Organoid Models and Next-Generation Sequencing for Bone Marrow and Related Disorders
Abstract
1. Introduction
2. The Multifaceted BM
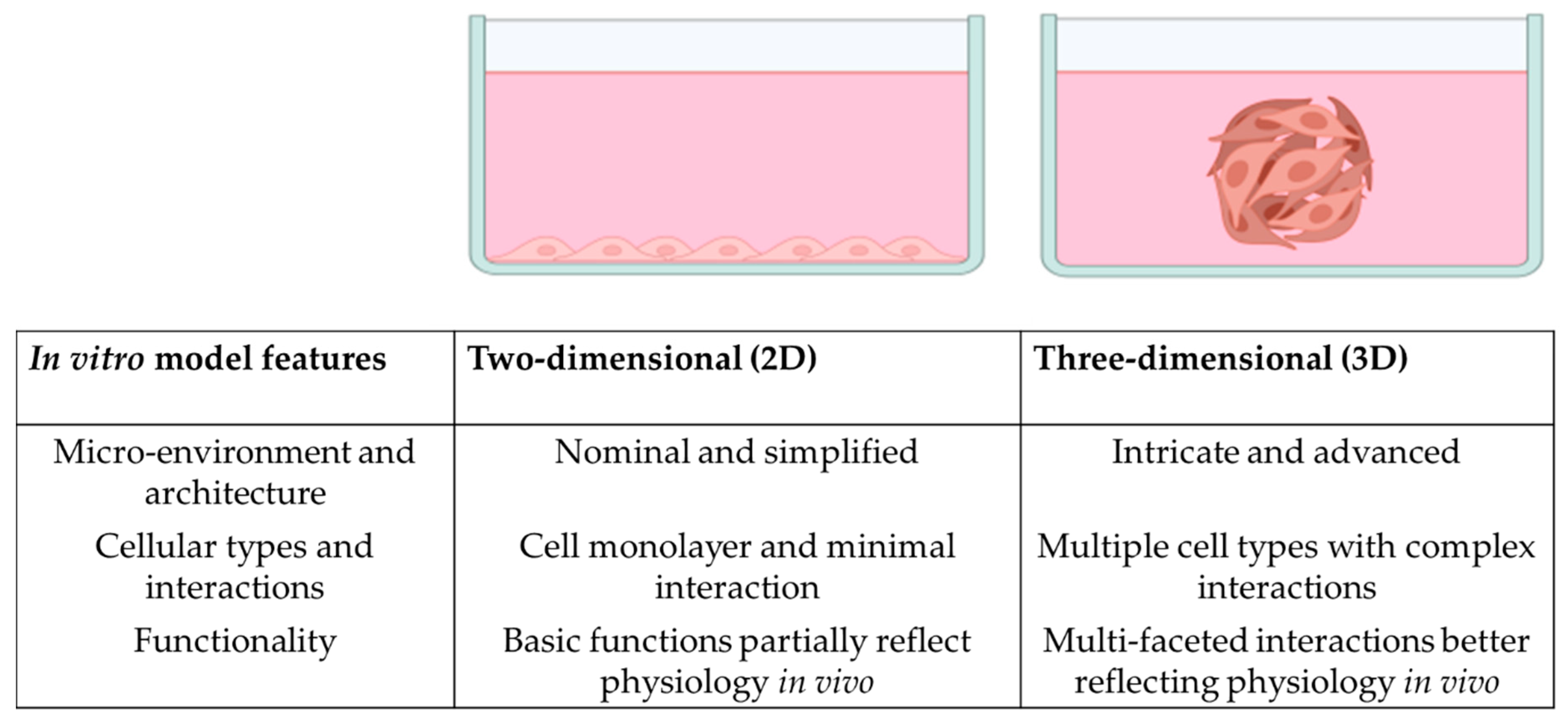
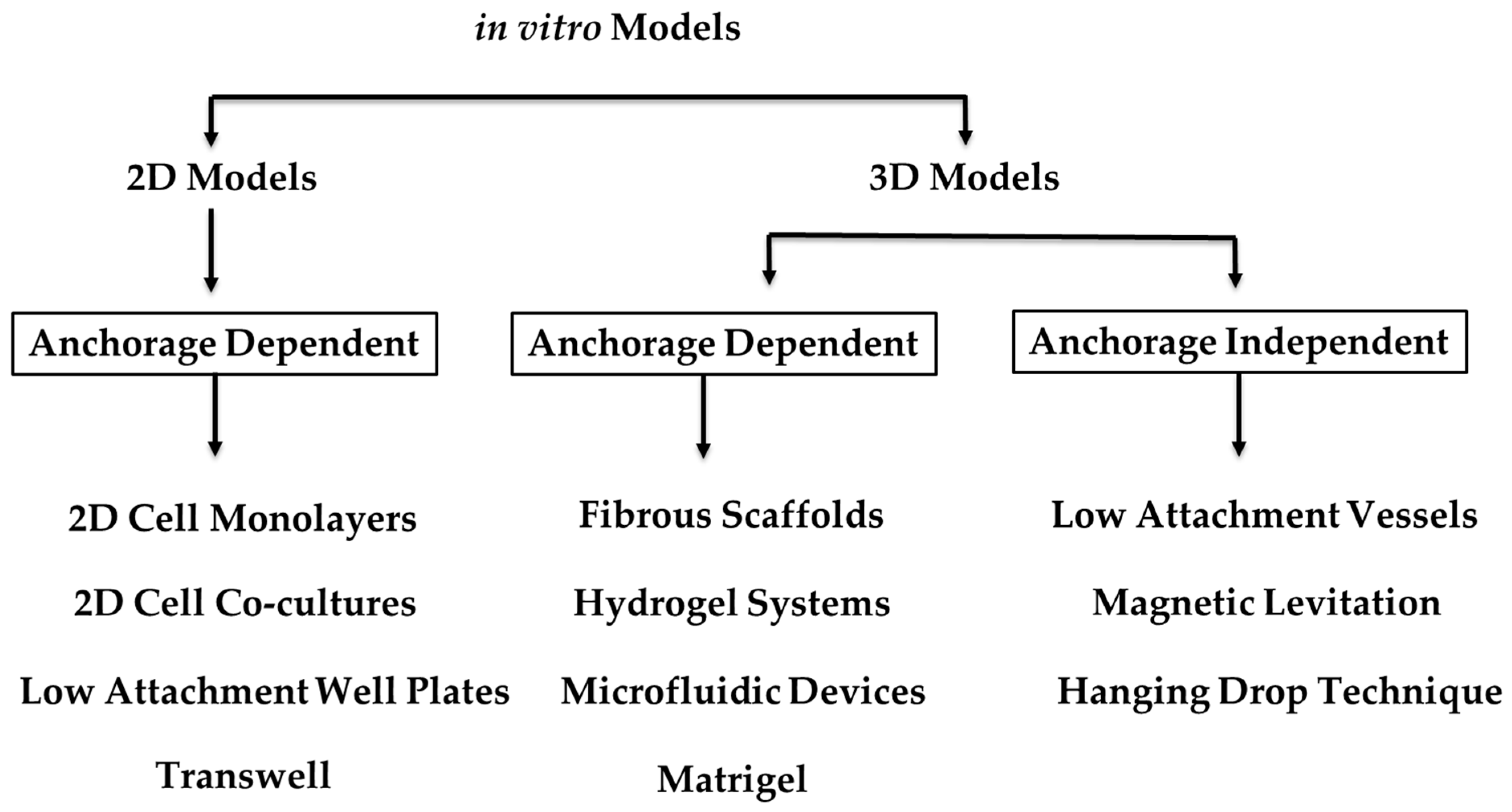
3. Organoids and 3D In Vitro Models
4. 3D Models and Organoids for the BM
4.1. Current In Vitro Models and Organoids
4.2. Scaffolds for Bone Regeneration
5. The Genetic Landscape in Organoids and 3D In Vitro Models
6. Challenges and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Safiri, S.; Kolahi, A.A.; Cross, M.; Hill, C.; Smith, E.; Carson-Chahhoud, K.; Mansournia, M.A.; Almasi-Hashiani, A.; Ashrafi-Asgarabad, A.; Kaufman, J. Prevalence, deaths, and disability-adjusted life years due to musculoskeletal disorders for 195 countries and territories 1990–2017. Arthritis Rheumatol. 2021, 73, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hendriks, M.; Chatzis, A.; Ramasamy, S.K.; Kusumbe, A.P. Bone vasculature and bone marrow vascular niches in health and disease. J. Bone Miner. Res. 2020, 35, 2103–2120. [Google Scholar] [CrossRef] [PubMed]
- Wolock, S.L.; Krishnan, I.; Tenen, D.E.; Matkins, V.; Camacho, V.; Patel, S.; Agarwal, P.; Bhatia, R.; Tenen, D.G.; Klein, A.M. Mapping distinct bone marrow niche populations and their differentiation paths. Cell Rep. 2019, 28, 302–311.e305. [Google Scholar] [CrossRef]
- Jones, E.; McGonagle, D. Human bone marrow mesenchymal stem cells in vivo. Rheumatology 2008, 47, 126–131. [Google Scholar] [CrossRef]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Moysidou, C.-M.; Barberio, C.; Owens, R.M. Advances in Engineering Human Tissue Models. Front. Bioeng. Biotechnol. 2021, 8, 620962. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Ning, B.; Zhao, Y.; Buza Iii, J.A.; Li, W.; Wang, W.; Jia, T. Surgically-induced mouse models in the study of bone regeneration: Current models and future directions (Review). Mol. Med. Rep. 2017, 15, 1017–1023. [Google Scholar] [CrossRef]
- Kleinschmidt, K.; Ploeger, F.; Nickel, J.; Glockenmeier, J.; Kunz, P.; Richter, W. Enhanced reconstruction of long bone architecture by a growth factor mutant combining positive features of GDF-5 and BMP-2. Biomaterials 2013, 34, 5926–5936. [Google Scholar] [CrossRef]
- Zhao, Y.-P.; Tian, Q.-Y.; Liu, C.-J. Progranulin deficiency exaggerates, whereas progranulin-derived Atsttrin attenuates, severity of dermatitis in mice. FEBS Lett. 2013, 587, 1805–1810. [Google Scholar] [CrossRef]
- Zhao, Y.-P.; Tian, Q.-Y.; Frenkel, S.; Liu, C.-J. The promotion of bone healing by progranulin, a downstream molecule of BMP-2, through interacting with TNF/TNFR signaling. Biomaterials 2013, 34, 6412–6421. [Google Scholar] [CrossRef]
- Ben-David, D.; Srouji, S.; Shapira-Schweitzer, K.; Kossover, O.; Ivanir, E.; Kuhn, G.; Müller, R.; Seliktar, D.; Livne, E. Low dose BMP-2 treatment for bone repair using a PEGylated fibrinogen hydrogel matrix. Biomaterials 2013, 34, 2902–2910. [Google Scholar] [CrossRef]
- Frescaline, G.; Bouderlique, T.; Mansoor, L.; Carpentier, G.; Baroukh, B.; Sineriz, F.; Trouillas, M.; Saffar, J.-L.; Courty, J.; Lataillade, J.-J. Glycosaminoglycan mimetic associated to human mesenchymal stem cell-based scaffolds inhibit ectopic bone formation, but induce angiogenesis in vivo. Tissue Eng. Part A 2013, 19, 1641–1653. [Google Scholar] [CrossRef]
- Gao, H.; Huang, J.; Wei, Q.; He, C. Advances in Animal Models for Studying Bone Fracture Healing. Bioengineering 2023, 10, 201. [Google Scholar] [CrossRef]
- Pobloth, A.-M.; Johnson, K.A.; Schell, H.; Kolarczik, N.; Wulsten, D.; Duda, G.N.; Schmidt-Bleek, K. Establishment of a preclinical ovine screening model for the investigation of bone tissue engineering strategies in cancellous and cortical bone defects. BMC Musculoskelet. Disord. 2016, 17, 111. [Google Scholar] [CrossRef]
- Barton, K.I.; Heard, B.J.; Sevick, J.L.; Martin, C.R.; Shekarforoush, S.M.; Chung, M.; Achari, Y.; Frank, C.B.; Shrive, N.G.; Hart, D.A. Posttraumatic osteoarthritis development and progression in an ovine model of partial anterior cruciate ligament transection and effect of repeated intra-articular methylprednisolone acetate injections on early disease. Am. J. Sport. Med. 2018, 46, 1596–1605. [Google Scholar] [CrossRef]
- Cone, S.G.; Warren, P.B.; Fisher, M.B. Rise of the pigs: Utilization of the porcine model to study musculoskeletal biomechanics and tissue engineering during skeletal growth. Tissue Eng. Part C Methods 2017, 23, 763–780. [Google Scholar] [CrossRef]
- Mäkelä, T.; Takalo, R.; Arvola, O.; Haapanen, H.; Yannopoulos, F.; Blanco, R.; Ahvenjärvi, L.; Kiviluoma, K.; Kerkelä, E.; Nystedt, J. Safety and biodistribution study of bone marrow–derived mesenchymal stromal cells and mononuclear cells and the impact of the administration route in an intact porcine model. Cytotherapy 2015, 17, 392–402. [Google Scholar] [CrossRef]
- Sparks, D.S.; Saifzadeh, S.; Savi, F.M.; Dlaska, C.E.; Berner, A.; Henkel, J.; Reichert, J.C.; Wullschleger, M.; Ren, J.; Cipitria, A. A preclinical large-animal model for the assessment of critical-size load-bearing bone defect reconstruction. Nat. Protoc. 2020, 15, 877–924. [Google Scholar] [CrossRef] [PubMed]
- McGovern, J.A.; Griffin, M.; Hutmacher, D.W. Animal models for bone tissue engineering and modelling disease. Dis. Model. Mech. 2018, 11, dmm033084. [Google Scholar] [CrossRef] [PubMed]
- Greek, R.; Menache, A.; Rice, M.J. Animal models in an age of personalized medicine. Pers. Med. 2012, 9, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Seifirad, S.; Haghpanah, V. Inappropriate modeling of chronic and complex disorders: How to reconsider the approach in the context of predictive, preventive and personalized medicine, and translational medicine. EPMA J. 2019, 10, 195–209. [Google Scholar] [CrossRef]
- Lo, Y.-H.; Karlsson, K.; Kuo, C.J. Applications of organoids for cancer biology and precision medicine. Nat. Cancer 2020, 1, 761–773. [Google Scholar] [CrossRef]
- El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.-H.; Matthes, T.; Preynat-Seauve, O. Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells 2023, 12, 1001. [Google Scholar] [CrossRef]
- Sereti, E.; Papapostolou, I.; Dimas, K. Pancreatic Cancer Organoids: An Emerging Platform for Precision Medicine? Biomedicines 2023, 11, 890. [Google Scholar] [CrossRef]
- Ganguly, P.; Toghill, B.; Pathak, S. Aging, bone marrow and next-generation sequencing (NGS): Recent advances and future perspectives. Int. J. Mol. Sci. 2021, 22, 12225. [Google Scholar] [CrossRef]
- Tikhonova, A.N.; Dolgalev, I.; Hu, H.; Sivaraj, K.K.; Hoxha, E.; Cuesta-Domínguez, Á.; Pinho, S.; Akhmetzyanova, I.; Gao, J.; Witkowski, M. The bone marrow microenvironment at single-cell resolution. Nature 2019, 569, 222–228. [Google Scholar] [CrossRef]
- Nelson, M.R.; Roy, K. Bone-marrow mimicking biomaterial niches for studying hematopoietic stem and progenitor cells. J. Mater. Chem. B 2016, 4, 3490–3503. [Google Scholar] [CrossRef]
- Reagan, M.R.; Rosen, C.J. Navigating the bone marrow niche: Translational insights and cancer-driven dysfunction. Nat. Rev. Rheumatol. 2016, 12, 154–168. [Google Scholar] [CrossRef]
- Wang, H.; Leng, Y.; Gong, Y. Bone marrow fat and hematopoiesis. Front. Endocrinol. 2018, 9, 694. [Google Scholar] [CrossRef]
- Ashammakhi, N.; GhavamiNejad, A.; Tutar, R.; Fricker, A.; Roy, I.; Chatzistavrou, X.; Hoque Apu, E.; Nguyen, K.-L.; Ahsan, T.; Pountos, I.; et al. Highlights on Advancing Frontiers in Tissue Engineering. Tissue Eng. Part B Rev. 2021, 28, 633–664. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Ho, W.J.; Pham, E.A.; Kim, J.W.; Ng, C.W.; Kim, J.H.; Kamei, D.T.; Wu, B.M. Incorporation of multicellular spheroids into 3-D polymeric scaffolds provides an improved tumor model for screening anticancer drugs. Cancer Sci. 2010, 101, 2637–2643. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic organ-on-a-chip technology for advancement of drug development and toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef]
- Sieber, S.; Wirth, L.; Cavak, N.; Koenigsmark, M.; Marx, U.; Lauster, R.; Rosowski, M. Bone marrow-on-a-chip: Long-term culture of human haematopoietic stem cells in a three-dimensional microfluidic environment. J. Tissue Eng. Regen. Med. 2018, 12, 479–489. [Google Scholar] [CrossRef]
- Kefallinou, D.; Grigoriou, M.; Boumpas, D.T.; Gogolides, E.; Tserepi, A. Fabrication of a 3D microfluidic cell culture device for bone marrow-on-a-chip. Micro Nano Eng. 2020, 9, 100075. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Qin, J. Microfluidic organs-on-a-chip for modeling human infectious diseases. Acc. Chem. Res. 2021, 54, 3550–3562. [Google Scholar] [CrossRef]
- Regmi, S.; Poudel, C.; Adhikari, R.; Luo, K.Q. Applications of microfluidics and organ-on-a-chip in cancer research. Biosensors 2022, 12, 459. [Google Scholar] [CrossRef]
- Satta, S.; Rockwood, S.J.; Wang, K.; Wang, S.; Mozneb, M.; Arzt, M.; Hsiai, T.K.; Sharma, A. Microfluidic organ-chips and stem cell models in the fight against COVID-19. Circ. Res. 2023, 132, 1405–1424. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hui, J.; Yang, P.; Mao, H. Microfluidic Organ-on-a-Chip System for Disease Modeling and Drug Development. Biosensors 2022, 12, 370. [Google Scholar] [CrossRef] [PubMed]
- de Souza, N. Organoids. Nat. Methods 2018, 15, 23. [Google Scholar] [CrossRef]
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; De Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019, 10, 5658. [Google Scholar] [CrossRef]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.-H.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.-G.; et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Caddeo, S.; Boffito, M.; Sartori, S. Tissue engineering approaches in the design of healthy and pathological in vitro tissue models. Front. Bioeng. Biotechnol. 2017, 5, 40. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, L.; Li, L.; Huang, C.; Shi, K.; Meng, X.; Wang, P.; Wu, M.; Li, L.; Cao, H. 3D-bioprinted BMSC-laden biomimetic multiphasic scaffolds for efficient repair of osteochondral defects in an osteoarthritic rat model. Biomaterials 2021, 279, 121216. [Google Scholar] [CrossRef]
- Lin, Y.-Y.; Kuan, C.-Y.; Chang, C.-T.; Chuang, M.-H.; Syu, W.-S.; Zhang, K.-L.; Lee, C.-H.; Lin, P.-C.; Dong, G.-C.; Lin, F.-H. 3D-Cultured Adipose-Derived Stem Cell Spheres Using Calcium-Alginate Scaffolds for Osteoarthritis Treatment in a Mono-Iodoacetate-Induced Rat Model. Int. J. Mol. Sci. 2023, 24, 7062. [Google Scholar] [CrossRef]
- Bai, H.; Cui, Y.; Wang, C.; Wang, Z.; Luo, W.; Liu, Y.; Leng, Y.; Wang, J.; Li, Z.; Liu, H. 3D printed porous biomimetic composition sustained release zoledronate to promote osteointegration of osteoporotic defects. Mater. Des. 2020, 189, 108513. [Google Scholar] [CrossRef]
- Hejazi, F.; Ebrahimi, V.; Asgary, M.; Piryaei, A.; Fridoni, M.J.; Kermani, A.A.; Zare, F.; Abdollahifar, M.-A. Improved healing of critical-size femoral defect in osteoporosis rat models using 3D elastin/polycaprolactone/nHA scaffold in combination with mesenchymal stem cells. J. Mater. Sci. Mater. Med. 2021, 32, 27. [Google Scholar] [CrossRef]
- Parente, R.; Possetti, V.; Schiavone, M.L.; Campodoni, E.; Menale, C.; Loppini, M.; Doni, A.; Bottazzi, B.; Mantovani, A.; Sandri, M. 3D co-cultures of osteoblasts and Staphylococcus aureus on biomimetic bone scaffolds as a tool to investigate the host–pathogen interface in osteomyelitis. Pathogens 2021, 10, 837. [Google Scholar] [CrossRef]
- Zhu, C.; He, M.; Sun, D.; Huang, Y.; Huang, L.; Du, M.; Wang, J.; Wang, J.; Li, Z.; Hu, B. 3D-Printed multifunctional polyetheretherketone bone scaffold for multimodal treatment of osteosarcoma and osteomyelitis. ACS Appl. Mater. Interfaces 2021, 13, 47327–47340. [Google Scholar] [CrossRef]
- Sitarski, A.M.; Fairfield, H.; Falank, C.; Reagan, M.R. 3D tissue engineered in vitro models of cancer in bone. ACS Biomater. Sci. Eng. 2018, 4, 324–336. [Google Scholar] [CrossRef]
- Posa, F.; Zerlotin, R.; Ariano, A.; Cosola, M.D.; Colaianni, G.; Fazio, A.D.; Colucci, S.; Grano, M.; Mori, G. Irisin Role in Chondrocyte 3D Culture Differentiation and Its Possible Applications. Pharmaceutics 2023, 15, 585. [Google Scholar] [CrossRef]
- Kawai, S.; Sunaga, J.; Nagata, S.; Nishio, M.; Fukuda, M.; Kamakura, T.; Sun, L.; Jin, Y.; Sakamoto, S.; Watanabe, A. 3D osteogenic differentiation of human iPSCs reveals the role of TGFβ signal in the transition from progenitors to osteoblasts and osteoblasts to osteocytes. Sci. Rep. 2023, 13, 1094. [Google Scholar] [CrossRef]
- Jeon, O.H.; Panicker, L.M.; Lu, Q.; Chae, J.J.; Feldman, R.A.; Elisseeff, J.H. Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci. Rep. 2016, 6, 26761. [Google Scholar] [CrossRef]
- Janagama, D.; Hui, S.K. 3-D Cell Culture Systems in Bone Marrow Tissue and Organoid Engineering, and BM Phantoms as In Vitro Models of Hematological Cancer Therapeutics—A Review. Materials 2020, 13, 5609. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Raic, A.; Riedel, S.; Kemmling, E.; Bieback, K.; Overhage, J.; Lee-Thedieck, C. Biomimetic 3D in vitro model of biofilm triggered osteomyelitis for investigating hematopoiesis during bone marrow infections. Acta Biomater. 2018, 73, 250–262. [Google Scholar] [CrossRef]
- Munoz-Garcia, J.; Jubelin, C.; Loussouarn, A.; Goumard, M.; Griscom, L.; Renodon-Cornière, A.; Heymann, M.-F.; Heymann, D. In vitro three-dimensional cell cultures for bone sarcomas. J. Bone Oncol. 2021, 30, 100379. [Google Scholar] [CrossRef] [PubMed]
- Jalilian, E.; Massoumi, H.; Bigit, B.; Amin, S.; Katz, E.A.; Guaiquil, V.H.; Anwar, K.N.; Hematti, P.; Rosenblatt, M.I.; Djalilian, A.R. Bone marrow mesenchymal stromal cells in a 3D system produce higher concentration of extracellular vesicles (EVs) with increased complexity and enhanced neuronal growth properties. Stem Cell Res. Ther. 2022, 13, 425. [Google Scholar] [CrossRef] [PubMed]
- Abraham, D.M.; Herman, C.; Witek, L.; Cronstein, B.N.; Flores, R.L.; Coelho, P.G. Self-assembling human skeletal organoids for disease modeling and drug testing. J. Biomed. Mater. Res. Part B Appl. Biomater. 2022, 110, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Sieberath, A.; Della Bella, E.; Ferreira, A.M.; Gentile, P.; Eglin, D.; Dalgarno, K. A comparison of osteoblast and osteoclast in vitro co-culture models and their translation for preclinical drug testing applications. Int. J. Mol. Sci. 2020, 21, 912. [Google Scholar] [CrossRef]
- Vernon, A.R.; Pemberton, R.M.; Morse, H.R. A novel in vitro 3D model of the human bone marrow to bridge the gap between in vitro and in vivo genotoxicity testing. Mutagenesis 2022, 37, 112–129. [Google Scholar] [CrossRef]
- Akiva, A.; Melke, J.; Ansari, S.; Liv, N.; van der Meijden, R.; van Erp, M.; Zhao, F.; Stout, M.; Nijhuis, W.H.; de Heus, C. An organoid for woven bone. Adv. Funct. Mater. 2021, 31, 2010524. [Google Scholar] [CrossRef]
- Visconti, R.J.; Kolaja, K.; Cottrell, J.A. A functional three-dimensional microphysiological human model of myeloma bone disease. J. Bone Miner. Res. 2021, 36, 1914–1930. [Google Scholar] [CrossRef]
- Ceccato, J.; Piazza, M.; Pizzi, M.; Manni, S.; Piazza, F.; Caputo, I.; Cinetto, F.; Pisoni, L.; Trojan, D.; Scarpa, R. A bone-based 3D scaffold as an in-vitro model of microenvironment–DLBCL lymphoma cell interaction. Front. Oncol. 2022, 12, 947823. [Google Scholar] [CrossRef]
- Suhito, I.R.; Kim, T.-H. Recent advances and challenges in organoid-on-a-chip technology. Organoid 2022, 2, e4. [Google Scholar] [CrossRef]
- .Han, J.J. FDA modernization act 2.0 allows for alternatives to animal testing. Artif. Organs 2023, 47, 449–450. [Google Scholar] [CrossRef]
- Mittal, R.; Woo, F.W.; Castro, C.S.; Cohen, M.A.; Karanxha, J.; Mittal, J.; Chhibber, T.; Jhaveri, V.M. Organ-on-chip models: Implications in drug discovery and clinical applications. J. Cell. Physiol. 2019, 234, 8352–8380. [Google Scholar] [CrossRef]
- León, I.E.; Cadavid-Vargas, J.F.; Resasco, A.; Maschi, F.; Ayala, M.A.; Carbone, C.; Etcheverry, S.B. In vitro and in vivo antitumor effects of the VO-chrysin complex on a new three-dimensional osteosarcoma spheroids model and a xenograft tumor in mice. JBIC J. Biol. Inorg. Chem. 2016, 21, 1009–1020. [Google Scholar] [CrossRef]
- Verrier, S.; Alini, M.; Alsberg, E.; Buchman, S.; Kelly, D.; Laschke, M.; Menger, M.; Murphy, W.; Stegemann, J.; Schütz, M. Tissue engineering and regenerative approaches to improving the healing of large bone defects. Eur. Cell. Mater. 2016, 32, 87–110. [Google Scholar] [CrossRef]
- Perez, J.R.; Kouroupis, D.; Li, D.J.; Best, T.M.; Kaplan, L.; Correa, D. Tissue Engineering and Cell-Based Therapies for Fractures and Bone Defects. Front. Bioeng. Biotechnol. 2018, 6, 105. [Google Scholar] [CrossRef]
- Qu, H.; Fu, H.; Han, Z.; Sun, Y. Biomaterials for bone tissue engineering scaffolds: A review. RSC Adv. 2019, 9, 26252–26262. [Google Scholar] [CrossRef]
- Filippi, M.; Born, G.; Chaaban, M.; Scherberich, A. Natural polymeric scaffolds in bone regeneration. Front. Bioeng. Biotechnol. 2020, 8, 474. [Google Scholar] [CrossRef]
- Iqbal, N.; Braxton, T.M.; Anastasiou, A.; Raif, E.M.; Chung, C.K.Y.; Kumar, S.; Giannoudis, P.V.; Jha, A. Dicalcium Phosphate Dihydrate Mineral Loaded Freeze-Dried Scaffolds for Potential Synthetic Bone Applications. Materials 2022, 15, 6245. [Google Scholar] [CrossRef]
- Pereira, H.F.; Cengiz, I.F.; Silva, F.S.; Reis, R.L.; Oliveira, J.M. Scaffolds and coatings for bone regeneration. J. Mater. Sci. Mater. Med. 2020, 31, 27. [Google Scholar] [CrossRef]
- Ganguly, P.; El-Jawhari, J.J.; Vun, J.; Giannoudis, P.V.; Jones, E.A. Evaluation of Human Bone Marrow Mesenchymal Stromal Cell (MSC) Functions on a Biomorphic Rattan-Wood-Derived Scaffold: A Comparison between Cultured and Uncultured MSCs. Bioengineering 2022, 9, 1. [Google Scholar] [CrossRef]
- Chen, G.; Lv, Y. Decellularized bone matrix scaffold for bone regeneration. Decellularized Scaffolds Organog. Methods Protoc. 2018, 1577, 239–254. [Google Scholar]
- Roseti, L.; Parisi, V.; Petretta, M.; Cavallo, C.; Desando, G.; Bartolotti, I.; Grigolo, B. Scaffolds for bone tissue engineering: State of the art and new perspectives. Mater. Sci. Eng. C 2017, 78, 1246–1262. [Google Scholar] [CrossRef] [PubMed]
- Maté-Sánchez de Val, J.E.; Mazón, P.; Calvo-Guirado, J.L.; Ruiz, R.A.D.; Ramírez Fernández, M.P.; Negri, B.; Abboud, M.; De Aza, P.N. Comparison of three hydroxyapatite/β-tricalcium phosphate/collagen ceramic scaffolds: An in vivo study. J. Biomed. Mater. Res. Part A 2014, 102, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Yıldızhan, Ş.; Çalık, A.; Ozcanli, M.; Serin, H. Bio-composite materials: A short review of recent trends, mechanical and chemical properties, and applications. Eur. Mech. Sci. 2018, 2, 83–91. [Google Scholar] [CrossRef]
- Kumari, S.; Katiyar, S.; Darshna; Anand, A.; Singh, D.; Singh, B.N.; Mallick, S.P.; Mishra, A.; Srivastava, P. Design strategies for composite matrix and multifunctional polymeric scaffolds with enhanced bioactivity for bone tissue engineering. Front. Chem. 2022, 10, 1051678. [Google Scholar] [CrossRef]
- Ansari, A.I.; Sheikh, N.A. A Review of Bone Regeneration Mechanisms and Bone Scaffold Fabrication Techniques (Conventional and Non-Conventional). J. Inst. Eng. (India) Ser. C 2022, 103, 1485–1513. [Google Scholar] [CrossRef]
- Safari, B.; Davaran, S.; Aghanejad, A. Osteogenic potential of the growth factors and bioactive molecules in bone regeneration. Int. J. Biol. Macromol. 2021, 175, 544–557. [Google Scholar] [CrossRef]
- Hodge, J.; Quint, C. The improvement of cell infiltration in an electrospun scaffold with multiple synthetic biodegradable polymers using sacrificial PEO microparticles. J. Biomed. Mater. Res. Part A 2019, 107, 1954–1964. [Google Scholar] [CrossRef]
- Ho-Shui-Ling, A.; Bolander, J.; Rustom, L.E.; Johnson, A.W.; Luyten, F.P.; Picart, C. Bone regeneration strategies: Engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials 2018, 180, 143–162. [Google Scholar] [CrossRef]
- Sadtler, K.; Wolf, M.T.; Ganguly, S.; Moad, C.A.; Chung, L.; Majumdar, S.; Housseau, F.; Pardoll, D.M.; Elisseeff, J.H. Divergent immune responses to synthetic and biological scaffolds. Biomaterials 2019, 192, 405–415. [Google Scholar] [CrossRef]
- Lee, Y.J.; Ryu, Y.H.; Lee, S.J.; Moon, S.-H.; Kim, K.J.; Jin, B.J.; Lee, K.-D.; Park, J.K.; Lee, J.W.; Lee, S.-J.; et al. Bone Regeneration with 3D-Printed Hybrid Bone Scaffolds in a Canine Radial Bone Defect Model. Tissue Eng. Regen. Med. 2022, 19, 1337–1347. [Google Scholar] [CrossRef]
- Kronemberger, G.S.; Dalmônico, G.M.; Rossi, A.L.; Leite, P.E.C.; Saraiva, A.M.; Beatrici, A.; Silva, K.R.; Granjeiro, J.M.; Baptista, L.S. Scaffold-and serum-free hypertrophic cartilage tissue engineering as an alternative approach for bone repair. Artif. Organs 2020, 44, E288–E299. [Google Scholar] [CrossRef]
- Anil-Inevi, M.; Yilmaz, E.; Sarigil, O.; Tekin, H.C.; Ozcivici, E. Single cell densitometry and weightlessness culture of mesenchymal stem cells using magnetic levitation. Stem Cell Nanotechnol. Methods Protoc. 2020, 2125, 15–25. [Google Scholar]
- Ganguly, P.; Jones, E.; Panagiotopoulou, V.; Jha, A.; Blanchy, M.; Antimisiaris, S.; Anton, M.; Dhuiège, B.; Marotta, M.; Marjanovic, N. Electrospun and 3D printed polymeric materials for one-stage critical-size long bone defect regeneration inspired by the Masquelet technique: Recent Advances. Injury 2022, 53, S2–S12. [Google Scholar] [CrossRef]
- Stevens, B.; Yang, Y.; Mohandas, A.; Stucker, B.; Nguyen, K.T. A review of materials, fabrication methods, and strategies used to enhance bone regeneration in engineered bone tissues. J. Biomed. Mater. Res. Part B Appl. Biomater. 2008, 85, 573–582. [Google Scholar] [CrossRef]
- Polo-Corrales, L.; Latorre-Esteves, M.; Ramirez-Vick, J.E. Scaffold design for bone regeneration. J. Nanosci. Nanotechnol. 2014, 14, 15–56. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Guo, J.; Tierney, E.G.; Curtin, C.M.; Malhotra, M.; Darcy, R.; O’Brien, F.J.; O’Driscoll, C.M. The use of collagen-based scaffolds to simulate prostate cancer bone metastases with potential for evaluating delivery of nanoparticulate gene therapeutics. Biomaterials 2015, 66, 53–66. [Google Scholar] [CrossRef]
- Dadwal, U.C.; Merkel, A.R.; Page, J.M.; Kwakwa, K.A.; Kessler, M.; Rhoades, J.A. 3D bone morphology alters gene expression, motility, and drug responses in bone metastatic tumor cells. Int. J. Mol. Sci. 2020, 21, 6913. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.; Liguori, M.J. Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Wu, H.; Wei, X.; Liu, Y.; Dong, H.; Tang, Z.; Wang, N.; Bao, S.; Wu, Z.; Shi, L.; Zheng, X. Dynamic degradation patterns of porous polycaprolactone/β-tricalcium phosphate composites orchestrate macrophage responses and immunoregulatory bone regeneration. Bioact. Mater. 2023, 21, 595–611. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, J.; Maevskaia, E.; Ghayor, C.; Bhattacharya, I.; Weber, F.E. Influence of Scaffold Microarchitecture on Angiogenesis and Regulation of Cell Differentiation during the Early Phase of Bone Healing: A Transcriptomics and Histological Analysis. Int. J. Mol. Sci. 2023, 24, 6000. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Gao, M.; Cavnar, M.J.; Kim, J. Utilizing gastric cancer organoids to assess tumor biology and personalize medicine. World J. Gastrointest. Oncol. 2019, 11, 509–517. [Google Scholar] [CrossRef] [PubMed]
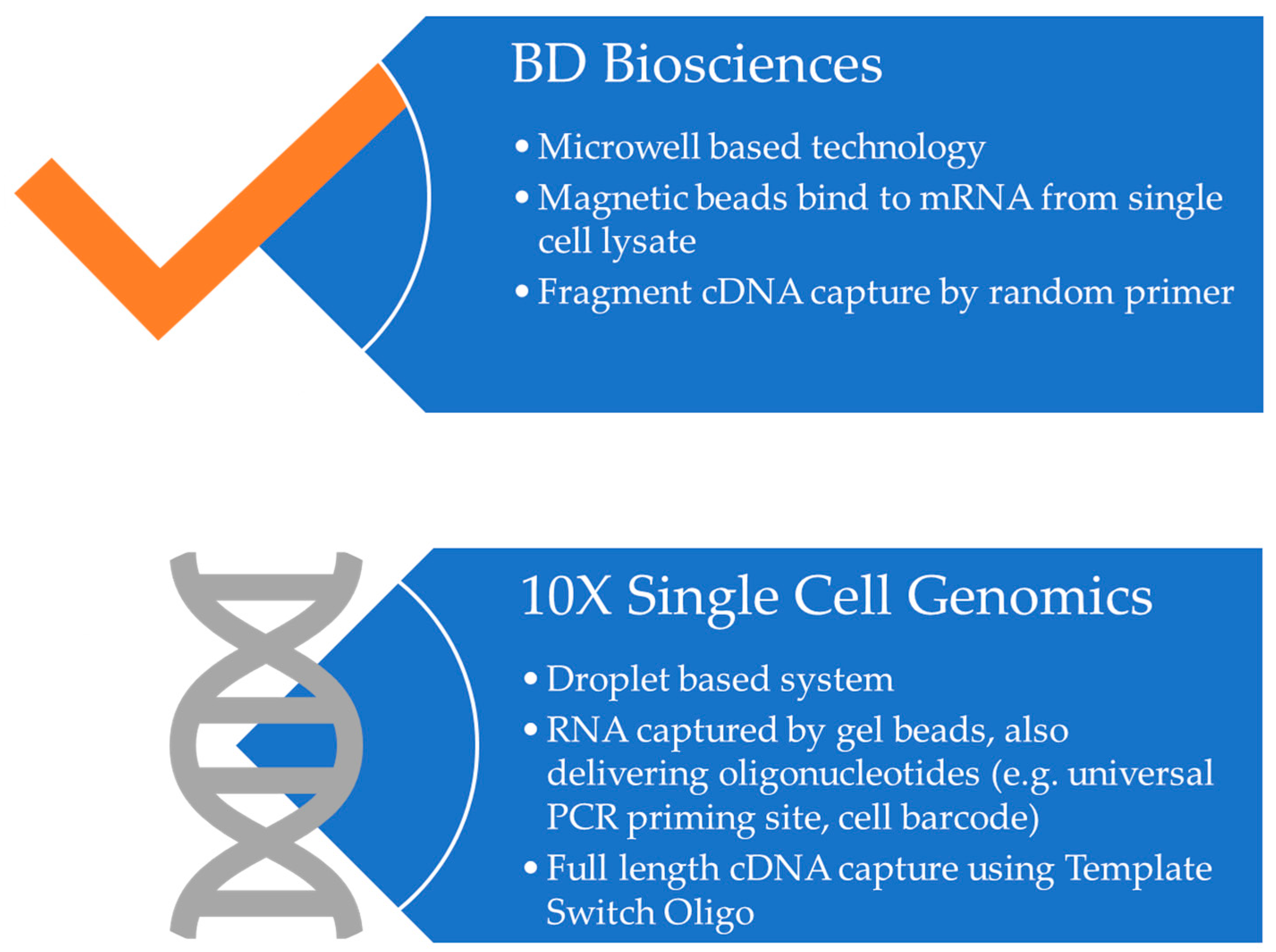
- Gao, C.; Zhang, M.; Chen, L. The Comparison of Two Single-cell Sequencing Platforms: BD Rhapsody and 10x Genomics Chromium. Curr. Genom. 2020, 21, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, J.; Lopez-Salmeron, V.; Gerrard, I. BD Rhapsody™ Single-Cell Analysis System Workflow: From Sample to Multimodal Single-Cell Sequencing Data. In Single Cell Transcriptomics; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2023; Volume 2584. [Google Scholar] [CrossRef]
- Khan, A.O.; Rodriguez-Romera, A.; Reyat, J.S.; Olijnik, A.-A.; Colombo, M.; Wang, G.; Wen, W.X.; Sousos, N.; Murphy, L.C.; Grygielska, B.; et al. Human Bone Marrow Organoids for Disease Modeling, Discovery, and Validation of Therapeutic Targets in Hematologic Malignancies. Cancer Discov. 2023, 13, 364–385. [Google Scholar] [CrossRef]
- Pathak, S.; Rowczenio, D.; Lara-Reyna, S.; Kacar, M.; Owen, R.; Doody, G.; Krause, K.; Lachmann, H.; Doffinger, R.; Newton, D.; et al. Evidence of B Cell Clonality and Investigation Into Properties of the IgM in Patients With Schnitzler Syndrome. Front. Immunol. 2020, 11, 569006. [Google Scholar] [CrossRef]
- Pathak, S.; Rowczenio, D.M.; Owen, R.G.; Doody, G.M.; Newton, D.J.; Taylor, C.; Taylor, J.; Cargo, C.; Hawkins, P.N.; Krause, K.; et al. Exploratory study of MYD88 L265P, rare NLRP3 variants and clonal hematopoiesis prevalence in patients with Schnitzler’s Syndrome. Arthritis Rheumatol. 2019, 71, 2121–2125. [Google Scholar] [CrossRef]
- Rowczenio, D.M.; Pathak, S.; Arostegui, J.I.; Mensa-Vilaro, A.; Omoyinmi, E.; Brogan, P.; Lipsker, D.; Scambler, T.; Owen, R.; Trojer, H.; et al. Molecular genetic investigation, clinical features, and response to treatment in 21 patients with Schnitzler syndrome. Blood 2018, 131, 974–981. [Google Scholar] [CrossRef]
- Wang, X.; He, Y.; Zhang, Q.; Ren, X.; Zhang, Z. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Genom. Proteom. Bioinform. 2021, 19, 253–266. [Google Scholar] [CrossRef]
- Bunpetch, V.; Zhang, X.; Li, T.; Lin, J.; Maswikiti, E.P.; Wu, Y.; Cai, D.; Li, J.; Zhang, S.; Wu, C. Silicate-based bioceramic scaffolds for dual-lineage regeneration of osteochondral defect. Biomaterials 2019, 192, 323–333. [Google Scholar] [CrossRef]
- Ji, C.; Qiu, M.; Ruan, H.; Li, C.; Cheng, L.; Wang, J.; Li, C.; Qi, J.; Cui, W.; Deng, L. Transcriptome analysis revealed the symbiosis niche of 3D scaffolds to accelerate bone defect healing. Adv. Sci. 2022, 9, 2105194. [Google Scholar] [CrossRef]
- McCray, T.; Moline, D.; Baumann, B.; Vander Griend, D.J.; Nonn, L. Single-cell RNA-Seq analysis identifies a putative epithelial stem cell population in human primary prostate cells in monolayer and organoid culture conditions. Am. J. Clin. Exp. Urol. 2019, 7, 123. [Google Scholar]
- Ma, Q.; Tao, H.; Li, Q.; Zhai, Z.; Zhang, X.; Lin, Z.; Ni Kuang, N.; Pan, J. OrganoidDB: A comprehensive organoid database for the multi-perspective exploration of bulk and single-cell transcriptomic profiles of organoids. Nucleic Acids Res. 2023, 51, D1086–D1093. [Google Scholar] [CrossRef]
- Correia, C.R.; Bjørge, I.M.; Zeng, J.; Matsusaki, M.; Mano, J.F. Liquefied Microcapsules as Dual-Microcarriers for 3D+ 3D Bottom-Up Tissue Engineering. Adv. Healthc. Mater. 2019, 8, 1901221. [Google Scholar] [CrossRef]
- Oliveira, C.S.; Nadine, S.; Gomes, M.C.; Correia, C.R.; Mano, J.F. Bioengineering the human bone marrow microenvironment in liquefied compartments: A promising approach for the recapitulation of osteovascular niches. Acta Biomater. 2022, 149, 167–178. [Google Scholar] [CrossRef]
- Wang, H.; Brown, P.C.; Chow, E.C.; Ewart, L.; Ferguson, S.S.; Fitzpatrick, S.; Freedman, B.S.; Guo, G.L.; Hedrich, W.; Heyward, S. 3D cell culture models: Drug pharmacokinetics, safety assessment, and regulatory consideration. Clin. Transl. Sci. 2021, 14, 1659–1680. [Google Scholar] [CrossRef]
- Marinucci, M.; Ercan, C.; Taha-Mehlitz, S.; Fourie, L.; Panebianco, F.; Bianco, G.; Gallon, J.; Staubli, S.; Soysal, S.D.; Zettl, A. Standardizing Patient-Derived Organoid Generation Workflow to Avoid Microbial Contamination From Colorectal Cancer Tissues. Front. Oncol. 2022, 11, 5605. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Vives, J.; Batlle-Morera, L. The challenge of developing human 3D organoids into medicines. Stem Cell Res. Ther. 2020, 11, 72. [Google Scholar] [CrossRef]
- Cahan, P.; Li, H.; Morris, S.A.; da Rocha, E.L.; Daley, G.Q.; Collins, J.J. CellNet: Network biology applied to stem cell engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef]
- Radley, A.H.; Schwab, R.M.; Tan, Y.; Kim, J.; Lo, E.K.W.; Cahan, P. Assessment of engineered cells using CellNet and RNA-seq. Nat. Protoc. 2017, 12, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Cahan, P. SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data Across Platforms and Across Species. Cell Syst. 2019, 9, 207–213.e2. [Google Scholar] [CrossRef] [PubMed]
- Lensink, M.A.; Jongsma, K.R.; Boers, S.N.; Noordhoek, J.J.; Beekman, J.M.; Bredenoord, A.L. Responsible use of organoids in precision medicine: The need for active participant involvement. Development 2020, 147, dev177972. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Cong, L.; Cong, X. Patient-derived organoids in precision medicine: Drug screening, organoid-on-a-chip and living organoid biobank. Front. Oncol. 2021, 11, 5625. [Google Scholar] [CrossRef] [PubMed]
- Papaccio, F.; Cabeza-Segura, M.; Garcia-Micò, B.; Tarazona, N.; Roda, D.; Castillo, J.; Cervantes, A. Will Organoids Fill the Gap towards Functional Precision Medicine? J. Pers. Med. 2022, 12, 1939. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Geng, Z.; Su, J. The horizon of bone organoid: A perspective on construction and application. Bioact. Mater. 2022, 18, 15–25. [Google Scholar] [CrossRef]
- Liu, Y.; Gan, Y.; AiErken, N.; Chen, W.; Zhang, S.; Ouyang, J.; Zeng, L.; Tang, D. Combining Organoid Models with Next-Generation Sequencing to Reveal Tumor Heterogeneity and Predict Therapeutic Response in Breast Cancer. J. Oncol. 2022, 2022, 9390912. [Google Scholar] [CrossRef]
- Agarwal, D.; Kuhns, R.; Dimitriou, C.N.; Barlow, E.; Wahlin, K.J.; Enke, R.A. Bulk RNA sequencing analysis of developing human induced pluripotent cell-derived retinal organoids. Sci. Data 2022, 9, 759. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rausch, M.; Iqbal, N.; Pathak, S.; Owston, H.E.; Ganguly, P. Organoid Models and Next-Generation Sequencing for Bone Marrow and Related Disorders. Organoids 2023, 2, 123-139. https://doi.org/10.3390/organoids2030010
Rausch M, Iqbal N, Pathak S, Owston HE, Ganguly P. Organoid Models and Next-Generation Sequencing for Bone Marrow and Related Disorders. Organoids. 2023; 2(3):123-139. https://doi.org/10.3390/organoids2030010
Chicago/Turabian StyleRausch, Magdalena, Neelam Iqbal, Shelly Pathak, Heather E. Owston, and Payal Ganguly. 2023. "Organoid Models and Next-Generation Sequencing for Bone Marrow and Related Disorders" Organoids 2, no. 3: 123-139. https://doi.org/10.3390/organoids2030010
APA StyleRausch, M., Iqbal, N., Pathak, S., Owston, H. E., & Ganguly, P. (2023). Organoid Models and Next-Generation Sequencing for Bone Marrow and Related Disorders. Organoids, 2(3), 123-139. https://doi.org/10.3390/organoids2030010








