3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy
Abstract
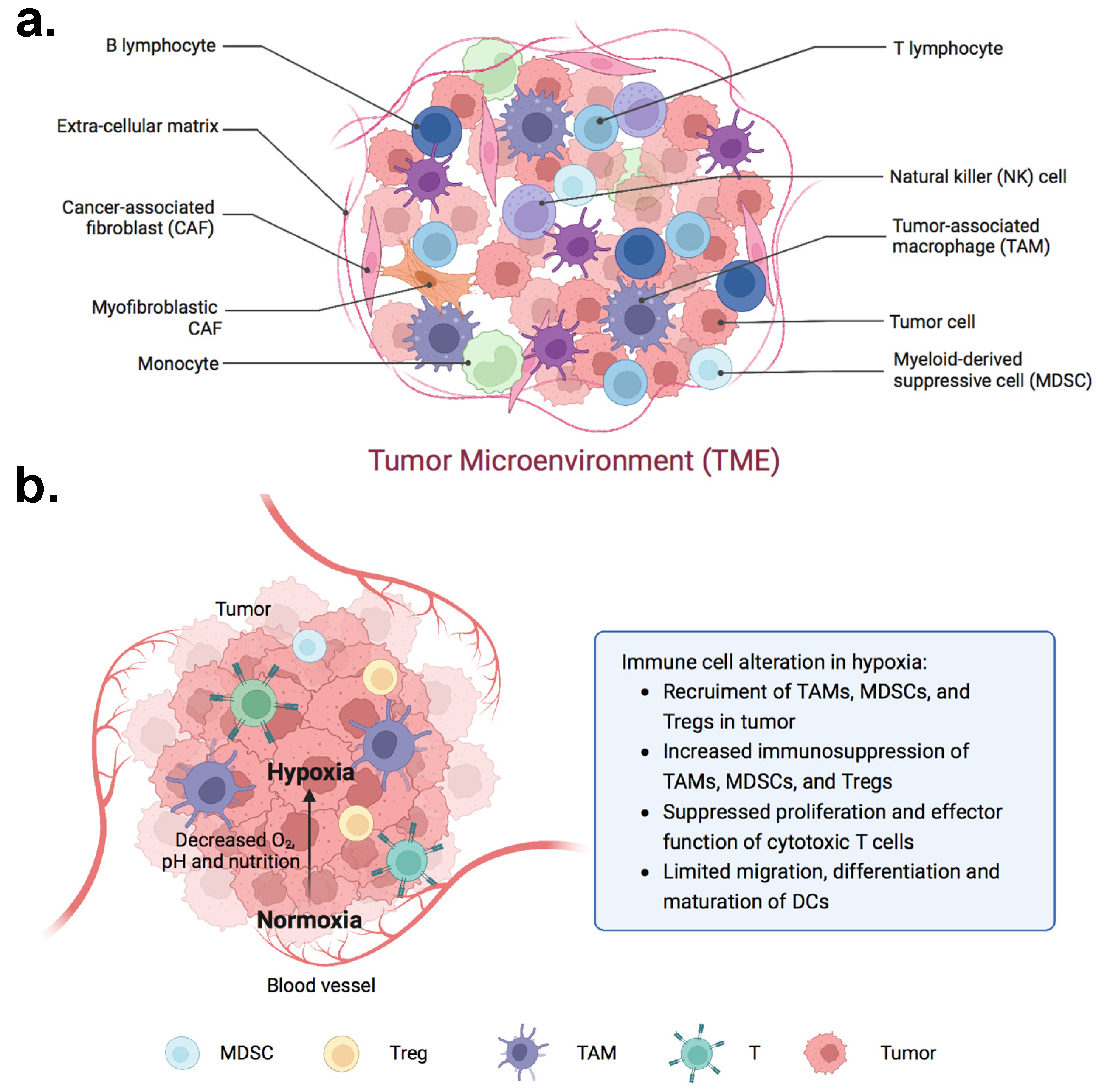
1. Introduction: The Tumor Microenvironment as a Barrier to Cancer Immunotherapy
2. 3D Modeling of the TME
3. Cell Line-Derived Tumor Spheroids
3.1. Culture Methods
3.2. TME Modeling Capabilities of Spheroids
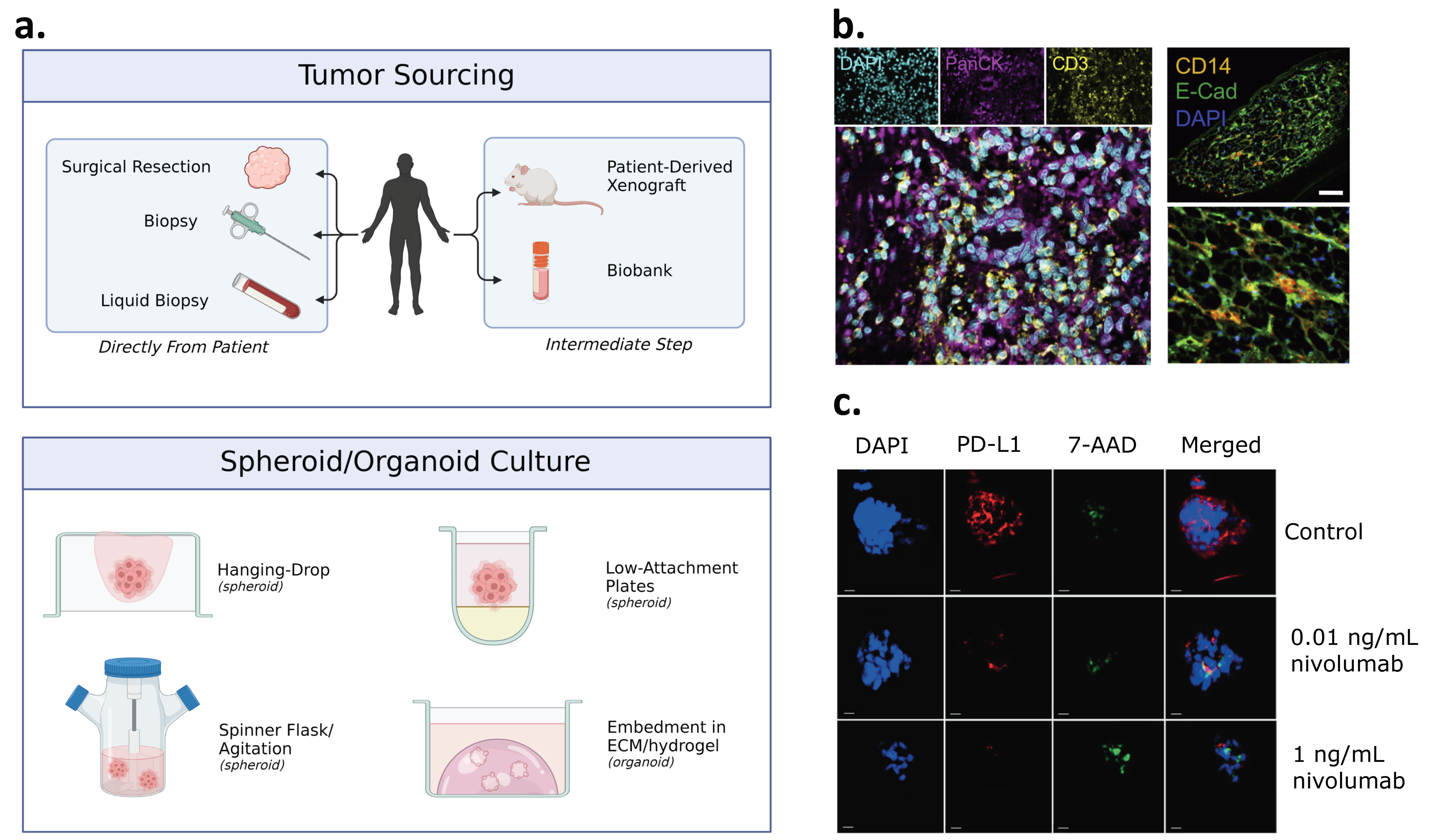
4. Patient-Derived Spheroid and Organoid Models
4.1. Patient-Derived Spheroids
4.2. Patient-Derived Organoids
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The Tumor Microenvironment and Its Role in Promoting Tumor Growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of Tumor Microenvironment in Tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New Horizons in Tumor Microenvironment Biology: Challenges and Opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef]
- Nagl, L.; Horvath, L.; Pircher, A.; Wolf, D. Tumor Endothelial Cells (TECs) as Potential Immune Directors of the Tumor Microenvironment—New Findings and Future Perspectives. Front. Cell Dev. Biol. 2020, 8, 766. [Google Scholar] [CrossRef]
- Hida, K.; Maishi, N.; Torii, C.; Hida, Y. Tumor Angiogenesis—Characteristics of Tumor Endothelial Cells. Int. J. Clin. Oncol. 2016, 21, 206–212. [Google Scholar] [CrossRef]
- Xing, F.; Saidou, J.; Watabe, K. Cancer Associated Fibroblasts (CAFs) in Tumor Microenvironment. Front. Biosci. Landmark Ed. 2010, 15, 166–179. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The Extracellular Matrix Modulates the Hallmarks of Cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in Cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Willumsen, N.; Thomsen, L.B.; Bager, C.L.; Jensen, C.; Karsdal, M.A. Quantification of Altered Tissue Turnover in a Liquid Biopsy: A Proposed Precision Medicine Tool to Assess Chronic Inflammation and Desmoplasia Associated with a Pro-Cancerous Niche and Response to Immuno-Therapeutic Anti-Tumor Modalities. Cancer Immunol. Immunother. 2017, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-R.; Brown, J.; Yu, Y.; Lee, D.; Zhou, K.; Dunn, Z.S.; Hon, R.; Wilson, M.; Kramer, A.; Zhu, Y.; et al. Targeting Immunosuppressive Tumor-Associated Macrophages Using Innate T Cells for Enhanced Antitumor Reactivity. Cancers 2022, 14, 2749. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Z.; Walpole, G.F.; Yu, Y. Kinetics of Phagosome Maturation Is Coupled to Their Intracellular Motility. Commun. Biol. 2022, 5, 1014. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, T.; Lawrence, T.; McNeish, I.; Charles, K.A.; Kulbe, H.; Thompson, R.G.; Robinson, S.C.; Balkwill, F.R. “Re-Educating” Tumor-Associated Macrophages by Targeting Nf-Kappab. J. Exp. Med. 2008, 205, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Doedens, A.L.; Stockmann, C.; Rubinstein, M.P.; Liao, D.; Zhang, N.; DeNardo, D.G.; Coussens, L.M.; Karin, M.; Goldrath, A.W.; Johnson, R.S. Macrophage Expression of Hypoxia-Inducible Factor-1 Alpha Suppresses T-Cell Function and Promotes Tumor Progression. Cancer Res. 2010, 70, 7465–7475. [Google Scholar] [CrossRef]
- Talmadge, J.E.; Gabrilovich, D.I. History of Myeloid-Derived Suppressor Cells. Nat. Rev. Cancer 2013, 13, 739–752. [Google Scholar] [CrossRef]
- Park, Y.-J.; Song, B.; Kim, Y.-S.; Kim, E.-K.; Lee, J.-M.; Lee, G.-E.; Kim, J.-O.; Kim, Y.-J.; Chang, W.-S.; Kang, C.-Y. Tumor Microenvironmental Conversion of Natural Killer Cells into Myeloid-Derived Suppressor Cells. Cancer Res. 2013, 73, 5669–5681. [Google Scholar] [CrossRef]
- Highfill, S.L.; Cui, Y.; Giles, A.J.; Smith, J.P.; Zhang, H.; Morse, E.; Kaplan, R.N.; Mackall, C.L. Disruption of CXCR2-Mediated MDSC Tumor Trafficking Enhances Anti-PD1 Efficacy. Sci. Transl. Med. 2014, 6, 237ra67. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, H.; Sun, M.; Deng, X.; Wu, X.; Ma, Y.; Li, M.; Shuoa, S.M.; You, Q.; Miao, L. Cxcl5/Cxcr2 Axis in Tumor Microenvironment as Potential Diagnostic Biomarker and Therapeutic Target. Cancer Commun. 2020, 40, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K. Vascular and Interstitial Barriers to Delivery of Therapeutic Agents in Tumors. Cancer Metastasis Rev. 1990, 9, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Tannock, I.F.; Lee, C.M.; Tunggal, J.K.; Cowan, D.S.; Egorin, M.J. Limited Penetration of Anticancer Drugs through Tumor Tissue: A Potential Cause of Resistance of Solid Tumors to Chemotherapy. Clin. Cancer Res. 2002, 8, 878–884. [Google Scholar] [PubMed]
- Tunggal, J.K.; Cowan, D.S.; Shaikh, H.; Tannock, I.F. Penetration of Anticancer Drugs through Solid Tissue: A Factor That Limits the Effectiveness of Chemotherapy for Solid Tumors. Clin. Cancer Res. 1999, 5, 1583–1586. [Google Scholar] [PubMed]
- Dunn, Z.S.; Li, Y.-R.; Yu, Y.; Lee, D.; Gibbons, A.; Kim, J.J.; Zhou, T.Y.; Li, M.; Nguyen, M.; Cen, X.; et al. Minimally Invasive Preclinical Monitoring of the Peritoneal Cavity Tumor Microenvironment. Cancers 2022, 14, 1775. [Google Scholar] [CrossRef]
- Khawar, I.A.; Kim, J.H.; Kuh, H.-J. Improving Drug Delivery to Solid Tumors: Priming the Tumor Microenvironment. J. Control. Release 2015, 201, 78–89. [Google Scholar] [CrossRef]
- Primeau, A.J.; Rendon, A.; Hedley, D.; Lilge, L.; Tannock, I.F.; Koido, S.; Hara, E.; Homma, S.; Torii, A.; Toyama, Y.; et al. The Distribution of the Anticancer Drug Doxorubicin in Relation to Blood Vessels in Solid Tumors. Clin. Cancer Res. 2005, 11, 8782–8788. [Google Scholar] [CrossRef]
- Murdoch, C.; Giannoudis, A.; Lewis, C.E. Mechanisms Regulating the Recruitment of Macrophages into Hypoxic Areas of Tumors and Other Ischemic Tissues. Blood 2004, 104, 2224–2234. [Google Scholar] [CrossRef]
- Facciabene, A.; Peng, X.; Hagemann, I.S.; Balint, K.; Barchetti, A.; Wang, L.P.; Gimotty, P.A.; Gilks, C.B.; Lal, P.; Zhang, L.; et al. Tumour Hypoxia Promotes Tolerance and Angiogenesis Via Ccl28 and T(Reg) Cells. Nature 2011, 475, 226–230. [Google Scholar] [CrossRef]
- Chiu, D.K.C.; Tse, A.P.W.; Xu, I.M.J.; Di Cui, J.; Lai, R.K.H.; Li, L.L.; Koh, H.Y.; Tsang, F.H.C.; Wei, L.L.; Wong, C.M.; et al. Hypoxia Inducible Factor Hif-1 Promotes Myeloid-Derived Suppressor Cells Accumulation through Entpd2/Cd39l1 in Hepatocellular Carcinoma. Nat. Commun. 2017, 8, 517. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of Hypoxia in Cancer Therapy by Regulating the Tumor Microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Hasmim, M.; Lequeux, A.; Xiao, M.; Duhem, C.; Chouaib, S.; Berchem, G.; Janji, B. Improving Cancer Immunotherapy by Targeting the Hypoxic Tumor Microenvironment: New Opportunities and Challenges. Cells 2019, 8, 1083. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Smallwood, C.A.; Siemens, D.R.; Graham, C.H. A Mechanism of Hypoxia-Mediated Escape from Adaptive Immunity in Cancer Cells. Cancer Res. 2014, 74, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 Is a Novel Direct Target of HIF-1α, and Its Blockade under Hypoxia Enhanced MDSC-Mediated T Cell Activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, Q.; Zhang, Y.; Liu, Z.; Zheng, Z.; Liu, S.; Meng, L.; Xin, Y.; Jiang, X. Targeting Hypoxia in the Tumor Microenvironment: A Potential Strategy to Improve Cancer Immunotherapy. J. Exp. Clin. Cancer Res. 2021, 40, 24. [Google Scholar] [CrossRef]
- Lu, Z.; Luo, R.Z.; Lu, Y.; Zhang, X.; Yu, Q.; Khare, S.; Kondo, S.; Kondo, Y.; Yu, Y.; Mills, G.B.; et al. The Tumor Suppressor Gene Arhi Regulates Autophagy and Tumor Dormancy in Human Ovarian Cancer Cells. J. Clin. Investig. 2008, 118, 3917–3929. [Google Scholar] [CrossRef]
- Viry, E.; Baginska, J.; Berchem, G.; Noman, M.Z.; Medves, S.; Chouaib, S.; Janji, B. Autophagic Degradation of Gzmb/Granzyme B: A New Mechanism of Hypoxic Tumor Cell Escape from Natural Killer Cell-Mediated Lysis. Autophagy 2014, 10, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Yang, M.; Li, J.; Gu, P.; Fan, X. The Application of Nanoparticles in Cancer Immunotherapy: Targeting Tumor Microenvironment. Bioact. Mater. 2020, 6, 1973–1987. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, Y.; Yu, Y. “Waltz” of Cell Membrane-Coated Nanoparticles on Lipid Bilayers: Tracking Single Particle Rotation in Ligand-Receptor Binding. ACS Nano 2018, 12, 11871–11880. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Yu, Y. Tracking Single Molecules in Biomembranes: Is Seeing Always Believing? Acs Nano 2019, 13, 10860–10868. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A.; Balkwill, F.; Bast, R.C.; Berek, J.S.; Kaye, A.; Boyd, J.A.; Mills, G.; Weinstein, J.N.; Woolley, K.; Workman, P. Opportunities and Challenges in Ovarian Cancer Research, a Perspective from the 11th Ovarian Cancer Action/HHMT Forum, Lake Como, March 2007. Gynecol. Oncol. 2008, 108, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2020, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Choi, N.; Kim, K.; Koo, H.-J.; Choi, J.; Kim, H.N. Chemoresistance of Cancer Cells: Requirements of Tumor Microenvironment-mimicking In Vitro Models in Anti-Cancer Drug Development. Theranostics 2018, 8, 5259–5275. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.J. Goodbye Flat Biology—Time for the 3rd and the 4th Dimensions. J. Cell Sci. 2017, 130, 3–5. [Google Scholar] [CrossRef]
- Stock, K.; Estrada, M.; Vidic, S.; Gjerde, K.; Rudisch, A.; Santo, V.E.; Barbier, M.; Blom, S.; Arundkar, S.C.; Selvam, I.; et al. Capturing Tumor Complexity In Vitro: Comparative Analysis of 2d and 3d Tumor Models for Drug Discovery. Sci. Rep. 2016, 6, 28951. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; De Melo-Diogo, D.M.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3d Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures-Examples of Methods of Preparation and the Most Important Application. nt. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef]
- Cowan, D.S.; Hicks, K.O.; Wilson, W.R. Multicellular Membranes as an In Vitro Model for Extravascular Diffusion in Tumours. Br. J. Cancer Suppl. 1996, 27, S28–S31. [Google Scholar]
- Chen, H.; Cheng, Y.; Wang, X.; Wang, J.; Shi, X.; Li, X.; Tan, W.; Tan, Z. 3d Printed In Vitro Tumor Tissue Model of Colorectal Cancer. Theranostics 2020, 10, 12127–12143. [Google Scholar] [CrossRef]
- Campbell, J.J.; Husmann, A.; Hume, R.D.; Watson, C.J.; Cameron, R.E. Development of Three-Dimensional Collagen Scaffolds with Controlled Architecture for Cell Migration Studies Using Breast Cancer Cell Lines. Biomaterials 2017, 114, 34–43. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Ingber. Microfluidic Organs-on-Chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Aung, A.; Kumar, V.; Theprungsirikul, J.; Davey, S.K.; Varghese, S. An Engineered Tumor-on-a-Chip Device with Breast Cancer–Immune Cell Interactions for Assessing T-cell Recruitment. Cancer Res. 2020, 80, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.A.; Mostafa, A.M.R.H.; Morton, J.P.; Hawinkels, L.J.A.C.; Prakash, J. Translating Complexity and Heterogeneity of Pancreatic Tumor: 3d In Vitro to In Vivo Models. Adv. Drug Deliv. Rev. 2021, 174, 265–293. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.P.; Roozitalab, R.; Gibson, S.V.; Grose, R.P. Tumour Microenvironment 3d-Modelling: Simplicity to Complexity and Back Again. Trends Cancer 2021, 7, 1033–1046. [Google Scholar] [CrossRef]
- Cekanova, M.; Rathore, K. Animal Models and Therapeutic Molecular Targets of Cancer: Utility and Limitations. Drug Des. Dev. Ther. 2014, 8, 1911–1921. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Kim, T.-H. Recent Advances in Multicellular Tumor Spheroid Generation for Drug Screening. Biosensors 2021, 11, 445. [Google Scholar] [CrossRef]
- Al-Hity, G.; Yang, F.; Campillo-Funollet, E.; Greenstein, A.E.; Hunt, H.; Mampay, M.; Intabli, H.; Falcinelli, M.; Madzvamuse, A.; Venkataraman, C.; et al. An Integrated Framework for Quantifying Immune-Tumour Interactions in a 3d Co-Culture Model. Commun. Biol. 2021, 4, 781. [Google Scholar] [CrossRef]
- Ryu, N.-E.; Lee, S.-H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef]
- Sant, S.; Johnston, P.A. The Production of 3d Tumor Spheroids for Cancer Drug Discovery. Drug Discov. Today Technol. 2017, 23, 27–36. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in Multicellular Spheroids Formation. J. R. Soc. Interface 2017, 14, 20160877. [Google Scholar] [CrossRef] [PubMed]
- Or, T.; Saem, S.; Esteve, A.; Osorio, D.A.; De France, K.J.; Vapaavuori, J.; Hoare, T.; Cerf, A.; Cranston, E.D.; Moran-Mirabal, J.M. Patterned Cellulose Nanocrystal Aerogel Films with Tunable Dimensions and Morphologies as Ultra-Porous Scaffolds for Cell Culture. ACS Appl. Nano Mater. 2019, 2, 4169–4179. [Google Scholar] [CrossRef]
- Feng, S.; Duan, X.; Lo, P.-K.; Liu, S.; Liu, X.; Chen, H.; Wang, Q. Expansion of Breast Cancer Stem Cells with Fibrous Scaffolds. Integr. Biol. 2013, 5, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Sci. Drug Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3d Tumor Spheroids as In Vitro Models to Mimic In Vivo Human Solid Tumors Resistance to Therapeutic Drugs. Biotechnol. Bioeng. 2018, 116, 206–226. [Google Scholar] [CrossRef]
- Han, S.J.; Kwon, S.; Kim, K.S. Challenges of Applying Multicellular Tumor Spheroids in Preclinical Phase. Cancer Cell Int. 2021, 21, 152. [Google Scholar] [CrossRef]
- Foty, R. A Simple Hanging Drop Cell Culture Protocol for Generation of 3D Spheroids. J. Vis. Exp. 2011, e2720. [Google Scholar] [CrossRef]
- Kwapiszewska, K.; Michalczuk, A.; Rybka, M.; Kwapiszewski, R.; Brzózka, Z. A Microfluidic-Based Platform for Tumour Spheroid Culture, Monitoring and Drug Screening. Lab Chip 2014, 14, 2096–2104. [Google Scholar] [CrossRef]
- Moshksayan, K.; Kashaninejad, N.; Warkiani, M.E.; Lock, J.; Moghadas, H.; Firoozabadi, B.; Saidi, M.S.; Nguyen, N.-T. Spheroids-on-a-Chip: Recent Advances and Design Considerations in Microfluidic Platforms for Spheroid Formation and Culture. Sens. Actuators B Chem. 2018, 263, 151–176. [Google Scholar] [CrossRef]
- Ruppen, J.; Cortes-Dericks, L.; Marconi, E.; Karoubi, G.; Schmid, R.A.; Peng, R.; Marti, T.M.; Guenat, O.T. A Microfluidic Platform for Chemoresistive Testing of Multicellular Pleural Cancer Spheroids. Lab Chip 2014, 14, 1198–1205. [Google Scholar] [CrossRef]
- Lim, W.; Park, S. A Microfluidic Spheroid Culture Device with a Concentration Gradient Generator for High-Throughput Screening of Drug Efficacy. Molecules 2018, 23, 3355. [Google Scholar] [CrossRef] [PubMed]
- Boedtkjer, E.; Pedersen, S.F. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu. Rev. Physiol. 2020, 82, 103–126. [Google Scholar] [CrossRef]
- Wojtkowiak, J.W.; Verduzco, D.; Schramm, K.J.; Gillies, R.J. Drug Resistance and Cellular Adaptation to Tumor Acidic pH Microenvironment. Mol. Pharm. 2011, 8, 2032–2038. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J.; Acker, H. Relations between Ph, Oxygen Partial Pressure and Growth in Cultured Cell Spheroids. Int. J. Cancer 1988, 42, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Swietach, P.; Hulikova, A.; Patiar, S.; Vaughan-Jones, R.D.; Harris, A. Importance of Intracellular pH in Determining the Uptake and Efficacy of the Weakly Basic Chemotherapeutic Drug, Doxorubicin. PLoS ONE 2012, 7, e35949. [Google Scholar] [CrossRef]
- Lee, S.H.; McIntyre, D.; Honess, D.; Hulikova, A.; Pacheco-Torres, J.; Cerdán, S.; Swietach, P.; Harris, A.L.; Griffiths, J.R. Carbonic Anhydrase Ix Is a Ph-Stat That Sets an Acidic Tumour Extracellular Ph In Vivo. Br. J. Cancer 2018, 119, 622–630. [Google Scholar] [CrossRef]
- Chiche, J.; Brahimi-Horn, M.C.; Pouysségur, J. Tumour Hypoxia Induces a Metabolic Shift Causing Acidosis: A Common Feature in Cancer. J. Cell. Mol. Med. 2010, 14, 771–794. [Google Scholar] [CrossRef]
- Leek, R.; Grimes, D.R.; Harris, A.L.; McIntyre, A. Methods: Using Three-Dimensional Culture (Spheroids) as an In Vitro Model of Tumour Hypoxia. Tumor Microenviron. 2016, 899, 167–196. [Google Scholar]
- Wigerup, C.; Påhlman, S.; Bexell, D. Therapeutic Targeting of Hypoxia and Hypoxia-Inducible Factors in Cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef]
- Wenzel, C.; Riefke, B.; Gründemann, S.; Krebs, A.; Christian, S.; Prinz, F.; Osterland, M.; Golfier, S.; Räse, S.; Ansari, N.; et al. 3d High-Content Screening for the Identification of Compounds That Target Cells in Dormant Tumor Spheroid Regions. Exp. Cell Res. 2014, 323, 131–1433. [Google Scholar] [CrossRef]
- Yoshii, Y.; Furukawa, T.; Waki, A.; Okuyama, H.; Inoue, M.; Itoh, M.; Zhang, M.-R.; Wakizaka, H.; Sogawa, C.; Kiyono, Y.; et al. High-Throughput Screening with Nanoimprinting 3d Culture for Efficient Drug Development by Mimicking the Tumor Environment. Biomaterials 2015, 51, 278–289. [Google Scholar] [CrossRef]
- Warburg, O. The Metabolism of Carcinoma Cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Khaitan, D.; Chandna, S.; Arya, M.; Dwarakanath, B. Establishment and Characterization of Multicellular Spheroids from a Human Glioma Cell Line; Implications for Tumor Therapy. J. Transl. Med. 2006, 4, 12. [Google Scholar] [CrossRef]
- Vidavsky, N.; Kunitake, J.A.M.R.; Diaz-Rubio, M.E.; Chiou, A.E.; Loh, H.-C.; Zhang, S.; Masic, A.; Fischbach, C.; Estroff, L.A. Mapping and Profiling Lipid Distribution in a 3D Model of Breast Cancer Progression. ACS Central Sci. 2019, 5, 768–780. [Google Scholar] [CrossRef]
- Wang, Q.; Beaumont, K.A.; Otte, N.J.; Font, J.; Bailey, C.G.; Van Geldermalsen, M.; Sharp, D.M.; Tiffen, J.C.; Ryan, R.M.; Jormakka, M.; et al. Targeting Glutamine Transport to Suppress Melanoma Cell Growth. Int. J. Cancer 2016, 135, 1060–1071. [Google Scholar] [CrossRef]
- Kunz-Schughart, L.A.; Kreutz, M.; Knuechel, R. Multicellular Spheroids: A Three-Dimensional In Vitro Culture System to Study Tumour Biology. Int. J. Exp. Pathol. 1998, 79, 1–23. [Google Scholar] [CrossRef]
- Guiet, R.; Van Goethem, E.; Cougoule, C.; Balor, S.; Valette, A.; Al Saati, T.; Lowell, C.A.; Le Cabec, V.; Maridonneau-Parini, I. The Process of Macrophage Migration Promotes Matrix Metalloproteinase-Independent Invasion by Tumor Cells. J. Immunol. 2011, 187, 3806–3814. [Google Scholar] [CrossRef]
- Li, Y.-R.; Yu, Y.; Kramer, A.; Hon, R.; Wilson, M.; Brown, J.; Yang, L. An Ex Vivo 3D Tumor Microenvironment-Mimicry Culture to Study TAM Modulation of Cancer Immunotherapy. Cells 2022, 11, 1583. [Google Scholar] [CrossRef]
- Yin, M.; Li, X.; Tan, S.; Zhou, H.J.; Ji, W.; Bellone, S.; Xu, X.; Zhang, H.; Santin, A.D.; Lou, G.; et al. Tumor-Associated Macrophages Drive Spheroid Formation During Early Transcoelomic Metastasis of Ovarian Cancer. J. Clin. Investig. 2016, 126, 4157–4173. [Google Scholar] [CrossRef]
- Rebelo, S.P.; Pinto, C.; Martins, T.R.; Harrer, N.; Estrada, M.F.; Loza-Alvarez, P.; Cabeçadas, J.; Alves, P.M.; Gualda, E.J.; Sommergruber, W.; et al. 3d-3-Culture: A Tool to Unveil Macrophage Plasticity in the Tumour Microenvironment. Biomaterials 2018, 163, 185–197. [Google Scholar] [CrossRef]
- Xu, S.; Yang, Y.; Dong, L.; Qiu, W.; Yang, L.; Wang, X.; Liu, L. Construction and Characteristics of an E-Cadherin-Related Three-Dimensional Suspension Growth Model of Ovarian Cancer. Sci. Rep. 2014, 4, 5646. [Google Scholar] [CrossRef] [PubMed]
- Luebke-Wheeler, J.L.; Nedredal, G.; Yee, L.; Amiot, B.P.; Nyberg, S.L. E-Cadherin Protects Primary Hepatocyte Spheroids from Cell Death by a Caspase-Independent Mechanism. Cell Transplant. 2009, 18, 1281–1287. [Google Scholar] [CrossRef]
- Kular, J.K.; Basu, S.; Sharma, R.I. The Extracellular Matrix: Structure, Composition, Age-Related Differences, Tools for Analysis and Applications for Tissue Engineering. J. Tissue Eng. 2014, 5, 2041731414557112. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Nederman, T.; Norling, B.; Glimelius, B.; Carlsson, J.; Brunk, U. Demonstration of an Extracellular Matrix in Multicellular Tumor Spheroids. Cancer Res. 1984, 44, 3090–3097. [Google Scholar] [PubMed]
- Tao, F.; Sayo, K.; Sugimoto, K.; Aoki, S.; Kojima, N. Development of a Tunable Method to Generate Various Three-Dimensional Microstructures by Replenishing Macromolecules Such as Extracellular Matrix Components and Polysaccharides. Sci. Rep. 2020, 10, 6567. [Google Scholar] [CrossRef] [PubMed]
- Bin Lee, Y.; Kim, E.M.; Byun, H.; Chang, H.-K.; Jeong, K.; Aman, Z.M.; Choi, Y.S.; Park, J.; Shin, H. Engineering Spheroids Potentiating Cell-Cell and Cell-Ecm Interactions by Self-Assembly of Stem Cell Microlayer. Biomaterials 2018, 165, 105–120. [Google Scholar]
- Ishiguro, T.; Ohata, H.; Sato, A.; Yamawaki, K.; Enomoto, T.; Okamoto, K. Tumor-Derived Spheroids: Relevance to Cancer Stem Cells and Clinical Applications. Cancer Sci. 2017, 108, 283–289. [Google Scholar] [CrossRef]
- Reynolds, D.S.; Tevis, K.M.; Blessing, W.A.; Colson, Y.L.; Zaman, M.H.; Grinstaff, M.W. Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment. Sci. Rep. 2017, 7, 10382. [Google Scholar] [CrossRef]
- Wright, M.H.; Calcagno, A.M.; Salcido, C.D.; Carlson, M.D.; Ambudkar, S.V.; Varticovski, L. Brca1 Breast Tumors Contain Distinct Cd44+/Cd24− and Cd133+ Cells with Cancer Stem Cell Characteristics. Breast Cancer Res 2008, 10, R10. [Google Scholar] [CrossRef]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and Challenges for Use of Tumor Spheroids as Models to Test Drug Delivery and Efficacy. J. Control. Release 2012, 164, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van Den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Guzzeloni, V.; Veschini, L.; Pedica, F.; Ferrero, E.; Ferrarini, M. 3D Models as a Tool to Assess the Anti-Tumor Efficacy of Therapeutic Antibodies: Advantages and Limitations. Antibodies 2022, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Foo, M.A.; You, M.; Chan, S.L.; Sethi, G.; Bonney, G.K.; Yong, W.-P.; Chow, E.K.-H.; Fong, E.L.S.; Wang, L.; Goh, B.-C. Clinical Translation of Patient-Derived Tumour Organoids- Bottlenecks and Strategies. Biomark. Res. 2022, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Kwon, S.-J.; Ku, B.; Lee, D.W.; Kim, J.; Dordick, J.S. 3D Tumor Spheroid Microarray for High-Throughput, High-Content Natural Killer Cell-Mediated Cytotoxicity. Commun. Biol. 2021, 4, 893. [Google Scholar] [CrossRef]
- Rae, C.; Amato, F.; Braconi, C. Patient-Derived Organoids as a Model for Cancer Drug Discovery. Int. J. Mol. Sci. 2021, 22, 3483. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Boucherit, N.; Gorvel, L.; Olive, D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front. Immunol. 2020, 11, 603640. [Google Scholar] [CrossRef]
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical Cancer Models in Tumor Biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef]
- Ma, R.; Mandell, J.; Lu, F.; Heim, T.; Schoedel, K.; Duensing, A.; Watters, R.J.; Weiss, K.R. Do Patient-Derived Spheroid Culture Models Have Relevance in Chondrosarcoma Research? Clin. Orthop. Relat. Res. 2021, 479, 477–490. [Google Scholar] [CrossRef]
- Bregenzer, M.E.; Davis, C.; Horst, E.N.; Mehta, P.; Novak, C.M.; Raghavan, S.; Snyder, C.; Mehta, G. Physiologic Patient Derived 3D Spheroids for Anti-neoplastic Drug Screening to Target Cancer Stem Cells. J. Vis. Exp. 2019, e59696. [Google Scholar] [CrossRef] [PubMed]
- Di Modugno, F.; Colosi, C.; Trono, P.; Antonacci, G.; Ruocco, G.; Nisticò, P. 3d Models in the New Era of Immune Oncology: Focus on T Cells, Caf and Ecm. J. Exp. Clin. Cancer Res. 2019, 38, 117. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.-B.; Richon, S.; Massonnet, G.; Guinebretière, J.-M.; Vacher, S.; Laurendeau, I.; Cottu, P.; Marangoni, E.; Nemati, F.; Validire, P.; et al. A Short-Term Colorectal Cancer Sphere Culture as a Relevant Tool for Human Cancer Biology Investigation. Br. J. Cancer 2013, 108, 1720–1731. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Theodoraki, M.A.; Rezende, C.O.R., Jr.; Chantarasriwong, O.; Corben, A.D.; Theodorakis, E.A.; Alpaugh, M.L. Spontaneously-Forming Spheroids as an In Vitro Cancer Cell Model for Anticancer Drug Screening. Oncotarget 2015, 6, 21255–21267. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.-B.; Richon, S.; Validire, P.; Briffod, M.; Lai-Kuen, R.; Cordelières, F.; Bertrand, F.; Dargere, D.; Massonnet, G.; Marangoni, E.; et al. Newly Characterised Ex Vivo Colospheres as a Three-Dimensional Colon Cancer Cell Model of Tumour Aggressiveness. Br. J. Cancer 2009, 101, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Li, G.; Zhang, C.; Jiang, T.; Li, Y.; Duan, X.; Zhong, G. B7-H3 as a Target for CAR-T Cell Therapy in Skull Base Chordoma. Front. Oncol. 2021, 11, 659662. [Google Scholar] [CrossRef]
- Appleton, K.M.; Elrod, A.K.; Lassahn, K.A.; Shuford, S.; Holmes, L.M.; DesRochers, T.M. Pd-1/Pd-L1 Checkpoint Inhibitors in Combination with Olaparib Display Antitumor Activity in Ovarian Cancer Patient-Derived Three-Dimensional Spheroid Cultures. Cancer Immunol. Immunother. 2021, 70, 843–856. [Google Scholar] [CrossRef]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Suzuki, T.; Okamoto, K.; Ichikawa, T.; Yano, A.; Kawakami, S.; Inoue, S. Aldh1a1 in Patient-Derived Bladder Cancer Spheroids Activates Retinoic Acid Signaling Leading to Tubb3 Overexpression and Tumor Progression. Int. J. Cancer 2019, 146, 1099–1113. [Google Scholar] [CrossRef]
- Hofmann, S.; Cohen-Harazi, R.; Maizels, Y.; Koman, I. Patient-Derived Tumor Spheroid Cultures as a Promising Tool to Assist Personalized Therapeutic Decisions in Breast Cancer. ransl. Cancer Res. 2022, 11, 134–147. [Google Scholar] [CrossRef]
- Velletri, T.; Villa, C.E.; Cilli, D.; Barzaghi, B.; Riso, P.L.; Lupia, M.; Luongo, R.; López-Tobón, A.; De Simone, M.; Bonnal, R.J.P.; et al. Single Cell-Derived Spheroids Capture the Self-Renewing Subpopulations of Metastatic Ovarian Cancer. Cell Death Differ. 2021, 29, 614–626. [Google Scholar] [CrossRef]
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of Primary Human Pancreatic Cancer Organoids, Matched Stromal and Immune Cells and 3d Tumor Microenvironment Models. BMC Cancer 2018, 18, 335. [Google Scholar] [CrossRef] [PubMed]
- Van Hemelryk, A.; van Weerden, W.M. Novel Patient-Derived 3d Culture Models to Guide Clinical Decision-Making in Prostate Cancer. Curr. Opin. Endocr. Metab. Res. 2020, 10, 7–15. [Google Scholar] [CrossRef]
- Sanchez-Fdez, A.; Sharma, A.K.; Tiriac, H.; Sicklick, J.K. Patient-Derived Sarcoma Organoids Offer a Novel Platform for Personalized Precision Medicine. Ann. Surg. Oncol. 2022, 29, 7239–7241. [Google Scholar] [CrossRef] [PubMed]
- Nyga, A.; Stamati, K.; Redondo, P.A.; Azimi, T.; Feber, A.; Neves, J.B.; Hamoudi, R.; Presneau, N.; El Sheikh, S.; Tran, M.G.B.; et al. Renal Tumouroids: Challenges of Manufacturing 3d Cultures from Patient Derived Primary Cells. J. Cell Commun. Signal. 2022. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef]
- Scognamiglio, G.; De Chiara, A.; Parafioriti, A.; Armiraglio, E.; Fazioli, F.; Gallo, M.; Aversa, L.; Camerlingo, R.; Cacciatore, F.; Colella, G.; et al. Patient-Derived Organoids as a Potential Model to Predict Response to Pd-1/Pd-L1 Checkpoint Inhibitors. Br. J. Cancer 2019, 121, 979–982. [Google Scholar] [CrossRef]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of Patient-Derived Cancer Organoids for Drug-Screening Applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Lee, S.; Burner, D.N.; Mendoza, T.R.; Muldong, M.T.; Arreola, C.; Wu, C.N.; Cacalano, N.A.; Kulidjian, A.A.; Kane, C.J.; Jamieson, C.A.M. Establishment and Analysis of Three-Dimensional (3D) Organoids Derived from Patient Prostate Cancer Bone Metastasis Specimens and their Xenografts. J. Vis. Exp. 2020, e60367. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; Van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; Van De Haar, J.; et al. Patient-Derived Organoids Can Predict Response to Chemotherapy in Metastatic Colorectal Cancer Patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Zou, F.; Tan, J.; Liu, T.; Liu, B.; Tang, Y.; Zhang, H.; Li, J. The Cd39+ Hbv Surface Protein-Targeted Car-T and Personalized Tumor-Reactive Cd8+ T Cells Exhibit Potent Anti-Hcc Activity. Mol. Ther. 2021, 29, 1794–1807. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26.e16. [Google Scholar] [CrossRef] [PubMed]
- Guillen, K.P.; Fujita, M.; Butterfield, A.J.; Scherer, S.D.; Bailey, M.H.; Chu, Z.; DeRose, Y.S.; Zhao, L.; Cortes-Sanchez, E.; Yang, C.-H.; et al. A Human Breast Cancer-Derived Xenograft and Organoid Platform for Drug Discovery and Precision Oncology. Nat. Cancer 2022, 3, 232–250. [Google Scholar] [CrossRef] [PubMed]
- Schnalzger, T.E.; De Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3d Model for Car-Mediated Cytotoxicity Using Patient-Derived Colorectal Cancer Organoids. EMBO J. 2019, 38, e100928. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e5. [Google Scholar] [CrossRef]
- Sokol, E.S.; Feng, Y.-X.; Jin, D.X.; Tizabi, M.D.; Miller, D.H.; Cohen, M.A.; Sanduja, S.; Reinhardt, F.; Pandey, J.; Superville, D.A.; et al. Smarce1 Is Required for the Invasive Progression of in Situ Cancers. Proc. Natl. Acad. Sci USA 2017, 114, 4153–4158. [Google Scholar] [CrossRef]
- Di Blasio, S.; Van Wigcheren, G.F.; Becker, A.; Van Duffelen, A.; Gorris, M.; Verrijp, K.; Stefanini, I.; Bakker, G.J.; Bloemendal, M.; Halilovic, A.; et al. The Tumour Microenvironment Shapes Dendritic Cell Plasticity in a Human Organotypic Melanoma Culture. Nat. Commun. 2020, 11, 2749. [Google Scholar] [CrossRef]
- Meng, Q.; Xie, S.; Gray, G.K.; Dezfulian, M.H.; Li, W.; Huang, L.; Akshinthala, D.; Ferrer, E.; Conahan, C.; Del Pino, S.P.; et al. Empirical Identification and Validation of Tumor-Targeting T Cell Receptors from Circulation Using Autologous Pancreatic Tumor Organoids. J. Immunother. Cancer 2021, 9, e003213. [Google Scholar] [CrossRef]
- Welti, J.; Sharp, A.; Yuan, W.; Dolling, D.; Rodrigues, D.N.; Figueiredo, I.; Gil, V.; Neeb, A.; Clarke, M.; Seed, G.; et al. Targeting Bromodomain and Extra-Terminal (BET) Family Proteins in Castration-Resistant Prostate Cancer (CRPC). Clin. Cancer Res. 2018, 24, 3149–3162. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Cleven, A.; Keramati, F.; Wezenaar, A.K.L.; van Vliet, E.J.; Puschhof, J.; Brazda, P.; Johanna, I.; Meringa, A.D.; et al. Uncovering the Mode of Action of Engineered T Cells in Patient Cancer Organoids. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Li, H.; Harrison, E.B.; Li, H.; Hirabayashi, K.; Chen, J.; Li, Q.-X.; Gunn, J.; Weiss, J.; Savoldo, B.; Parker, J.S.; et al. Targeting Brain Lesions of Non-Small Cell Lung Cancer by Enhancing Ccl2-Mediated Car-T Cell Migration. Nat. Commun. 2022, 13, 2154. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Niu, C.; Wang, B.; Zhao, S.; Roex, G.; Qian, J.; Qie, J.; Chen, L.; Yi, C.; et al. Chimeric Antigen Receptor Clustering via Cysteines Enhances T-Cell Efficacy against Tumor. Cancer Immunol. Immunother. 2022, 71, 2801–2814. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhao, H.; Zhang, W.; Wang, J.; Liu, Y.; Cao, Y.; Zheng, H.; Hu, Z.; Wang, S.; Zhu, Y.; et al. An Automated Organoid Platform with Inter-Organoid Homogeneity and Inter-Patient Heterogeneity. Cell Rep. Med. 2020, 1, 100161. [Google Scholar] [CrossRef]
- Gong, L.; Petchakup, C.; Shi, P.; Tan, P.L.; Tan, L.P.; Tay, C.Y.; Hou, H.W. Direct and Label-Free Cell Status Monitoring of Spheroids and Microcarriers Using Microfluidic Impedance Cytometry. Small 2021, 17, 2007500. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, K.; Hana, D.; Chou, J.T.-T.; Singh, C.; Mackiewicz, A.; Kaczmarek, M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021, 12, 656364. [Google Scholar] [CrossRef]
- Croix, C.M.S.; Shand, S.H.; Watkins, S.C. Confocal Microscopy: Comparisons, Applications, and Problems. BioTechniques 2005, 39, S2–S5. [Google Scholar] [CrossRef]
- Wu, H.; Yang, Y.; Bagnaninchi, P.O.; Jia, J. Electrical Impedance Tomography for Real-Time and Label-Free Cellular Viability Assays of 3d Tumour Spheroids. Analyst 2018, 143, 4189–4198. [Google Scholar] [CrossRef]
- Gomes, A.; Russo, A.; Vidal, G.; Demange, E.; Pannetier, P.; Souguir, Z.; Lagarde, J.-M.; Ducommun, B.; Lobjois, V. Evaluation by Quantitative Image Analysis of Anticancer Drug Activity on Multicellular Spheroids Grown in 3d Matrices. Oncol. Lett. 2016, 12, 4371–4376. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Ma, N.; Sun, X.; Li, Q.; Zeng, Y.; Chen, F.; Sun, S.; Xu, J.; Zhang, J.; Ye, H.; et al. Automated Evaluation of Tumor Spheroid Behavior in 3d Culture Using Deep Learning-Based Recognition. Biomaterials 2021, 272, 120770. [Google Scholar] [CrossRef]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-Chip: From Bioinspired Design to Biomedical Application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef]
| Spheroids or Organoids | Culture Methods | Advantages | Disadvantages |
|---|---|---|---|
| Cell-line spheroids | Ultra-low attachment plates [59], Agitation-based method, Liquid overlay, Hanging drop, Microfluidics [65] | Low cost, Capability to model hypoxia and metabolic environment, Mimic cell heterogeneity and cell–cell interaction, Platform for drug screening [47] | Variability in production, Lengthy formation time [66], Lack of control on architecture [54] |
| Patient-derived spheroids | Identical to cell-line spheroids [43,144] | Accurate modeling of in vivo gene expression [43,110], Enrichment for CSCs [109], Easy handling [119] | Inconsistent formation efficiency [110], Lack of properly defined ECM [111] |
| Patient-derived organoids | Embedment of primary tissue into ECM/hydrogel [4,102,123] | Apicobasal polarity [121], Organlike heterogeneity [43], Defined ECM [55,121] | Cell density limit [111], Unwanted interaction with antigens present in matrix [43,108] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Kang, E.; Wilson, M.; Basso, T.; Chen, E.; Yu, Y.; Li, Y.-R. 3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy. Organoids 2022, 1, 149-167. https://doi.org/10.3390/organoids1020012
Zhu Y, Kang E, Wilson M, Basso T, Chen E, Yu Y, Li Y-R. 3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy. Organoids. 2022; 1(2):149-167. https://doi.org/10.3390/organoids1020012
Chicago/Turabian StyleZhu, Yichen, Elliot Kang, Matthew Wilson, Taylor Basso, Evelynn Chen, Yanqi Yu, and Yan-Ruide Li. 2022. "3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy" Organoids 1, no. 2: 149-167. https://doi.org/10.3390/organoids1020012
APA StyleZhu, Y., Kang, E., Wilson, M., Basso, T., Chen, E., Yu, Y., & Li, Y.-R. (2022). 3D Tumor Spheroid and Organoid to Model Tumor Microenvironment for Cancer Immunotherapy. Organoids, 1(2), 149-167. https://doi.org/10.3390/organoids1020012








