-DA and deprived of oxygen during some parts of droplet generation and gel formation show lower viability than cells encapsulated into PEG-norbornene, which can be photopolymerized [6,7] in the presence of oxygen.
PEG-based polymers can also be converted into hydrogels through click chemistry [8] or ion-mediated complexation. These methods use microfluidic devices to combine reagents immediately prior to droplet formation. In one report, a cell-containing stream of poly(ethylene glycol)-dicyclooctyne was merged with dendritic poly(glycerol azide) upstream of a drop-making junction to create cell-laden hydrogels with pH-controlled degradation [9]. In a complexation-based approach, microfluidic pico-injection was used to add iron (II) ions to droplets containing cells and bipyridine-modified PEG polymers, causing gelation. The addition of a chelator, disodium ethylenediaminetetraacetate dihydrate, disrupts the gels and releases viable cells [10].
The naturally derived polysaccharide polymers alginate and agarose are routinely used for cell encapsulation. Alginate rapidly gels upon the addition of divalent ions such as Ca2+. Microfluidic devices, together with methods to limit ion accessibility (e.g., the use of chelated ions or nanoparticle calcium), provide gelation control and allow for the generation of hydrogels with uniform physical properties [11,12]. Low-melt agarose remains a liquid at ~25–37 °C and solidifies at a reduced temperature. Cells survive encapsulation, and the gel beads are robust and compatible with Fluorescence Activated Cell Sorting (FACS) instruments [13]. Agarose [14] and alginate (for a review, see [15]) can be modified, e.g., by the chemical addition of peptides, to improve cell adhesion and add functionality. Researchers also mix polysaccharide polymers with protein-based polymers, e.g., agarose and collagen [16], or agarose and fibrin [17], to create hydrogels that are robust and provide a favorable cell environment. However, the polysaccharide-based polymers cannot be completely degraded by cell-secreted proteases; in some cases, this will limit the cells’ ability to elongate, migrate, and organize into higher order structures.
Hydrogel beads generated from naturally derived protein-based polymers, for example gelatin and Matrigel, are excellent substrates for cell growth and are degraded by proteases. These materials are often physically weak [18] and can be modified to improve hydrogel strength. Gelatin methacrylate (gelMA) [18], created by conjugating the methacrylate group to gelatin, is cross-linked into a robust gel [19] that is an excellent substrate for cell growth. However, it, and similar semi-synthetic hydrogels [20], are gelled through exposure to UV light and a photo-initiator, which may alter the cell phenotype.
Purified collagen [21] is a promising natural protein for cell encapsulation. It can be maintained as a liquid at low pH and low temperature and is converted to hydrogel via pH neutralization followed by incubation at 37 °C. Collagen hydrogels permit the diffusion of macromolecules such as proteins and cell nutrients, are degraded by cell-secreted proteases, and can fully support cell growth and differentiation [22,23]. However, care must be taken to prevent cell exposure to liquid collagen at low pHs.
Here, we introduce a simple microfluidics method to generate cell-laden collagen hydrogels that are sufficiently robust to be sorted via FACS. We inject acidic liquid collagen, neutral-pH HEPES buffer, and cells in growth medium into a microfluidic drop-making device. These fluids merge and mix, and the resulting stream is cut into droplets using a perpendicular stream of fluorinated oil. Incubation at 37 °C converts the aqueous droplets into hydrogel beads. The microfluidic devices can be fabricated to generate droplets with a wide size range, as demonstrated by production of both small (~55 µm diameter) and large (~300 µm diameter) collagen gels. All hydrogels are sufficiently robust to allow sorting using FACS. Moreover, high cell viability is maintained throughout the process.
2. Methods
2.1. Microfluidic Device Design and Fabrication
We used AutoCAD software (version 2022, Autodesk, San Francisco, CA, USA) to design the microfluidic devices. In this study, we primarily used devices to make collagen droplets of ~55 μm and ~300 μm diameter (see Figure 1 and Figure S1 for device details; corresponding AutoCad files are provided in the “Supplementary Files” folder). We have used similar devices to make collagen gels as small as ~40 µm and as large as ~500 µm. Microfluidic devices were fabricated via standard soft lithography, as previously described [24]. Note: devices can be designed to include features to disrupt cell aggregates if desired (Figure S1 in reference [25]).
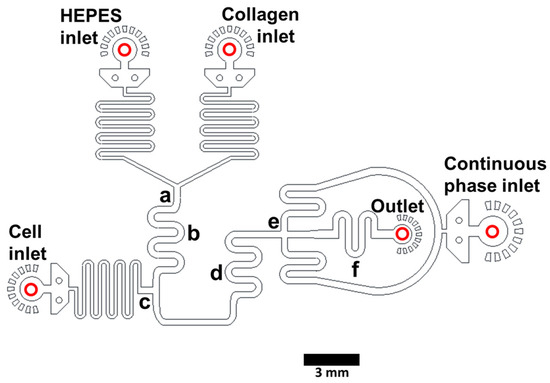
Figure 1.
Collagen hydrogel drop-making device [25]. Liquid acidic collagen and HEPES-based neutralizing buffer (pH ~7.3) are injected into the indicated inlets and the merged stream (a) is mixed in the serpentine region (b). Cells in growth medium are added (c). The stream is mixed (d) and cut into droplets in the continuous phase (HFE7500 oil with 2% fluorosurfactant) (e). Droplets pass through a mixing region (f), exit the device (Outlet) and are incubated off-chip to complete gelation. Channel depth for a device to make ~300 µm diameter droplets is 100 µm. Scale bar: 3 mm. A video panning through each feature of a device making 300 µm droplets is included as Supporting information (“SM1—making 300 µm droplet gels”).
2.2. Culture of Cell Lines
We cultured HepG2/C3A cells (epithelial-like human hepatocellular carcinoma cells—ATCC, Manassas, VA, USA, HB-8065) in complete DMEM (Dulbecco’s modified Eagle medium, Mediatech, cat. no. 10-013-CV; with 1.0% (v/v) penicillin–streptomycin, (Life Technologies, Carlsbad, CA, USA, cat. no. 15140-122); and 10% (v/v) fetal bovine serum (Life Technologies, cat. no. 10313-039), in a cell culture incubator (37 °C and 5% CO2). We passaged cells every 2–3 days using trypsin-EDTA solution (Sigma-Aldrich, Saint Louis, MO, USA, cat. no. T4549) to detach cells from flasks.
2.3. Generation of Collagen Gels and Transfer to Growth Medium
We used syringe pumps (Harvard Apparatus, Holliston, MA, USA, 2000/2200, cat. no. 702001) to pump fluids from syringes (Beckton-Dickinson, Franklin Lakes, NJ, USA, cat. no. 309646) into the microfluidic devices. Syringes were connected to the microfluidic device via blunt needles (Beckton-Dickinson, Franklin Lakes, NJ, USA, PrecisionGlide, cat. no. D929_SS27x0.5) and PE-2 tubing (Scientific Commodities, Lake Havasu City, AZ USA, cat. no. BB31695). We monitored droplet formation via a high-speed camera (FASTEC Imaging, San Diego, CA, USA, HiSpec1 4G Mono) mounted on a bright-field inverted microscope (Nikon, 803743, Melville, NY, USA).
We injected acidic liquid collagen (Corning, Glendale, AZ, USA, high concentration collagen type 1, cat. no. 354249, diluted to 4 mg/mL with sterile water) and neutralizing buffer (100 mM HEPES in 3× final concentration PBS, pH ~7.3; made by combining HEPES, Corning cat. no. 25-060-CI, and 10× PBS, Lifetech, Carlsbad, CA, USA, cat. no. 10010-023) into two separate inlets. The streams merged and mixed, and the neutralized collagen flowed past an inlet that adds cells suspended in complete DMEM supplemented with 16% Optiprep (Sigma-Aldrich, cat. no. D1556) to prevent cell settling. This aqueous stream was cut into droplets using a stream of continuous phase HFE7500 (3M, cat. no. 98-0212-2928-5) containing 2% surfactant (RAN Scientific, Beverly, MA, USA, cat. no. 033-007). We observed the aqueous stream merging regions to ensure that flows were equilibrated (the fluids have different refractive indices—see drop-making video in Supporting Information “SM1—Making 300 µm droplet gels”). We then collected droplets off-chip onto ice. To maximize viability of delicate cells in experiments requiring many gels, we recommend collecting several batches of droplets, rather than one large batch. This minimizes the time cells spend at high concentration in the syringe and within nutrient-impermeable water-in-oil droplets. We incubated collected droplets in a capped tube (e.g., 15 mL conical tube) at 37 °C for 45 min to allow collagen gelation. We added 5 mL growth medium and removed the HFE7500/surfactant from the bottom of the tube, as much as possible without disturbing the gelled droplets. We then added 1 mL HFE7500 (neat) and mixed gently. We allowed the droplet emulsion to cream to the top of the HFE7500 and we removed the HFE7500 without disturbing the emulsion. We repeated this “surfactant-removal step” a total of three times. We merged the droplets with the growth medium by adding 1 mL of HFE7500 containing 20% (final concentration, v/v) 1H,2H,2H-perfluoro-1-octanol (PFO; Sigma-Aldrich, cat. no. 370533). We inverted gently to mix and then incubated ~five min at room temperature while the gel-containing droplets merged with the growth medium. Note: PFO can reduce the viability of some cell types, so it is best to transfer the released gels to fresh growth medium soon after droplet-merging. We then transferred the gel-containing growth medium to culture plates or a fresh tube. Prior to using gels in sorting, we completely removed the remaining HFE7500 continuous phase. We poured the gels onto a 40 µm cell strainer (Thermo Fisher Scientific, Waltham, MA, USA, Falcon, cat. no. 165811), rinsed with one mL growth medium, and then collected the gels by inverting the strainer over a 50 mL conical tube and rinsing with several mL growth medium. To generate large (~300 μm diameter) droplets, we used the device in Figure 1 with flow rates 800, 400, 200, and 200 µL/h for the oil, collagen, HEPES buffer, and cell phases, respectively. To generate small (~55 µm diameter) droplets, we used the device in Figure S1 with flow rates 800, 400, 200, and 200 µL/h for the oil, collagen, HEPES buffer, and cell phases. Note: when necessary, gel-aggregates can be removed by straining gel suspension through a filter with a mesh size larger than the gels. We used 70 μm and 350 μm micron-sized mesh strainers to remove aggregates from the 55 mm and 300 mm gels, respectively.
There is some leeway in the conditions that produce robust collagen gels. We titrated pH, finding that robust gels are formed at final pHs (after HEPES is added) of 6.2, 7.2, and 8.5, while no gels are formed with final pH 11 (Figure S2) [26,27]. We also titrated collagen concentrations and found that we could not generate gels at final concentrations of 0.125 and 0.25 mg/mL; gels with incomplete edges were produced at 0.5 mg/mL, and robust gels were formed using final collagen concentrations of 0.75, 1, and 1.5 mg/mL (Figure S3) [28]. Further, we performed the gel-making method at various temperatures and found that gels are best formed by generating droplets at room temperature, collecting just-formed droplets on ice, then shifting to 25 °C or 37 °C for gelation.
2.4. Characterization of Collagen Gels
Characterization was carried out via staining with anti-collagen antibody. We added 5 μL anti-collagen antibody (Abcam, Waltham, MA, USA, cat. no. ab34710) to 250 μL complete growth medium containing large (~300 μm) collagen hydrogels and incubated overnight at room temperature with rolling. To wash, we allowed the gels to settle due to gravity for 5 min, removed the supernatant, added 500 μL complete growth medium, and again allowed the gels to settle with gravity. We then removed the medium and repeated this wash 4 times. During these washes, we removed as much supernatant as possible without disturbing the gels. We then resuspended the gels in 500 µL complete growth medium and added 2 μL Cy3-labeled fluorescent secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA, cat. no. 111-165-008). After incubation at room temperature with rolling for >1 h, we washed with 500 μL medium, resuspended in 500 μL growth medium, and observed the hydrogels via confocal fluorescence microscopy (Leica Microsystems Inc., Deerfield, IL, USA, Leica TCS SP5).
To perform confocal sectioning of hydrogels, we collected images in 10 μm steps. We assembled these images into videos (see Supporting Information folder “SM2a-SM2d—Unstained cells cultured in 300 μm collagen gels”) using Leica LASX software, version 3.7.4.23463 (Leica Microsystems Inc.). Typically, we imaged gels in culture medium in 96-well plates. We used confocal reflection microscopy, at 488 nm excitation, to reveal the collagen fiber network (false-colored in green). We used Leica LASX software to determine hydrogel dimensions and analyze fluorescence.
2.5. Measurement of Cell Viability
We encapsulated HepG2/C3A cells into ~300 μm diameter collagen gels at a density of ~80 cells/gel. We transferred gels from the continuous phase HFE7500 to growth medium and incubated at 37 °C for the indicated periods. At each time point, we swirled the culture flask and removed 1 mL of gel-containing medium. We pelleted the gels via centrifugation (1000× g for 2 min), removed 800 μL supernatant, then resuspended gels to 1 mL final volume. We added Calcein AM solution and ethidium homodimer-1 solution to 100 nM and 8 μM final concentrations, respectively (Thermo Fisher Scientific, Waltham, MA, USA, LIVE/DEAD® Viability/Cytotoxicity kit, cat. no. L3224). We incubated samples for 15 min at room temperature. As a control to demonstrate the staining of dead cells, we treated some gels from each day with 70% methanol for 30 min to kill the cells prior to staining. We analyzed the stained cells via confocal fluorescence microscopy using 488 nm excitation and measuring the green fluorescence emission for Calcein AM (i.e., 530/30 bandpass) and red fluorescence emission for ethidium homodimer-1 (i.e., 610/20 bandpass). We performed confocal microscopy sectioning of collagen hydrogels by imaging in 10 μm steps (see Supporting Information folder “SM3—Cells in gels incubated with live dead dyes”). The live cells showed green fluorescence and dead cells showed red fluorescence.
2.6. Gel Sorting
We used a Beckman CoulterMoFlo Astrios EQ Cell Sorter (Beckman Coulter, Inc., Brea, CA, USA fitted with a 200 µm nozzle to sort small (<60 μm diameter) collagen gels. We generated ~55 μm diameter collagen droplets containing ~1–3 cells per gel, then incubated at 37 °C to complete hydrogel formation, and then transferred the gel emulsion to growth medium. To completely remove the remaining HFE7500 continuous phase, we poured the gels onto a 40 µm cell strainer (Falcon, cat. no. 165811), then collected the gels by inverting the strainer over a 50 mL conical tube and rinsing with several mL growth medium. We divided these gel sets into two fractions, stained with either CellTracker™ Green (ThermoFisher #C7025) or Calcein Red-Orange, AM (Thermo Fisher ™ C34851), and confirmed staining via confocal fluorescence microscopy. To set the Astrios drop delay setting, we used 10 µm diameter Beckman Flow-Check Fluorospheres (product number 6605359) for initial calibration; we then used a sample of the gels, which are much larger than the Flow Check beads, to fine-tune the drop delay. We used a Union Biometrica BioSorter® (Holliston, MA, USA) to sort large (~300 µm diameter) collagen hydrogels. We generated ~300 μm diameter gels containing ~80 cells/gel. We transferred the gels to growth medium and washed as above but used a 100 μm cell strainer to capture the gels. We used Union Biometrica Control Particles (part #310-5071-001) to calibrate the Biosorter, which was fitted with a 1000 µm Fluidics and Optics Core Assembly. We used fluorescence microscopy to analyze the Astrios and the Biosorter sorting outputs. After counting the gels that contained green cells (“green gels”) and the gels that contained red cells (“red gels”), we calculated the “Purity” as the # of desired gels/total # of gels in the output. “Enrichment” was calculated as the % sorted gels of correct color/% correct-color gels in the input. “Recovery” was calculated as the number of correct gels (determined via microscopy analysis of output)/number of target gels in the input (cytometry data). The data files for Figure 6 sorting are included in the Supporting Information in the folder “Biosorter Raw Data—Figure 6”.
3. Results and Discussion
3.1. Microfluidic Device Design and Fabrication and Collagen Hydrogel Generation
We use PDMS (polydimethylsiloxane)-on-glass devices to generate cell-laden collagen hydrogels droplets. We inject acidic liquid collagen and HEPES buffer (pH ~7.3) into separate inlets and the merged stream (Figure 1a) is mixed in the serpentine region (Figure 1b). Cells in growth medium are added to the pH-neutralized collagen (Figure 1c). This stream passes through a second mixing region (Figure 1d) and is cut into droplets via the perpendicular continuous phase (HFE7500 containing surfactant) (Figure 1e). Droplets exit the device (Figure 1, Outlet) and are collected off-chip and incubated at 37 °C for 45 min to allow for complete gelation (shorter incubation times are likely sufficient but have not been tested). Collagen hydrogel spheres are then released from the droplets and transferred to cell culture medium. The use of three aqueous inlets allows for the controlled gelation of collagen and eliminates cell exposure to low pH, ensuring high viability. Devices can be fabricated to generate droplets with a wide size range; we describe devices to generate gels of ~300 μm (Figure 1) and ~55 μm diameter (Supporting Information “Figure S1: Collagen hydrogel droplet making device”). Corresponding AutoCad files are in the Supporting Information (“Device files”). The droplet generation speed is dependent on flow rates. We typically generate 300 µm droplets at ~4 Hz and 55 μm droplets at ~4 kHz. A video demonstrating the generation of 300 µm diameter cell-containing collagen droplets is included in the Supporting Information (“SM1—making 300 μm droplet gels”).
We performed experiments to define the parameters required to produce robust collagen gels. We performed the gel-making method at various temperatures and found that gels are best formed by collecting the just-formed droplets on ice, and then moving them to 25 °C or 37 °C for gelation. We titrated pH, finding that robust gels are formed at a final pH (after HEPES is added) of 6.2, 7.2, and 8.5, while no gels are formed with a final pH of11 (Figure S2). We also titrated collagen concentrations and found that we could not generate gels at final concentrations of 0.125, 0.25, 0.5 mg/mL; gels with incomplete edges are produced at 0.75 mg/mL, and robust gels are formed using final collagen concentrations of 1 and 1.5 mg/mL (Figure S3).
3.2. Collagen Hydrogel Characterization
We used microscopy to confirm that the microfluidics method generates spherical collagen hydrogels with fibrous networks. Brightfield imaging revealed that the large-diameter (~300 μm) collagen hydrogels are homogeneous, indicating the complete mixing of aqueous fluids within the droplets prior to gelation. In addition, the gels have smooth, rounded edges with no loosely connected regions that might fail during mechanically stressful manipulations such as sorting (Figure 2A). We used reflection mode imaging [29] to highlight the collagen fibrils [30] (Figure 2B,C). Finally, we used an anti-collagen antibody and appropriate Cy3-labeled fluorescent secondary antibody to confirm that the collagen forms an evenly distributed network within these gels (shown in red in Figure 2D). These structures are consistent with the research published by others, showing collagen networks assembled from purified collagen [31]. To examine the collagen hydrogel structure in three dimensions, we imaged unstained gels and gels labeled with fluorescent anti-collagen antibody in 10 μm steps using brightfield, reflection mode, and fluorescence microscopy. We assembled the images into videos which clearly show a regular fibrous network throughout the hydrogel sphere (see movies in the folder “SM2a-SM2d—Unstained cells cultured in 300 μm collagen gels” in Supporting Information).
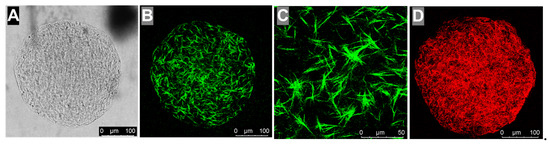
Figure 2.
Microscopy characterization of collagen hydrogel spheres. Brightfield image (A). Collagen hydrogel imaged using reflection mode to reveal collagen fiber network, false-colored in green (B,C) is a region of image (B) at higher magnification. Collagen gel incubated with anti-collagen antibody and Cy3-labeled fluorescent secondary antibody (D). Scale bars: 100 µm (A,B,D) and 50 µm (C).
3.3. Cells Divide within Collagen Hydrogels
To evaluate the cell growth in collagen gels, we generated ~300 μm diameter spherical collagen hydrogels that initially contain ~80 HepG2/C3A cells per gel. Immediately after encapsulation, most cells are distributed throughout the gels and have a distinct, rounded morphology (Figure 3, Day 0; dotted arrows indicate individual cells). After one day of incubation, some cells maintained the rounded morphology, while others began to display rough edges (Figure 3, Day 1; solid arrows indicate cell clusters). The cells began to form mini colonies after one day, and the cells within these colonies continued to divide, eventually filling the hydrogel spheres with large structures of densely packed cells (Figure 3, Days 2–5; note that Day 3, Day 4, and Day 5 images are of the same gel). Videos assembled from confocal microscopy sectioning of collagen hydrogels are included as Supporting Information in the folder “SM2a-SM2d—Unstained cells cultured in 300 μm collagen gels”.
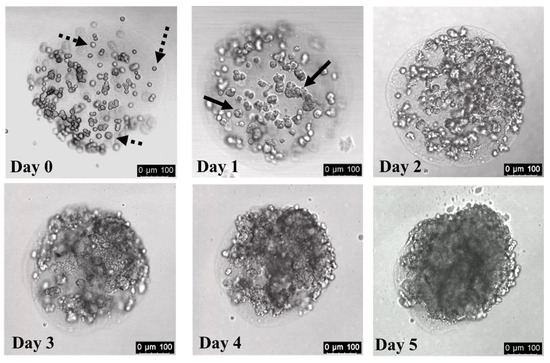
Figure 3.
HepG2/C3A cells expand in number during culture in collagen hydrogel spheres. Cell-laden collagen droplets were generated using microfluidics devices and incubated at 37 °C for 45 min to gel the collagen. Gels were transferred from the continuous phase oil to growth medium and incubated at 37 °C. Gels were imaged after the indicated incubation period. The Day 3, Day 4, and Day 5 images are of the same gel. Dashed arrows indicate representative single cells; solid black arrows indicate representative cell clusters that arise after 1–2 days of culture in collagen gels. Scale bars: 100 µm.
3.4. Cells Remain Viable within Collagen Hydrogels for Several Days
The experiments summarized in Figure 3 and Figure S3 clearly show that cells can divide and form dense structures in collagen gels. To assess the viability of the cells deep within these structures, we generate collagen hydrogels containing HepG2/C3A cells, incubate them in growth medium for the indicated lengths of time, and then add Calcein AM, which stains live cells green, and ethidium homodimer, which stains the nuclei of dead cells red. At each time point, we treat a fraction of the gels with 70% methanol prior to staining to generate a reference sample with dead cells. Cells stained immediately after encapsulation show bright Calcein AM fluorescence, and almost no cells show any red signal from ethidium homodimer, indicating that the encapsulation of cells into collagen gels and the transfer from the continuous-phase HFE7500/surfactant into growth medium has no negative effect on cell viability (Figure 4, Day 0, untreated panel). The Day 0 methanol-treated cells are brightly stained with ethidium-homodimer, confirming that these staining conditions clearly distinguish between living and dead cells. Over the course of 6 days, we remove gels from the culture and assess their viability. The percentage of cells that are clearly dead (brightly stained with ethidium homodimer) does not dramatically increase during the six-day incubation, indicating that HepG2/C3A cells, even those deep within cell clusters, maintain high viability during multi-day culture in collagen hydrogels (Figure 4, days 1–6). To further illustrate this, we include videos assembled from the confocal fluorescence microscopy sectioning of collagen hydrogels as Supporting Information in the folder “SM3—Cells in gels incubated with live dead dyes”. In initial experiments to encapsulate Pluripotent Stem Cells (iPSCs), more delicate than the HegG2/3CA used cells for the bulk of this work, a significant fraction of the iPSCs survive the encapsulation, collagen gelling, and gel-transfer into growth medium (Figure S4). However, we observe limited cell division during several days of in-gel culture, and encapsulated cells generally die within 4–5 days after transfer into Pluripotent Stem Cell SFM XF/FF medium or DMEM. Protocols for optimal viability are likely to be specific to the cells used and the cell-differentiation goals. In the Supporting Information (file “Supplemental Figure S4 with methods”), we suggest strategies to improve viability, such as initiating differentiation prior to encapsulation and the inclusion of additional reagents to enhance cell survival, e.g., the small molecule cocktail CEPT [32].
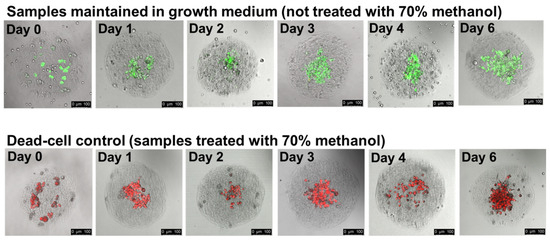
Figure 4.
HepG2/C3A cells remain viable throughout 6 days of incubation in collagen hydrogels. We encapsulated cells into collagen hydrogel droplets and then transferred gelled droplets from the continuous phase into cell culture medium. We cultured the cell-containing gels for the indicated periods. We assessed cell viability via staining with Calcein AM, which stains live cells green, and ethidium homodimer, which stains the nuclei of dead cells red. As a control to demonstrate the staining of dead cells, some gels from each day were treated with 70% methanol for 30 min prior to staining. Each image is an overlay of signal from green fluorescence, red fluorescence, and brightfield channels. Scale bar: 100 µm.
3.5. Sorting of Collagen Hydrogels
Research on topics such as tissue engineering and organoid development will benefit from methods to identify and select the desired cell clusters that can then be assembled into precise 3D structures. Hydrogel spheres are promising assembly units; they provide high cell viability, are permeable to growth and differentiation factors, and they protect multicellular structures during manipulations. To further develop collagen gel spheres as “bio-bricks”, we use standard FACS instruments to select desired cell-laden collagen gels.
We use a Beckman Coulter MoFlo Astrios EQ Cell Sorter to sort smaller gels, ~55 μm in diameter, at rates of a thousand events per second. We encapsulate HepG2/C3A cells into collagen gels (microfluidic device shown in Figure S1) and incubate the gels with CellTracker™ green or Calcein-red-orange to generate gels with bright green cells and gels with bright red cells (hereafter, “green gels” and “red gels”, respectively). We confirm cell-staining via fluorescence microscopy (see insets in Figure 5A,B). To determine the appropriate FACS settings to sort mixed populations of gels, we separately analyze and sort collagen gel populations containing either green- or red-stained cells. The resulting scatterplots show a bright green population (Figure 5A, indicated by a green box) and a bright red population (Figure 5B, indicated by a red box) that are easily distinguished from each other and from the population that contains debris, gels with very poorly stained cells, and empty gels (regions bounded by dotted lines in Figure 5A–C; we discard this population when we characterize sorting. Note: we expect a significant number of empty gels. The cell concentration during gel droplet formation is ~two per gel, and cells load into gel droplets according to Poisson distribution). We use these results to set gates to sort green gels and red gels into separate collection tubes. We then perform fluorescence microscopy on a fraction of the sorted, collected gels to determine enrichment and recovery. Sort #1 and Sort #2 inputs contain red gels in vast excess to green gels, and we focus on the enrichment and recovery of green gels. In Sort #1, cytometer data characterizes the input as ~99.4% red gels and ~0.6% green gels, and from this sample 251 “green-high” (above threshold) FACS sort signals are generated. After sorting, we observe 204 gels via fluorescence microscopy in the “green high” sorted sample, indicating a recovery rate of ~80% (204/251). Further, 98% (200/204) of the gels sorted as “green-high” contain the expected bright green cells, and four gels contain bright red cells, giving a sorting enrichment of ~163-fold. In Sort #2, the input is ~99.8 red gels and ~0.2% green gels. Although the cytometer detects only 27 “green-high” gels, we recover 14 gels from the “green-high” collection tube, and all 14 contained bright green cells, as judged via fluorescence microscopy (a 52% recovery rate and ~500-fold enrichment). The sort #3 input is 0.8% red-cell gels and 99.2% green-cell gels, and we focus on the enrichment and recovery of red gels. We recover 131 of the 201 cytometer-detected red-high gels (~54%). Of these, 109 had bright red cells, while 22 contained green cells, giving an enrichment of ~104-fold. These results clearly demonstrate that we can sort, with high accuracy and good recovery, low-abundance desired gels from a large excess of undesired gels. For each sort, we also analyze 200 gels from the collection tube containing the high-abundance gels sorted from the mixed population. In each case, 100% of the gels contain correct-color cells; this is expected because these gels comprised over 99% of the total cell-containing gels in each input.
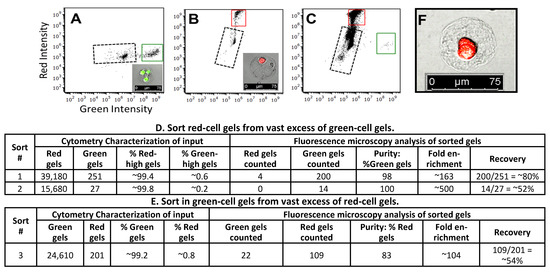
Figure 5.
Astrios EQ FACS instrument was used to analyze and sort cell-laden 55 µm diameter collagen gels. Gel characterization: We incubated gels containing HepG2/C3A cells with either CellTracker Green or Calcein-red-orange (these dyes are retained in living cells) to create gels containing green or red cells (“Green gels” and “Red gels”, respectively). We characterized the gel populations into (A,B) green- and red-boxed populations, respectively; insets are images of representative gels). Sorting: We mixed green and red gels to create sorting inputs. Flow cytometry data collected during sorting show the proportion of green and red gels within each input ((C), from sort #2, is representative of the other sorts). We sorted into separate tubes and used fluorescence microscopy to characterize each sorting output. “Purity”, “Enrichment”, and “Recovery” are calculated as described in Materials and Methods and summarized in (D,E). The input composition is shown in the “Cytometry Characterization of input” section. The Gel in (F) was sorted as “red” and imaged post-sorting. Image scale bars are 75 µm. Black dotted boxes in panels (A–C) indicate debris.
For large gel sorting, we use a Union Biometrica Biosorter® that directs the selected gels into a collection tube and sends the undesired gels into waste. To ensure that the differently stained gel populations are sorted according to fluorescence and not some other difference, we perform sorts in both “directions”: green gels from an excess of red gels, and red gels from an excess of green.
We use microfluidic devices (Figure 1) to generate ~300 μm diameter gels containing ~80 HepG2/C3A cells per gel. We divide the gels into two groups, incubate with CellTracker™ Green or Calcein-red-orange to create “red” and “green” gels, and confirm cell staining via fluorescence microscopy (see images inserted into Figure 6A,B, respectively). We analyze these separate gel populations via BioSorter® and plot green vs. red fluorescence. The green gels and red gels are grouped into distinct populations (Figure 6A,B, indicated by green and red boxes, respectively) and help to define the gates for sorting experiments.
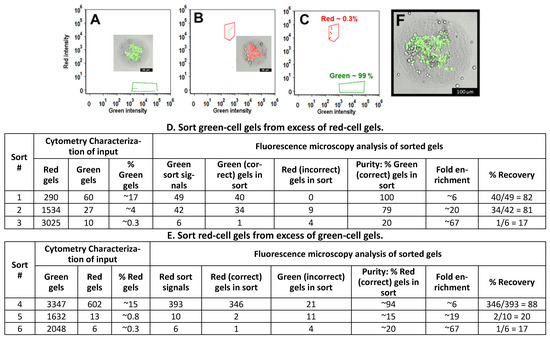
Figure 6.
Union Biometrica BioSorter instrument used to analyze and sort 300 µm diameter cell-laden gels. ~300 µm diameter collagen gels containing HepG2 cells (~80 cells/gel) were incubated with either CellTracker Green or Calcein-red-orange to create populations of “Green gels” and “Red gels”, respectively. We separately characterized these green gel and red gel populations ((A,B); the insets are images of representative gels). We mixed these gels to create the inputs used for Sort #s 1–6. Cytometry data collected during sort #6 is shown in (C) and is representative of Sort #s 1–6. Six sorting experiments are summarized in (D,E). The “Cytometry characterization of Input” gives the gel composition of each input. The “Green sort signals” and “Red sort signals” indicate the number of above-sorting-threshold gels detected while collection tubes were in place. We used fluorescence microscopy to analyze the sorting outputs. “Purity”, “Enrichment”, and “Recovery” were calculated as described in Materials and Methods. The gel in (F) was sorted as “green” and imaged post-sorting (scale bars: 100 µm).
We mix red and green gels to generate sorting samples and then use the Biosorter® to analyze the inputs and select gels with an above-threshold fluorescence signal. Flow cytometry data for Sort #3 is shown in Figure 6C. We use fluorescence microscopy to observe the sorted gels and determine sorting enrichment and recovery. In Sort #1, the input gel composition was 17% green gels and 83% red gels. During the sort, 49 sorting signals were generated. We recovered 40 gels; all contained only bright green cells, as expected for this target-rich input population (Figure 6D, Sort #1). In Sort #2, with an input composition of 4% green gels and 96% red gels, 42 sorting signals were generated. We recovered 43 gels; 34 contained green cells and 9 contained red cells, giving an approximate enrichment of 20-fold (Figure 6D, Sort #2), and a purity of 79% (34 green gels/43 total gels recovered). In Sort #3, green gels comprised only ~0.3% of the input gels. Six sorting signals were generated; one of the five recovered gels contained green cells, and four contained red cells, giving an enrichment of ~67-fold (this number is inexact, as few gels were isolated due to the low abundance of green gels in the input—Figure 6D, Sort #3). The recoveries for Sort #s 1–3 were 82%, 81%, and 17%, respectively. We generated similar results when sorting red gels from an excess of green gels. The Sort #4 (Figure 6E) input gel composition was 15% red gels and 85% green gels. The Biosorter® detected 393 above-threshold signals. We recovered 346 red gels and 21 green gels. In Sort #s 5 and 6, red cell gels were rare in the inputs, ~0.8 and ~0.3%, respectively. In Sort #5, we recovered two red-cell gels and 11 green-cell gels; enrichment was ~19-fold. In sort #6, we recovered one red-cell gel and four green-cell gels, giving an enrichment of ~67-fold. The recoveries for Sort #s 4–6 were 88%, 20%, and 17%, respectively.
Although our Biosorter® is relatively slow and we process only a limited number of gels during each sort, we always recover numerous sorted gels. We find that the desired gels are efficiently selected and that gels and the encapsulated cells are not visibly damaged during the sorting (Figure 6F is an image of a representative post-sorting gel). These results demonstrate that BioSorter® can accurately sort ~300 μm diameter collagen hydrogels, based on the fluorescence of the encapsulated cells.
4. Conclusions
Here, we demonstrate an on-device pH-change as a simple and easily adopted method to encapsulate cells into collagen hydrogels. We inject acidic liquid collagen, HEPES buffer (pH ~7.3), and cells in growth medium from three separate inlets to ensure that cells are always kept at a physiological pH. After hydrogel formation, droplets are transferred from continuous phase oil to cell growth medium. Encapsulated cells maintain high viability throughout these steps and cells divide within these gels (Figure 3 and Figure 4 and Supporting Information folders “SM2a-SM2d—Unstained cells cultured in 300 μm collagen gels” and “SM3—Cells in gels incubated with live dead dyes”.
Gels made through on-device pH changes are physically robust and compatible with commercial flow cytometry sorting instruments. We use the Beckman Coulter MoFlo Astrios EQ Cell Sorter and Union Biometrica Biosorter® instruments, respectively, to analyze and sort mixtures of small (~55 µm diameter) or large (~300 µm diameter) gels that contain either red-fluorescent or green-fluorescent cells. Both sorting instruments generate outputs highly enriched with the desired gels (Figure 5 and Figure 6). These results confirm that gels survive the sorting and that gels do not significantly stick to each other during manipulations.
We anticipate that these techniques to rapidly generate FACS-compatible collagen hydrogels will facilitate a “building block” process to reproducibly produce complex three-dimensional multicellular structures. For example, cells or cell clusters can be encapsulated into hydrogels, then cultured to induce differentiation into desired cell states. Hydrogels that contain cells with the desired phenotype will be identified by, e.g., state-specific antibodies, and isolated via FACS. This process can create discrete populations of gels, each containing cells/cell clusters with specific characteristics. The gels could then be combined in precise ratios to reproducibly create three-dimensional structures with defined cell types; the gels could be placed in specific layers and patterns if required.
With further development, these methods would facilitate, for example, the rapid generation of reproducible organoids, perhaps enabling organoid-based drug screening. Similarly, these gels should be useful in tumor microenvironment engineering, leading to tumor models that better mimic tumor initiation and progression in patients [33,34]. We note that this new method is straightforward and can easily be modified. Devices to make gels droplets smaller than ~55 µm diameter and larger than ~300 µm diameter can easily be designed, and the gels are likely compatible with a wide variety of commercial sorting instruments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/organoids2040016/s1. The “Supplementary Files” folder contains the file “Supplemental Figures S1–S3” (comprising “Figure S1: Collagen hydrogel droplet making device”; “Figure S2: Effect of pH on in-droplet collagen gelation”; and “Figure S3: Effect of collagen concentration”); the file “Supplemental Figure S4 with methods” (comprising “Figure S4: Live-dead staining of iPSCs cultured in collagen gel beads”, and the related methods); and the subfolder “Biosorter Raw Data—Figure 6” (containing sorting data files used in Figure 6: “Figure 6A green only”, “Figure 6B red only”, “Figure 6C green from mix”, and “Figure 6D red from mix”). The AutoCad files for the gel-making devices are included as “Device files”. Videos/movies are in the “Supplementary Movies” folder. The file “SM1—Making 300 µm droplet gels” shows a microfluidic device in use. The folder “SM2a-SM2d—Unstained cells cultured in 300 µm collagen gels” contains the movies “SM2a—Scan through 300 µm gel bright field”; “SM2b—Scan through 300 µm gel reflection mode”; “SM2c—Magnified Scan through 300 µm gel reflection mode”; and “SM2d—Scan through 300 µm gel collagen ab stain”. The folder “SM3—Cells in gels incubated with live dead dyes” contains the movies “Day 1 cells in 300 µm gels—methanol treated”, “Day 1 cells in 300 µm gels—no methanol”, “Day 3 cells in 300 µm gels—methanol treated”, “Day 3 cells in 300 µm gels—no methanol”, “Day 6 cells in 300 µm gels—methanol treated”, and “Day 6 cells in 300 µm gels—no methanol”. The folder “Scans through iPSC-containing gels” contains the movies “SM4 iPSCs No Methanol BrField Green Red Blue overlay”, “SM4.1 iPSCs No methanol Green Red Blue overlay”, “SM5 iPSCs Meth treated BrField Green Red Blue overlay”, and “SM5.1 iPSCs Meth treated Green Red Blue overlay”.
Author Contributions
Investigation (lead) and writing (original draft and review and editing), Y.X.; investigation (supporting), J.W.C., J.B., Q.H., J.A.N. and Z.N.; conceptualization, A.O.; funding acquisition and supervision, F.Y.; conceptualization, funding acquisition, supervision, and writing (review and editing), D.A.W.; conceptualization, investigation (supporting), funding acquisition, supervision, and writing (original draft and review and editing), J.A.H. All authors have read and agreed to the published version of the manuscript.
Funding
Y.X. was supported by the National Natural Science Foundation of China (81803720),the Natural Science Foundation of Hunan Province (2019JJ50383), and Harvard University; Q.H. was supported by the National Natural Science Foundation of China (11904301) and the Higher Education Discipline Innovation Project (B16029). J.A.H. and J.W.C. were supported by the Massachusetts Life Sciences Center Award (Drop-MINT: A high-throughput platform for Droplet-based Microbial Inhibition Testing). J.A.H. was supported by The Foundation for Aids Research (amfAR) award# WEIT171L1. J.B. was supported by The Foundation for Aids Research (amfAR) award # 109326-59-RGRL and by Ragon Institute Innovation Proposal #226618. This work was partially supported by the Health@InnoHK program of the Innovation and Technology Commission of the Hong Kong SAR Government. F.Y. was supported by the National Key Research and Development Program of China (2022YFA1405002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data are contained within the article or Supporting Information. Raw flow cytometry data are available upon request.
Acknowledgments
We thank Sijie Sun for his microscopy and file management expertise; we thank Kayla Keepseagle for device fabrication.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bajaj, P.; Schweller, R.M.; Khademhosseini, A.; West, J.L.; Bashir, R. 3D biofabrication strategies for tissue engineering and regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 247–276. [Google Scholar] [CrossRef] [PubMed]
- LeSavage, B.L.; Suhar, R.A.; Broguiere, N.; Lutolf, M.P.; Heilshorn, S.C. Next-generation cancer organoids. Nat. Mater. 2022, 21, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Barrs, R.W.; Jia, J.; Ward, M.; Richards, D.J.; Yao, H.; Yost, M.J.; Mei, Y. Engineering a Chemically Defined Hydrogel Bioink for Direct Bioprinting of Microvasculature. Biomacromolecules 2021, 22, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Dawson, C.; Lamb, M.; Mueller, E.; Stefanek, E.; Akbari, M.; Hoare, T. Hydrogels for Tissue Engineering: Addressing Key Design Needs Toward Clinical Translation. Front. Bioeng. Biotechnol. 2022, 10, 849831. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.T.; Morimoto, Y.; Takeuchi, S. Molding Cell Beads for Rapid Construction of Macroscopic 3D Tissue Architecture. Adv. Mater. 2011, 23, H90–H94. [Google Scholar] [CrossRef]
- Xia, B.; Krutkramelis, K.; Oakey, J. Oxygen-Purged Microfluidic Device to Enhance Cell Viability in Photopolymerized PEG Hydrogel Microparticles. Biomacromolecules 2016, 17, 2459–2465. [Google Scholar] [CrossRef]
- Jiang, Z.; Xia, B.; McBride, R.; Oakey, J. A microfluidic-based cell encapsulation platform to achieve high long-term cell viability in photopolymerized PEGNB hydrogel microspheres. J. Mater. Chem. B 2017, 5, 173–180. [Google Scholar] [CrossRef]
- Rossow, T.; Heyman, J.A.; Ehrlicher, A.J.; Langhoff, A.; Weitz, D.A.; Haag, R.; Seiffert, S. Controlled Synthesis of Cell-Laden Microgels by Radical-Free Gelation in Droplet Microfluidics. J. Am. Chem. Soc. 2012, 134, 4983–4989. [Google Scholar] [CrossRef]
- Steinhilber, D.; Rossow, T.; Wedepohl, S.; Paulus, F.; Seiffert, S.; Haag, R. A Microgel Construction Kit for Bioorthogonal Encapsulation and pH-Controlled Release of Living Cells. Angew. Chem. Int. Ed. 2013, 52, 13538–13543. [Google Scholar] [CrossRef]
- Rossow, T.; Bayer, S.; Albrecht, R.; Tzschucke, C.C.; Seiffert, S. Supramolecular Hydrogel Capsules Based on PEG: A Step Toward Degradable Biomaterials with Rational Design. Macromol. Rapid Commun. 2013, 34, 1401–1407. [Google Scholar] [CrossRef]
- Mao, A.S.; Shin, J.-W.; Utech, S.; Wang, H.; Uzun, O.; Li, W.; Cooper, M.; Hu, Y.; Zhang, L.; Weitz, D.A.; et al. Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery. Nat. Mater. 2016, 16, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.M.; Koshy, S.T.; Hilderbrand, S.A.; Mooney, D.J.; Joshi, N.S. Versatile click alginate hydrogels crosslinked via tetrazine–norbornene chemistry. Biomaterials 2015, 50, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Yanakieva, D.; Elter, A.; Bratsch, J.; Friedrich, K.; Becker, S.; Kolmar, H. FACS-Based Functional Protein Screening via Microfluidic Co-encapsulation of Yeast Secretor and Mammalian Reporter Cells. Sci. Rep. 2020, 10, 10182. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, G.; Musicò, A.; Frigerio, R.; Strada, A.; Pizzi, A.; Talone, B.; Ghezzi, J.; Gautieri, A.; Chiari, M.; Metrangolo, P.; et al. Composite Peptide–Agarose Hydrogels for Robust and High-Sensitivity 3D Immunoassays. ACS Appl. Mater. Interfaces 2022, 14, 4811–4822. [Google Scholar] [CrossRef]
- Rosiak, P.; Latanska, I.; Paul, P.; Sujka, W.; Kolesinska, B. Modification of Alginates to Modulate Their Physic-Chemical Properties and Obtain Biomaterials with Different Functional Properties. Molecules 2021, 26, 7264. [Google Scholar] [CrossRef] [PubMed]
- Lund, A.W.; Bush, J.A.; Plopper, G.E.; Stegemann, J.P. Osteogenic differentiation of mesenchymal stem cells in defined protein beads. J. Biomed. Mater. Res. B Appl. Biomater. 2008, 87, 213–221. [Google Scholar] [CrossRef]
- Mulas, C.; Hodgson, A.C.; Kohler, T.N.; Agley, C.C.; Humphreys, P.; Kleine-Brüggeney, H.; Hollfelder, F.; Smith, A.; Chalut, K.J. Microfluidic platform for 3D cell culture with live imaging and clone retrieval. Lab Chip 2020, 20, 2580–2591. [Google Scholar] [CrossRef]
- Loessner, D.; Meinert, C.; Kaemmerer, E.; Martine, L.C.; Yue, K.; Levett, P.A.; Klein, T.J.; Melchels, F.P.W.; Khademhosseini, A.; Hutmacher, D.W. Functionalization, preparation and use of cell-laden gelatin methacryloyl–based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016, 11, 727–746. [Google Scholar] [CrossRef]
- Zhang, Y.; Ouyang, H.; Lim, C.T.; Ramakrishna, S.; Huang, Z.-M. Electrospinning of gelatin fibers and gelatin/PCL composite fibrous scaffolds. J. Biomed. Mater. Res. 2005, 72B, 156–165. [Google Scholar] [CrossRef]
- Ullah, F.; Othman, M.B.H.; Javed, F.; Ahmad, Z.; Akil, H.M. Classification, processing and application of hydrogels: A review. Mater. Sci. Eng. C 2015, 57, 414–433. [Google Scholar] [CrossRef]
- Li, Z.; Leung, M.; Hopper, R.; Ellenbogen, R.; Zhang, M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials 2010, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, B.; Engelbrecht, L.K.; Fernandez, P.; Hutterer, F.P.; Raich, M.K.; Scheel, C.H.; Bausch, A.R. Mechanical plasticity of collagen directs branch elongation in human mammary gland organoids. Nat. Commun. 2021, 12, 2759. [Google Scholar] [CrossRef] [PubMed]
- Rajan, N.; Habermehl, J.; Coté, M.-F.; Doillon, C.J.; Mantovani, D. Preparation of ready-to-use, storable and reconstituted type I collagen from rat tail tendon for tissue engineering applications. Nat. Protoc. 2006, 1, 2753–2758. [Google Scholar] [CrossRef] [PubMed]
- Mazutis, L.; Gilbert, J.; Ung, W.L.; Weitz, D.A.; Griffiths, A.D.; Heyman, J.A. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2013, 8, 870–891. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Hung, K.-C.; Mitra, A.; Ung, L.W.; Lightwood, D.; Tu, R.; Starkie, D.; Cai, L.; Mazutis, L.; Chong, S.; et al. Rapid isolation of antigen-specific B-cells using droplet microfluidics. RSC Adv. 2020, 10, 27006–27013. [Google Scholar] [CrossRef]
- Yadavalli, V.K.; Svintradze, D.V.; Pidaparti, R.M. Nanoscale measurements of the assembly of collagen to fibrils. Int. J. Biol. Macromol. 2010, 46, 458–464. [Google Scholar] [CrossRef]
- Jiang, F.; Hörber, H.; Howard, J.; Müller, D.J. Assembly of collagen into microribbons: Effects of pH and electrolytes. J. Struct. Biol. 2004, 148, 268–278. [Google Scholar] [CrossRef]
- Yan, M.; Li, B.; Zhao, X.; Qin, S. Effect of concentration, pH and ionic strength on the kinetic self-assembly of acid-soluble collagen from walleye pollock (Theragra chalcogramma) skin. Food Hydrocoll. 2012, 29, 199–204. [Google Scholar] [CrossRef]
- Cheung, A.S.; Zhang, D.K.; Koshy, S.T.; Mooney, D.J. Scaffolds that mimic antigen-presenting cells enable ex vivo expansion of primary T cells. Nat. Biotechnol. 2018, 36, 160–169. [Google Scholar] [CrossRef]
- Brightman, A.; Rajwa, B.; Sturgis, J.; McCallister, M.; Robinson, J.; Voytik-Harbin, S. Time-lapse confocal reflection microscopy of collagen fibrillogenesis and extracellular matrix assembly in vitro. Biopolym. Orig. Res. Biomol. 2000, 54, 222–234. [Google Scholar] [CrossRef]
- Camacho, P.; Fainor, M.; Seims, K.B.; Tolbert, J.W.; Chow, L.W. Fabricating spatially functionalized 3D-printed scaffolds for osteochondral tissue engineering. J. Biol. Methods 2021, 8, e146. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tristan, C.A.; Chen, L.; Jovanovic, V.M.; Malley, C.; Chu, P.-H.; Ryu, S.; Deng, T.; Ormanoglu, P.; Tao, D.; et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods 2021, 18, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Grolman, J.M.; Zhang, D.; Smith, A.M.; Moore, J.S.; Kilian, K.A. Rapid 3D Extrusion of Synthetic Tumor Microenvironments. Adv. Mater. 2015, 27, 5512–5517. [Google Scholar] [CrossRef]
- Tian, Y.F.; Ahn, H.; Schneider, R.S.; Yang, S.N.; Roman-Gonzalez, L.; Melnick, A.M.; Cerchietti, L.; Singh, A. Integrin-specific hydrogels as adaptable tumor organoids for malignant B and T cells. Biomaterials 2015, 73, 110–119. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).