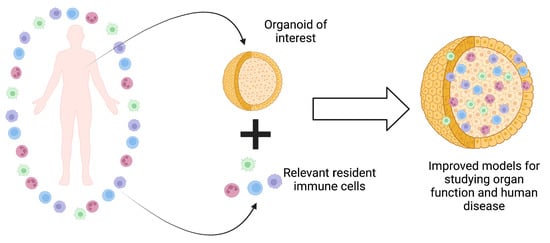
Incorporating Immune Cells into Organoid Models: Essential for Studying Human Disease
Abstract
1. Introduction
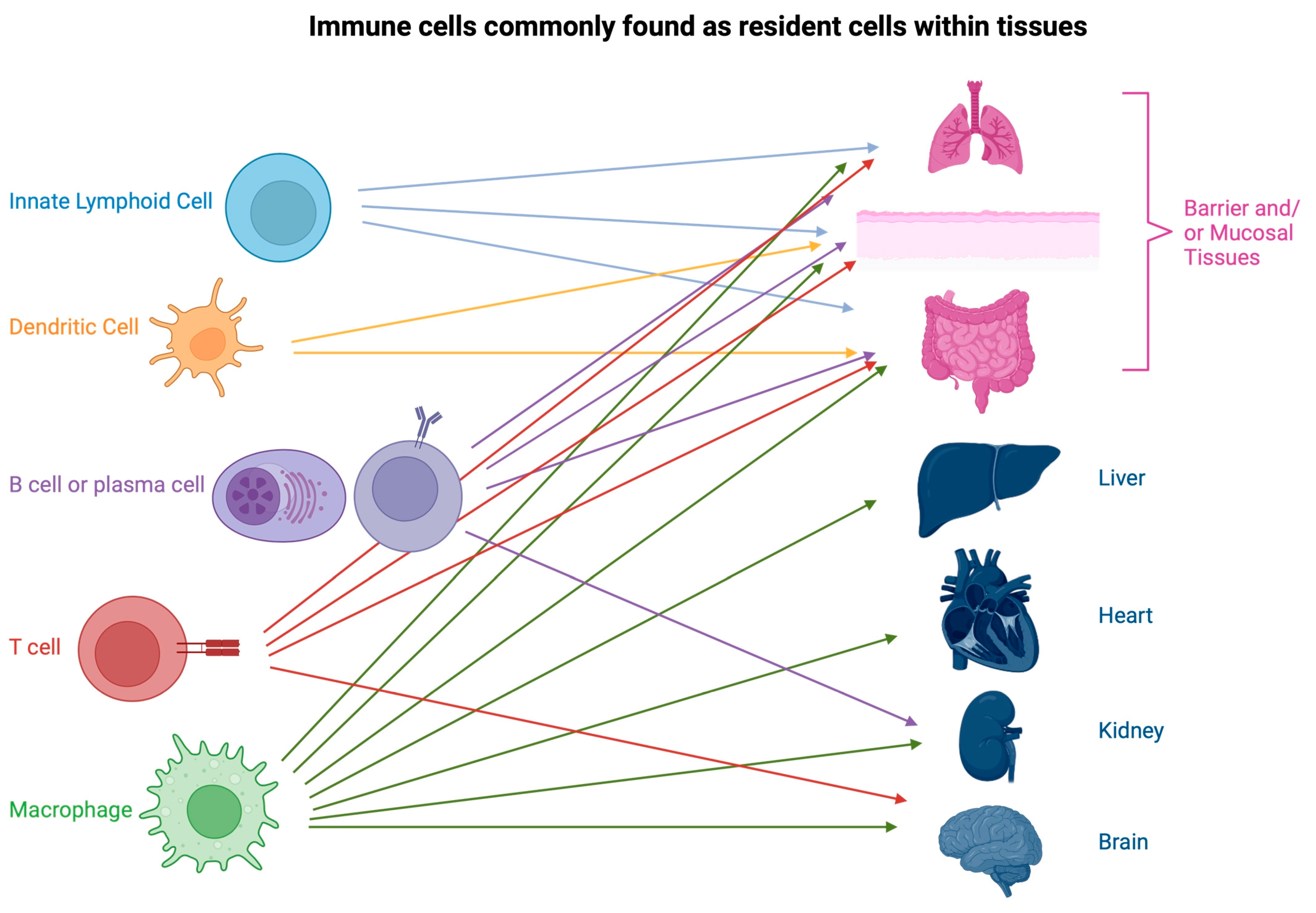
2. Resident Immune Cells
2.1. Macrophages
2.2. Dendritic Cells
2.3. T Cells
2.4. Innate Lymphoid Cells
2.5. Resident B Cells
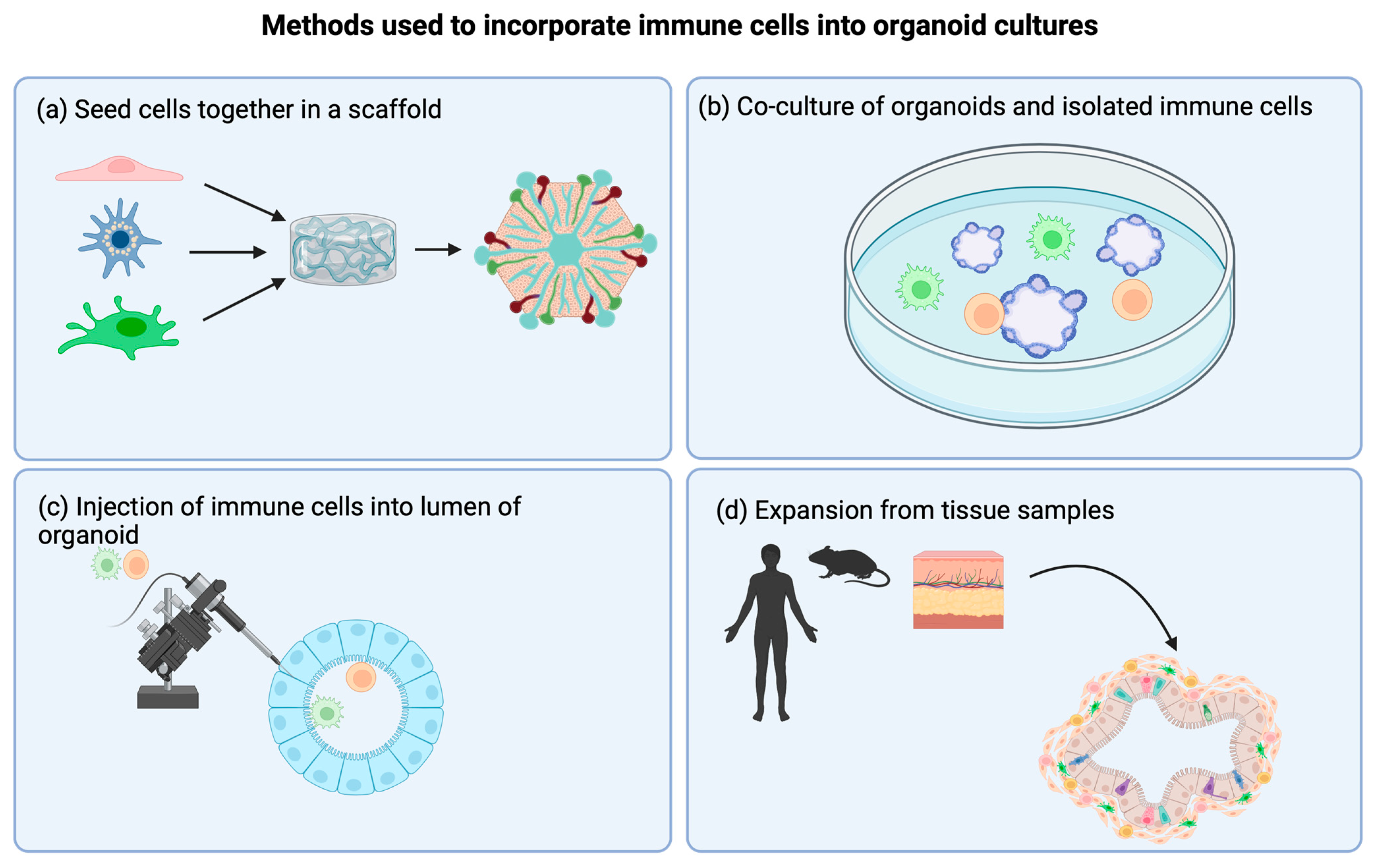
3. Incorporating Immune Cells into Organoids: Methods and Discoveries
3.1. Scaffold Systems
3.2. Co-Culture Models
3.3. Direct Injection of Immune Cells into Organoids
4. Organoids to Study Tumor Immunology
4.1. Co-Culture Models
4.2. Expansion of the Tissue Sample
5. Sources of Immune Cells
6. Future Outlook
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fogel, D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 2018, 11, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Nair, R.R.; Fisher, E.M.C.; Cunningham, T.J. Humanising the mouse genome piece by piece. Nat. Commun. 2019, 10, 1845. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Papapetrou, E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016, 22, 1392–1401. [Google Scholar] [CrossRef]
- Min, S.; Kim, S.; Cho, S.-W. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp. Mol. Med. 2020, 52, 227–237. [Google Scholar] [CrossRef]
- Kong, J.; Wen, S.; Cao, W.; Yue, P.; Xu, X.; Zhang, Y.; Luo, L.; Chen, T.; Li, L.; Wang, F.; et al. Lung organoids, useful tools for investigating epithelial repair after lung injury. Stem Cell Res. Ther. 2021, 12, 95. [Google Scholar] [CrossRef]
- Romero-Guevara, R.; Ioannides, A.; Xinaris, C. Kidney Organoids as Disease Models: Strengths, Weaknesses and Perspectives. Front. Physiol. 2020, 11, 563981. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Aguirre, A. Heart Organoids and Engineered Heart Tissues: Novel Tools for Modeling Human Cardiac Biology and Disease. Biomolecules 2021, 11, 1277. [Google Scholar] [CrossRef]
- Harrison, S.P.; Baumgarten, S.F.; Verma, R.; Lunov, O.; Dejneka, A.; Sullivan, G.J. Liver Organoids: Recent Developments, Limitations and Potential. Front. Med. 2021, 8, 574047. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, R.A.; Leopoldi, A.; Aichinger, M.; Wick, N.; Hantusch, B.; Novatchkova, M.; Taubenschmid, J.; Hämmerle, M.; Esk, C.; Bagley, J.A.; et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019, 565, 505–510. [Google Scholar] [CrossRef]
- Coffaro, K.A.; Hinegardner, R.T. Immune Response in the Sea Urchin Lytechinus pictus. Science 1977, 197, 1389–1390. [Google Scholar] [CrossRef]
- Rankin, L.C.; Artis, D. Beyond Host Defense: Emerging Functions of the Immune System in Regulating Complex Tissue Physiology. Cell 2018, 173, 554–567. [Google Scholar] [CrossRef]
- Thompson, E.C. Focus issue: Structure and function of lymphoid tissues. Trends Immunol. 2012, 33, 255. [Google Scholar] [CrossRef]
- Kabashima, K.; Honda, T.; Ginhoux, F.; Egawa, G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019, 19, 19–30. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Konkel, J.E. Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends Immunol. 2018, 39, 276–287. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, E.Y.; Lauzon-Joset, J.-F.; Debley, J.S.; Ziegler, S.F. Cross-Talk Between Alveolar Macrophages and Lung Epithelial Cells is Essential to Maintain Lung Homeostasis. Front. Immunol. 2020, 11, 583042. [Google Scholar] [CrossRef]
- Robinson, M.W.; Harmon, C.; O’farrelly, C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016, 13, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J. Immune topology of the human kidney. Nat. Rev. Nephrol. 2019, 15, 729. [Google Scholar] [CrossRef]
- Sansonetti, M.; Waleczek, F.J.G.; Jung, M.; Thum, T.; Perbellini, F. Resident cardiac macrophages: Crucial modulators of cardiac (patho)physiology. Basic Res. Cardiol. 2020, 115, 77. [Google Scholar] [CrossRef] [PubMed]
- Bogoslowski, A.; Wijeyesinghe, S.; Lee, W.-Y.; Chen, C.-S.; Alanani, S.; Jenne, C.; Steeber, D.A.; Scheiermann, C.; Butcher, E.C.; Masopust, D.; et al. Neutrophils Recirculate through Lymph Nodes to Survey Tissues for Pathogens. J. Immunol. 2020, 204, 2552–2561. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2018, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Hamidzadeh, K.; Goncalves, R. Macrophages and the maintenance of homeostasis. Cell. Mol. Immunol. 2021, 18, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Red Eagle, A.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma; controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef]
- Oishi, Y.; Manabe, I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. [Google Scholar] [CrossRef]
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449. [Google Scholar] [CrossRef]
- Gentek, R.; Molawi, K.; Sieweke, M.H. Tissue macrophage identity and self-renewal. Immunol. Rev. 2014, 262, 56–73. [Google Scholar] [CrossRef]
- Epelman, S.; LaVine, K.J.; Randolph, G.J. Origin and Functions of Tissue Macrophages. Immunity 2014, 41, 21–35. [Google Scholar] [CrossRef]
- Maus, U.A.; Koay, M.A.; Delbeck, T.; Mack, M.; Ermert, M.; Ermert, L.; Blackwell, T.S.; Christman, J.W.; Schlöndorff, D.; Seeger, W.; et al. Role of resident alveolar macrophages in leukocyte traffic into the alveolar air space of intact mice. Am. J. Physiol. Cell. Mol. Physiol. 2002, 282, L1245–L1252. [Google Scholar] [CrossRef]
- Carey, B.; Trapnell, B.C. The molecular basis of pulmonary alveolar proteinosis. Clin. Immunol. 2010, 135, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Macrophages and Systemic Iron Homeostasis. J. Innate Immun. 2012, 4, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Nose, M.; Niida, S.; Yamasaki, A. Essential role of macrophage colony-stimulating factor in the osteoclast differentiation supported by stromal cells. J. Exp. Med. 1991, 173, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.-Y.; Yoshida, H.; Sarosi, I.; Tan, H.-L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-Dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Nicolás-Ávila, J.A.; Hidalgo, A.; Ballesteros, I. Specialized functions of resident macrophages in brain and heart. J. Leukoc. Biol. 2018, 104, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.W. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009, 9, 259–270. [Google Scholar] [CrossRef]
- Lipscomb, M.F.; Masten, B.J. Dendritic Cells: Immune Regulators in Health and Disease. Physiol. Rev. 2002, 82, 97–130. [Google Scholar] [CrossRef] [PubMed]
- Matta, B.M.; Castellaneta, A.; Thomson, A.W. Tolerogenic plasmacytoid DC. Eur. J. Immunol. 2010, 40, 2667–2676. [Google Scholar] [CrossRef]
- Chen, K.; Wang, J.M.; Yuan, R.; Yi, X.; Li, L.; Gong, W.; Yang, T.; Li, L.; Su, S. Tissue-resident dendritic cells and diseases involving dendritic cell malfunction. Int. Immunopharmacol. 2016, 34, 1–15. [Google Scholar] [CrossRef]
- Merad, M.; Manz, M.G. Dendritic cell homeostasis. Blood 2009, 113, 3418–3427. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Masopust, D. Tissue-resident memory T cells. Immunity 2014, 41, 886–897. [Google Scholar] [CrossRef]
- Paik, D.H.; Farber, D.L. Anti-viral protective capacity of tissue resident memory T cells. Curr. Opin. Virol. 2021, 46, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, A.; Mnich, M.E.; Clemente, B.; Cruz, A.R.; Tavarini, S.; Bagnoli, F.; Soldaini, E. Staphylococcus aureus-Specific Tissue-Resident Memory CD4+ T Cells Are Abundant in Healthy Human Skin. Front. Immunol. 2021, 12, 642711. [Google Scholar] [CrossRef]
- Glennie, N.D.; Volk, S.W.; Scott, P. Skin-resident CD4+ T cells protect against Leishmania major by recruiting and activating inflammatory monocytes. PLoS Pathog. 2017, 13, e1006349. [Google Scholar] [CrossRef]
- Mueller, S.N.; Gebhardt, T.; Carbone, F.R.; Heath, W.R. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 2013, 31, 137–161. [Google Scholar] [CrossRef]
- Clark, R.A. Resident memory T cells in human health and disease. Sci. Transl. Med. 2015, 7, 269rv1. [Google Scholar] [CrossRef] [PubMed]
- Sasson, S.C.; Gordon, C.L.; Christo, S.N.; Klenerman, P.; Mackay, L.K. Local heroes or villains: Tissue-resident memory T cells in human health and disease. Cell. Mol. Immunol. 2020, 17, 113–122. [Google Scholar] [CrossRef]
- Dhodapkar, K.M. Role of Tissue-Resident Memory in Intra-Tumor Heterogeneity and Response to Immune Checkpoint Blockade. Front. Immunol. 2018, 9, 1655. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, D.; Min, B. Tissue Resident Foxp3+ Regulatory T Cells: Sentinels and Saboteurs in Health and Disease. Front. Immunol. 2022, 13, 865593. [Google Scholar] [CrossRef]
- Feuerer, M.; Herrero, L.; Cipolletta, D.; Naaz, A.; Wong, J.; Nayer, A.; Lee, J.; Goldfine, A.B.; Benoist, C.; Shoelson, S.; et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009, 15, 930–939. [Google Scholar] [CrossRef]
- Eller, K.; Kirsch, A.; Wolf, A.M.; Sopper, S.; Tagwerker, A.; Stanzl, U.; Wolf, D.; Patsch, W.; Rosenkranz, A.R.; Eller, P. Potential Role of Regulatory T Cells in Reversing Obesity-Linked Insulin Resistance and Diabetic Nephropathy. Diabetes 2011, 60, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Deng, Z.; Benadjaoud, F.; Yang, D.; Yang, C.; Shi, G.P. Regulatory T cells promote adipocyte beiging in subcutaneous adipose tissue. FASEB J. 2020, 34, 9755–9770. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumar, A.; Chisanga, D.; Blume, J.; Gloury, R.; Britt, K.; Henstridge, D.C.; Zhan, Y.; Torres, S.V.; Liene, S.; Collins, N. Sex-specific adipose tissue imprinting of regulatory T cells. Nature 2020, 579, 581–585. [Google Scholar] [CrossRef]
- Ali, N.; Zirak, B.; Rodriguez, R.S.; Pauli, M.L.; Truong, H.-A.; Lai, K.; Ahn, R.; Corbin, K.; Lowe, M.M.; Scharschmidt, T.C.; et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017, 169, 1119–1129. [Google Scholar] [CrossRef]
- Nosbaum, A.; Prevel, N.; Truong, H.-A.; Mehta, P.; Ettinger, M.; Scharschmidt, T.C.; Ali, N.H.; Pauli, M.L.; Abbas, A.K.; Rosenblum, M.D. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J. Immunol. 2016, 196, 2010–2014. [Google Scholar] [CrossRef]
- Shemer, A.; Scheyltjens, I.; Frumer, G.R.; Kim, J.-S.; Grozovski, J.; Ayanaw, S.; Dassa, B.; Van Hove, H.; Chappell-Maor, L.; Boura-Halfon, S.; et al. Interleukin-10 Prevents Pathological Microglia Hyperactivation following Peripheral Endotoxin Challenge. Immunity 2020, 53, 1033–1049. [Google Scholar] [CrossRef]
- Dansokho, C.; Ahmed, D.A.; Aid, S.; Toly-Ndour, C.; Chaigneau, T.; Calle, V.; Cagnard, N.; Holzenberger, M.; Piaggio, E.; Aucouturier, P.; et al. Regulatory T cells delay disease progression in Alzheimer-like pathology. Brain 2016, 139, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mao, K.; Germain, R.N. Thinking differently about ILCs-Not just tissue resident and not just the same as CD4+ T-cell effectors. Immunol. Rev. 2018, 286, 160–171. [Google Scholar] [CrossRef]
- Miller, M.M.; Reinhardt, R.L. The Heterogeneity, Origins, and Impact of Migratory iILC2 Cells in Anti-helminth Immunity. Front. Immunol. 2020, 11, 1594. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Clevers, H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 2021, 29, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, A.; Wang, Y.; Sun, Y.; Han, J.; Chen, W.; Wang, S.; Wu, Y.; Lu, Y. Innate Lymphoid Cells: Regulators of Gut Barrier Function and Immune Homeostasis. J. Immunol. Res. 2019, 2019, 2525984. [Google Scholar] [CrossRef]
- Woodruff, M.C.; Heesters, B.A.; Herndon, C.N.; Groom, J.R.; Thomas, P.G.; Luster, A.D.; Turley, S.J.; Carroll, M.C. Trans-nodal migration of resident dendritic cells into medullary interfollicular regions initiates immunity to influenza vaccine. J. Exp. Med. 2014, 211, 1611–1621. [Google Scholar] [CrossRef]
- Sitaru, C.; Mihai, S.; Zillikens, D. The relevance of the IgG subclass of autoantibodies for blister induction in autoimmune bullous skin diseases. Arch. Dermatol. Res. 2007, 299, 1–8. [Google Scholar] [CrossRef]
- Allie, S.R.; Randall, T.D. Resident Memory B Cells. Viral Immunol. 2020, 33, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Fujimoto, M.; Ishiura, N.; Kuwano, Y.; Nakashima, H.; Yazawa, N.; Okochi, H.; Sato, S.; Tedder, T.F.; Tamaki, K. CD19 expression in B cells is important for suppression of contact hypersensitivity. Am. J. Pathol. 2007, 171, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Solchaga, L.A.; Tognana, E.; Penick, K.; Baskaran, H.; Goldberg, V.M.; Caplan, A.I.; Welter, J.F. A rapid seeding technique for the assembly of large cell/scaffold composite construct. Tissue Eng. 2006, 12, 1851–1863. [Google Scholar] [CrossRef]
- Rana, D.; Arulkumar, S.; Vishwakarma, A.; Ramalingam, M. Considerations on Designing Scaffold for Tissue Engineering. In Stem Cell Biology and Tissue Engineering in Dental Sciences; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- de L’hortet, A.C.; Takeishi, K.; Guzman-Lepe, J.; Morita, K.; Achreja, A.; Popovic, B.; Wang, Y.; Handa, K.; Mittal, A.; Meurs, N.; et al. Generation of Human Fatty Livers Using Custom-Engineered Induced Pluripotent Stem Cells with Modifiable SIRT1 Metabolism. Cell Metab. 2019, 30, 385–401. [Google Scholar] [CrossRef]
- Rogoz, A.; Reis, B.S.; Karssemeijer, R.A.; Mucida, D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J. Immunol. Methods 2015, 421, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, K.; Mochizuki, W.; Matsumoto, Y.; Matsumoto, T.; Fukuda, M.; Mizutani, T.; Watanabe, M.; Nakamura, T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016, 51, 206–213. [Google Scholar] [CrossRef]
- Popova, G.; Soliman, S.S.; Kim, C.N.; Keefe, M.G.; Hennick, K.M.; Jain, S.; Li, T.; Tejera, D.; Shin, D.; Chhun, B.B.; et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell 2021, 28, 2153–2166. [Google Scholar] [CrossRef]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef]
- Vazquez-Armendariz, A.I.; Heiner, M.; El Agha, E.; Salwig, I.; Hoek, A.; Hessler, M.C.; Shalashova, I.; Shrestha, A.; Carraro, G.; Mengel, J.P.; et al. Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J. 2020, 39, e103476. [Google Scholar] [CrossRef]
- Seo, H.-R.; Han, H.-J.; Lee, Y.; Noh, Y.-W.; Cho, S.-J.; Kim, J.-H. Human Pluripotent Stem Cell-Derived Alveolar Organoid with Macrophages. Int. J. Mol. Sci. 2022, 23, 9211. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.T.; Alférez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinská, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Huang, L.; Bockorny, B.; Paul, I.; Akshinthala, D.; Frappart, P.-O.; Gandarilla, O.; Bose, A.; Sanchez-Gonzalez, V.; Rouse, E.E.; Lehoux, S.D.; et al. PDX-derived organoids model in vivo drug response and secrete biomarkers. J. Clin. Investig. 2020, 5, e135544. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Xia, T.; Du, W.; Chen, X.; Zhang, Y. Organoid models of the tumor microenvironment and their applications. J. Cell. Mol. Med. 2021, 25, 5829–5841. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, J.; Holokai, L.; Syu, L.; Steele, N.G.; Chang, J.; Wang, J.; Ahmed, S.; Dlugosz, A.; Zavros, Y. Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget 2018, 9, 37439–37457. [Google Scholar] [CrossRef]
- Holokai, L.; Chakrabarti, J.; Lundy, J.; Croagh, D.; Adhikary, P.; Richards, S.S.; Woodson, C.; Steele, N.; Kuester, R.; Scott, A.; et al. Murine- and Human-Derived Autologous Organoid / Immune Cell Co-Cultures as Pre-Clinical Models of Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 3816. [Google Scholar] [CrossRef] [PubMed]
- Koh, V.; Chakrabarti, J.; Torvund, M.; Steele, N.; Hawkins, J.A.; Ito, Y.; Ito, Y.; Wang, J.; Helmrath, M.A.; Merchant, J.L.; et al. Hedgehog transcriptional effector GLI mediates mTOR-Induced PD-L1 expression in gastric cancer organoids. Cancer Lett. 2021, 518, 59–71. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598. [Google Scholar] [CrossRef]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, I.V.A.W.; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Farma, J.M.; Abbas, A.E.-S.; et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988. [Google Scholar] [CrossRef]
- Chun, Y.S.; Byun, K.; Lee, B. Induced pluripotent stem cells and personalized medicine: Current progress and future perspectives. Anat. Cell Biol. 2011, 44, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Jaenisch, R. Technical Challenges in Using Human Induced Pluripotent Stem Cells to Model Disease. Cell Stem Cell 2009, 5, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Nianias, A.; Themeli, M. Induced Pluripotent Stem Cell (iPSC)–Derived Lymphocytes for Adoptive Cell Immunotherapy: Recent Advances and Challenges. Curr. Hematol. Malig. Rep. 2019, 14, 261–268. [Google Scholar] [CrossRef]
- Raulf-Heimsoth, M. T Cell—Primary Culture from Peripheral Blood. Allergy Methods Protoc. 2008, 138, 17–30. [Google Scholar] [CrossRef]
- Raab, S.; Klingenstein, M.; Liebau, S.; Linta, L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014, 2014, 768391. [Google Scholar] [CrossRef] [PubMed]
- Autengruber, A.; Gereke, M.; Hansen, G.; Hennig, C.; Bruder, D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur. J. Microbiol. Immunol. 2012, 2, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Blanter, M.; Gouwy, M.; Struyf, S. Studying Neutrophil Function in vitro: Cell Models and Environmental Factors. J. Inflamm. Res. 2021, 14, 141–162. [Google Scholar] [CrossRef]
- Little, M.H.; Combes, A.N. Kidney organoids: Accurate models or fortunate accidents. Genes Dev. 2019, 33, 1319–1345. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Wang, N.; Sugimura, R. Give them vasculature and immune cells—How to fill the gap of organoids. Cells Tissues Organs 2023, 1–14. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; et al. Organoids. Nat. Rev. Methods Prim. 2022, 2, 94. [Google Scholar] [CrossRef] [PubMed]
| Organoid System (Target Organoid/Scaffold) | Immune Cells Used | Method for Incorporation of Immune Cells | Outcome of Adding Immune Cells | Reference |
|---|---|---|---|---|
| Liver | Macrophages | Scaffold | Increase in inflammatory factors in a “NASH”-like model | [66] |
| Gut (enteroids) | T cells | Co-culture | Observation of T cell migration in organoid | [67] |
| Gut (enteroids) | T cells | Co-culture | Ex vivo system to study motility differences between αβ T cells and γδ T cells | [68] |
| Brain (cerebroids) | Microglia | Co-culture | Microglia modulate the gene expression of glia and neurons | [69] |
| Gut (enteroids converted to monolayer) | Macrophages | Co-culture (monolayer) | Macrophages enhance barrier function and the phagocytosis of bacteria | [70] |
| Lung (bronchioalveolar organoids) | Macrophages | Direct injection | Cytokine secretion in response to influenza A exposure | [71] |
| Lung (bronchioalveolar organoids) | Macrophages | Direct injection | Cytokine secretion in response to lipopolysaccharide injection | [72] |
| Tumor organoid (gastic cancer) | Dendritic cells and cytotoxic T cells | Co-culture | Identified role for hedgehog signaling in gastric cancer progression | [73] |
| Tumor organoid (pancreatic ductal adenocarcinoma) | Myeloid derived suppressor cells (MDSC) and cytotoxic T cells | Co-culture | Demonstrated MDSC inhibition of cytotoxic T cells | [74] |
| Tumor organoid (gastric cancer) | Myeloid derived suppressor cells | Co-culture | Demonstrated enhanced efficacy for checkpoint inhibitors with cabozantinib treatment | [75] |
| Tumor organoid | T cells | Co-culture | Enrichment of tumor-specific T cells | [76] |
| Tumor organoid | All existing tumour microenvironment immune cells | Expansion of tissue | Prediction of an immune response to checkpoint blockade | [77] |
| Tumor organoid (lung and colorectal cancers) | All existing tumor microenvironment immune cells | Expansion of tissue | High-throughput drug testing model | [78] |
| Tumor organoid | All existing tumor microenvironment immune cells | Expansion of tissue | T-cell repertoire was conserved | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogoslowski, A.; An, M.; Penninger, J.M. Incorporating Immune Cells into Organoid Models: Essential for Studying Human Disease. Organoids 2023, 2, 140-155. https://doi.org/10.3390/organoids2030011
Bogoslowski A, An M, Penninger JM. Incorporating Immune Cells into Organoid Models: Essential for Studying Human Disease. Organoids. 2023; 2(3):140-155. https://doi.org/10.3390/organoids2030011
Chicago/Turabian StyleBogoslowski, Ania, Meilin An, and Josef M. Penninger. 2023. "Incorporating Immune Cells into Organoid Models: Essential for Studying Human Disease" Organoids 2, no. 3: 140-155. https://doi.org/10.3390/organoids2030011
APA StyleBogoslowski, A., An, M., & Penninger, J. M. (2023). Incorporating Immune Cells into Organoid Models: Essential for Studying Human Disease. Organoids, 2(3), 140-155. https://doi.org/10.3390/organoids2030011