3DNA: A Tool for Sculpting Brick-Based DNA Nanostructures †
Abstract
:1. Introduction
2. Methods and Implementation
- (a)
option navigates the user to the 3D canvas.
- (b)
option allows the user to save the output DNA sequence for visualization and eventual synthesis of DNA oligonucleotide sets.
- (c)
provides an estimation of the cost per DNA base in USD for a particular model.
- (d)
navigates the user to the Advanced panel.
- (e)
option allows user to visualize the DNA nanostructure.
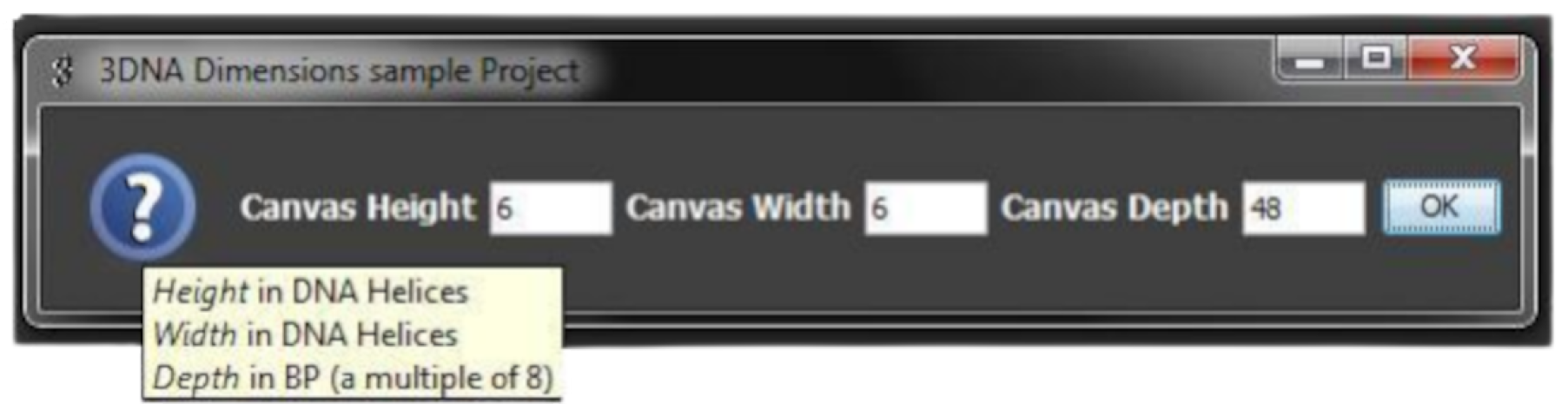
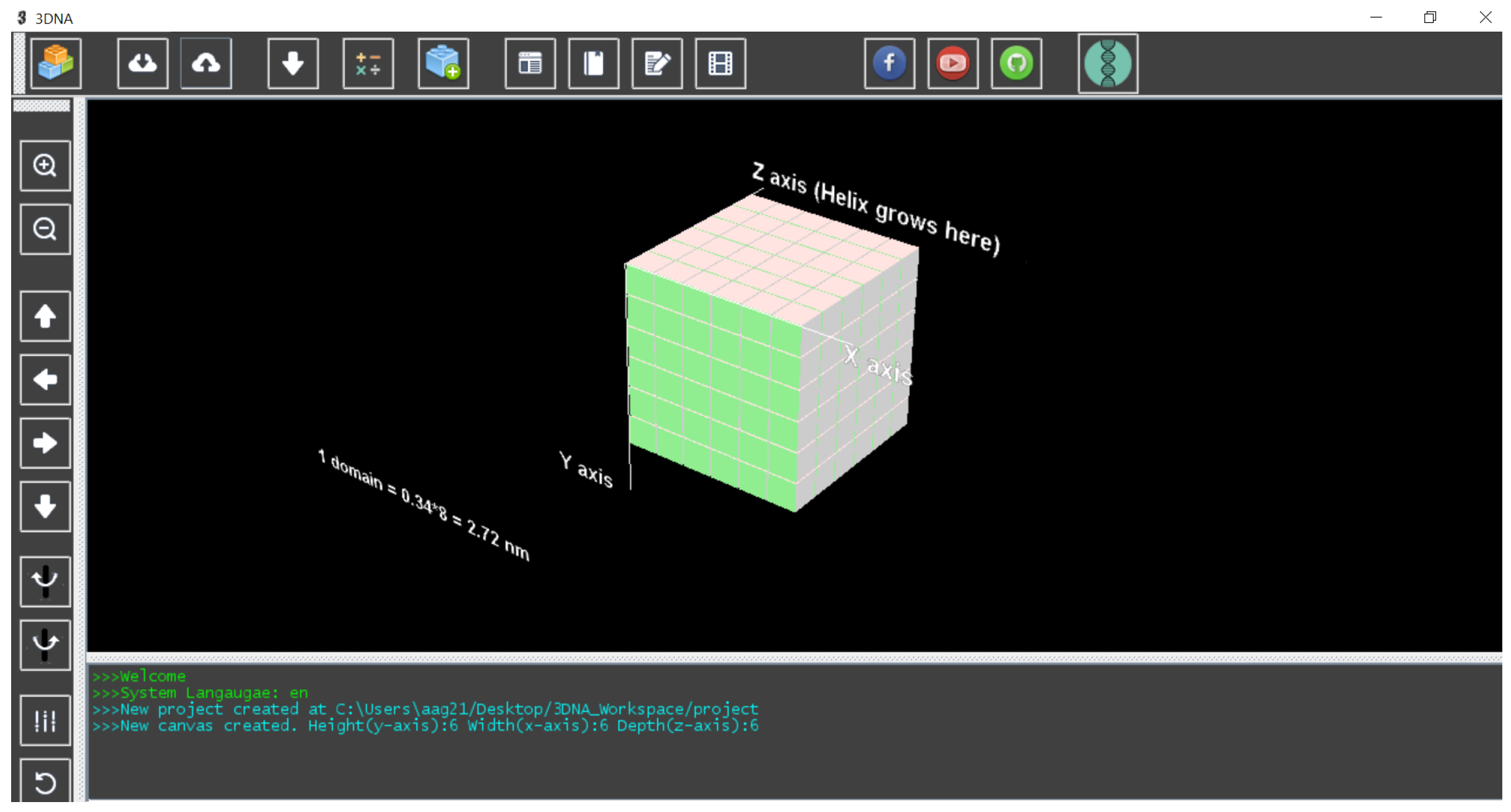
2.1. 3D Molecular Canvas
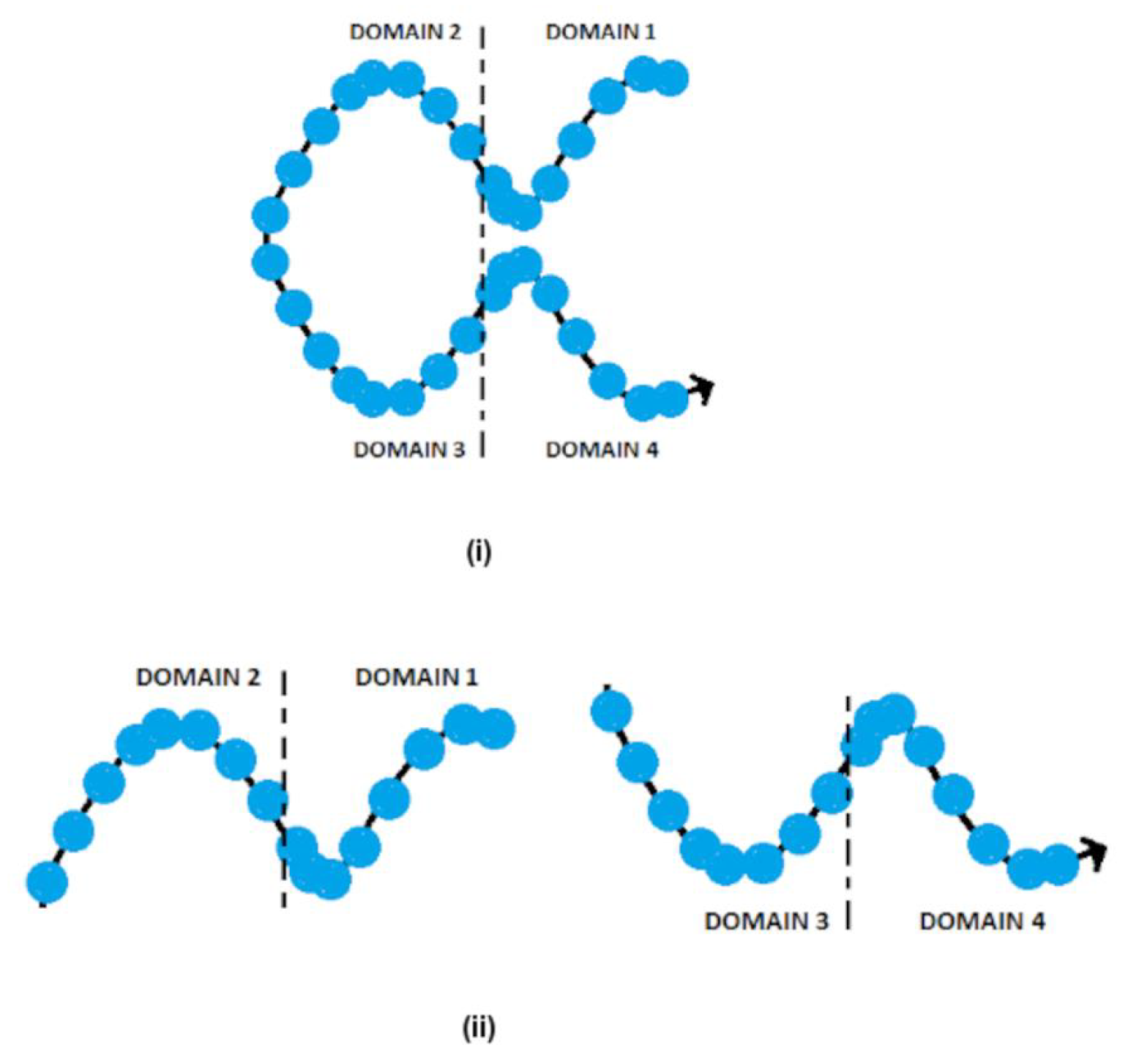
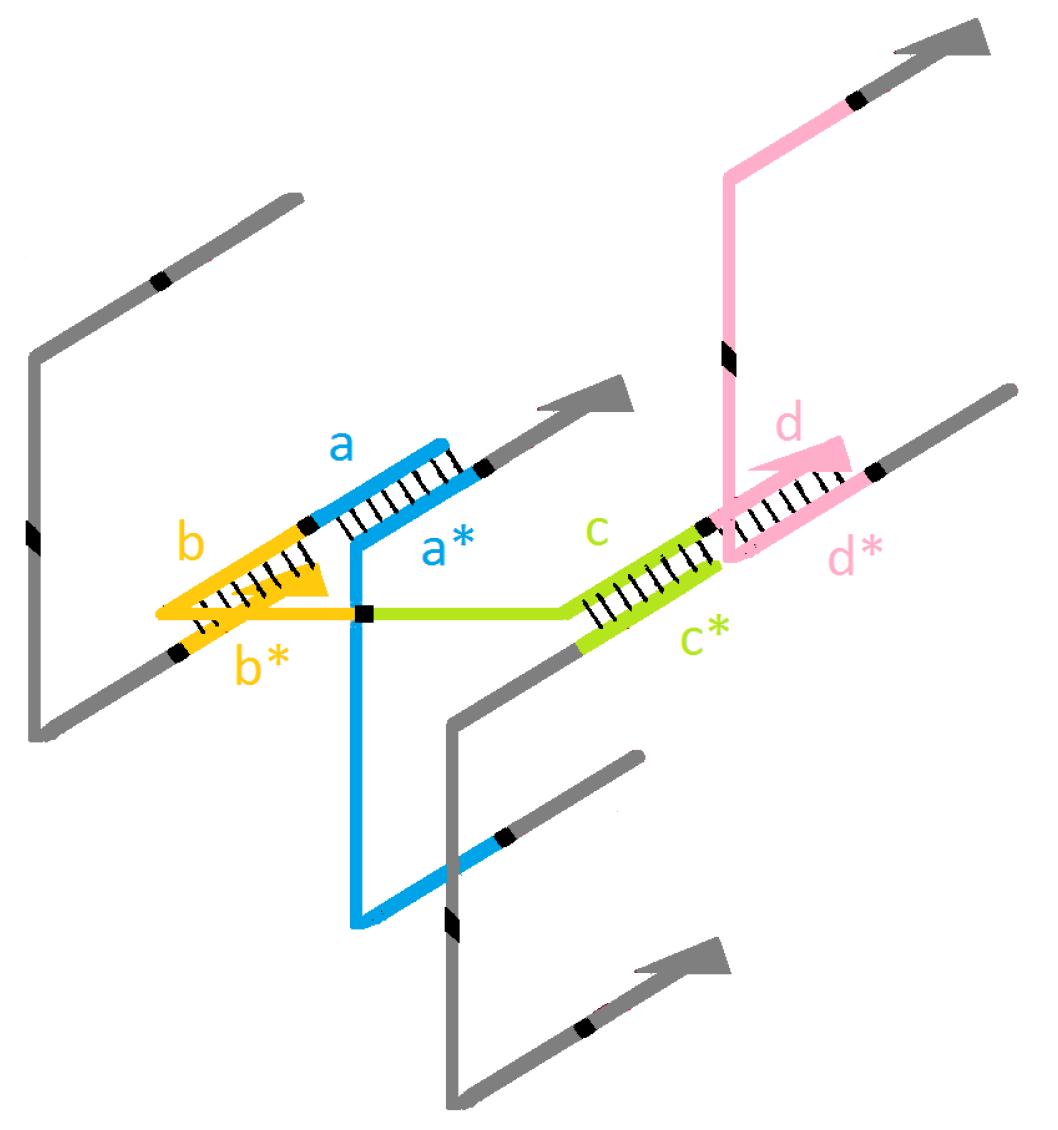
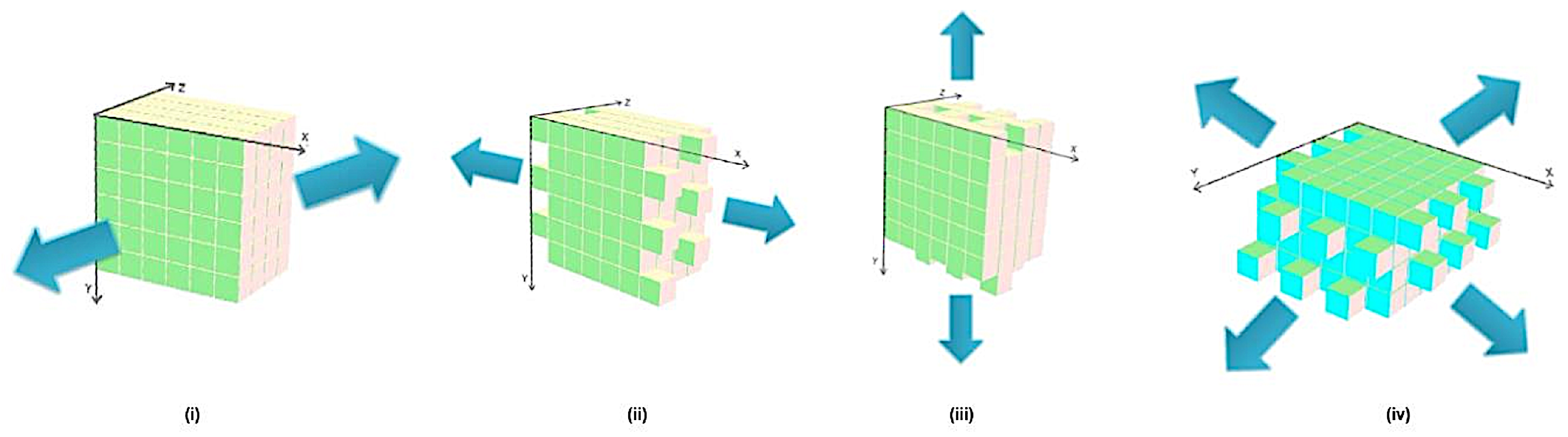
2.2. Modeling/Sculpting Shapes
2.3. Advanced Panel
2.4. Implementation
2.5. Visualization
2.6. Silent Features
- Import and Export: To achieve more adaptability, the option of exporting a current project is available to the users. The current project will be stored as a .3dna file by availing the Export button. This feature also enables importing an existing project exported previously with the .3dna extension, which can be opened and modified.
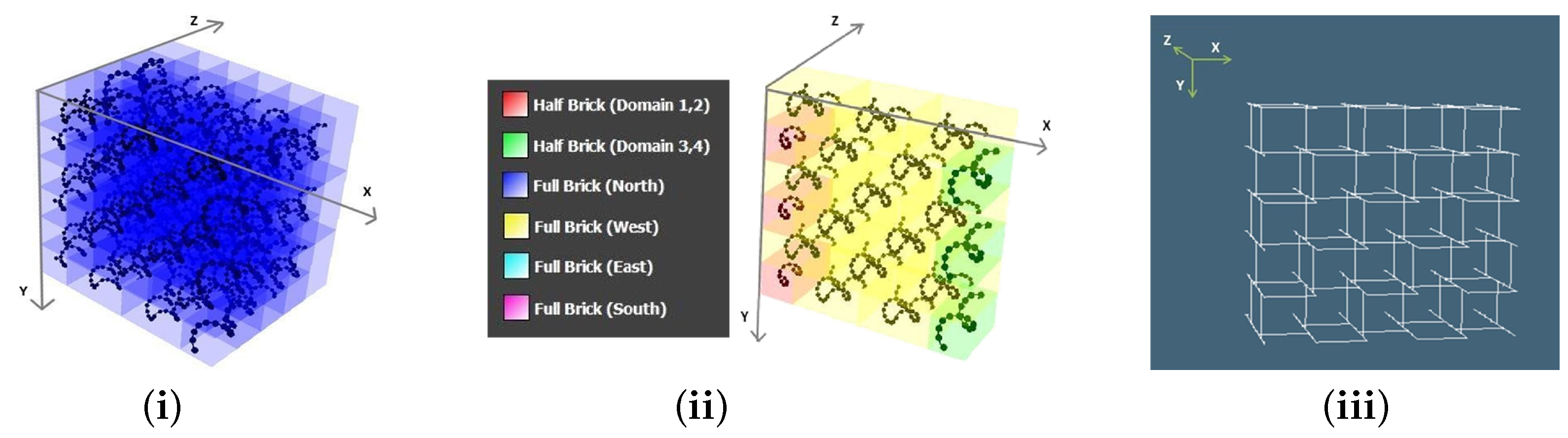
- Importing user’s DNA sequence: The user also has the ability to import a set of DNA sequences in .csv format to create shapes. On a brief technical note, the “Import sequence module” can take 3 .csv files to complete the molecular canvas, each one for half bricks (16 nt .csv file), full bricks (32 nt .csv file), and boundary bricks (48 nt .csv file) in specific formats.
- Structure filtration for stability: A developed module is attached that avoids DNA structures, allowing improper folding to test the topological connectivity for stability using the Graph Theory approach.
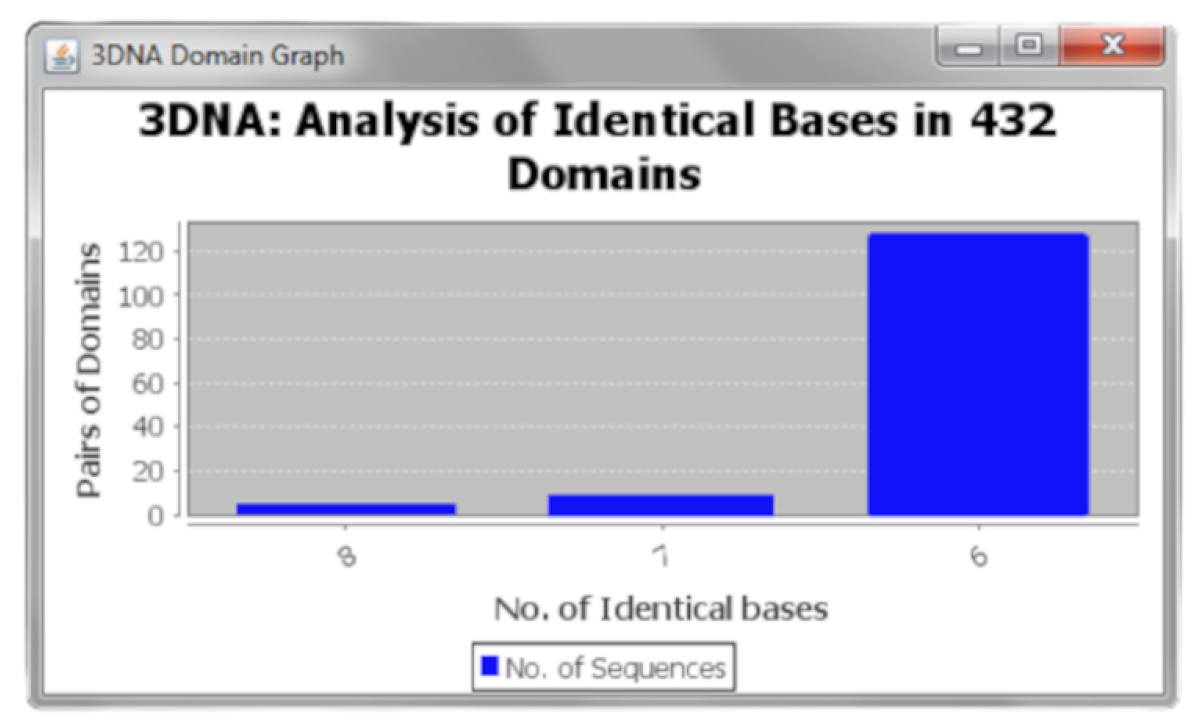
- Output analysis: Further statistical analysis on the output set of sequences can be done using the “Graphical Analysis option”, which can be useful for conducting laboratory-based experiments. It also introduces an abrupt idea about the structural tolerance and stability of a self-assembly DNA nanostructure containing indistinguishable bases in domain sequences. The option represents the frequency of pairs of 8-base domains classified by the factor of containing 8, 7, or 6 identical bases among the sample space of the 432 domains. Figure 10 shows the analysis of the 6 H H BP cuboid (Figure 6) in 432 domains.
- Cost estimator: 3DNA also has an inbuilt estimator function that evaluates the experimental cost in USD by considering the number of nucleotide bases used in the process.
- While generating the structure, the user can undo the last step using the UNDO button available in the software.
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, N.; Liedl, T. DNA-assembled advanced plasmonic architectures. Chem. Rev. 2018, 118, 3032–3053. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Bai, R.; Liu, H. DNA-Based Nanofabrication for Nanoelectronics. Adv. Funct. Mater. 2022, 32, 2112331. [Google Scholar] [CrossRef]
- Smith, D.M.; Keller, A. DNA nanostructures in the fight against infectious diseases. Adv. NanoBiomed Res. 2021, 1, 2000049. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2018, 119, 6459–6506. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Gothelf, K.V. LEGO-like DNA structures. Science 2012, 338, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Ong, L.L.; Hanikel, N.; Yaghi, O.K.; Grun, C.; Strauss, M.T.; Bron, P.; Lai-Kee-Him, J.; Schueder, F.; Wang, B.; Wang, P.; et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 2017, 552, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ong, L.L.; Sun, W.; Song, J.; Dong, M.; Shih, W.M.; Yin, P. DNA brick crystals with prescribed depths. Nat. Chem. 2014, 6, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.M.; Reinhardt, A.; Frenkel, D. Communication: Theoretical prediction of free-energy landscapes for complex self-assembly. J. Chem. Phys. 2015, 142, 021101. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.; Romano, F.; Schreck, J.S.; Ouldridge, T.E.; Doye, J.P.; Louis, A.A. Multi-scale coarse-graining for the study of assembly pathways in DNA-brick self-assembly. J. Chem. Phys. 2018, 148, 134910. [Google Scholar] [CrossRef]
- Reinhardt, A.; Frenkel, D. DNA brick self-assembly with an off-lattice potential. Soft Matter 2016, 12, 6253–6260. [Google Scholar] [CrossRef]
- Reinhardt, A.; Frenkel, D. Numerical evidence for nucleated self-assembly of DNA brick structures. Phys. Rev. Lett. 2014, 112, 238103. [Google Scholar] [CrossRef]
- Wales, D.J. Atomic clusters with addressable complexity. J. Chem. Phys. 2017, 146, 054306. [Google Scholar] [CrossRef]
- Jacobs, W.M.; Frenkel, D. Self-assembly protocol design for periodic multicomponent structures. Soft Matter 2015, 11, 8930–8938. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Frenkel, D.; Reinhardt, A. Investigating the role of boundary bricks in DNA brick self-assembly. Soft Matter 2017, 13, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.M.; Reinhardt, A.; Frenkel, D. Rational design of self-assembly pathways for complex multicomponent structures. Proc. Natl. Acad. Sci. USA 2015, 112, 6313–6318. [Google Scholar] [CrossRef] [PubMed]
- Madge, J.; Miller, M.A. Design strategies for self-assembly of discrete targets. J. Chem. Phys. 2015, 143, 044905. [Google Scholar] [CrossRef] [PubMed]
- Madge, J.; Miller, M.A. Optimising minimal building blocks for addressable self-assembly. Soft Matter 2017, 13, 7780–7792. [Google Scholar] [CrossRef] [PubMed]
- Slone, S.M.; Li, C.Y.; Yoo, J.; Aksimentiev, A. Molecular mechanics of DNA bricks: In situ structure, mechanical properties and ionic conductivity. New J. Phys. 2016, 18, 055012. [Google Scholar] [CrossRef]
- Sajfutdinow, M.; Jacobs, W.M.; Reinhardt, A.; Schneider, C.; Smith, D.M. Direct observation and rational design of nucleation behavior in addressable self-assembly. Proc. Natl. Acad. Sci. USA 2018, 115, E5877–E5886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Reinhardt, A.; Wang, P.; Song, J.; Ke, Y. Programming the nucleation of DNA brick self-assembly with a seeding strand. Angew. Chem. Int. Ed. 2020, 59, 8594–8600. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Chen, C.; Li, X.; Song, T.; Chen, Z.; Zhang, Z.; Wang, Y. Size-controllable DNA nanoribbons assembled from three types of reusable brick single-strand DNA tiles. Soft Matter 2015, 11, 8484–8492. [Google Scholar] [CrossRef]
- Sun, S.; Yang, Y.; Li, D.; Zhu, J. Large chiral nanotubes self-assembled by DNA bricks. J. Am. Chem. Soc. 2019, 141, 19524–19528. [Google Scholar] [CrossRef]
- Sun, S.; Wang, M.; Zhang, F.; Zhu, J. DNA polygonal cavities with tunable shapes and sizes. Chem. Commun. 2015, 51, 16247–16250. [Google Scholar] [CrossRef]
- Scheible, M.B.; Ong, L.L.; Woehrstein, J.B.; Jungmann, R.; Yin, P.; Simmel, F.C. A compact DNA cube with side length 10 nm. Small 2015, 11, 5200–5205. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, D.; Kou, B.; Shen, L.; Li, H.; Wang, P. Designer Structures Assembled from Modular DNA Superbricks. ACS Appl. Bio Mater. 2019, 3, 2850–2853. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yin, P. Enhancing biocompatible stability of DNA nanostructures using dendritic oligonucleotides and brick motifs. Angew. Chem. 2020, 132, 710–713. [Google Scholar] [CrossRef]
- Schmidt, T.L.; Beliveau, B.J.; Uca, Y.O.; Theilmann, M.; Da Cruz, F.; Wu, C.T.; Shih, W.M. Scalable amplification of strand subsets from chip-synthesized oligonucleotide libraries. Nat. Commun. 2015, 6, 8634. [Google Scholar] [CrossRef]
- Cannon, B.L.; Kellis, D.L.; Davis, P.H.; Lee, J.; Kuang, W.; Hughes, W.L.; Graugnard, E.; Yurke, B.; Knowlton, W.B. Excitonic AND Logic Gates on DNA Brick Nanobreadboards. ACS Photonics 2015, 2, 398–404. [Google Scholar] [CrossRef]
- Sun, W.; Shen, J.; Zhao, Z.; Arellano, N.; Rettner, C.; Tang, J.; Cao, T.; Zhou, Z.; Ta, T.; Streit, J.K.; et al. Precise pitch-scaling of carbon nanotube arrays within three-dimensional DNA nanotrenches. Science 2020, 368, 874–877. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, C.; Zhu, Z.; Sun, W. Suppressing high-dimensional crystallographic defects for ultra-scaled DNA arrays. Nat. Commun. 2022, 13, 2707. [Google Scholar] [CrossRef]
- Glaser, M.; Deb, S.; Seier, F.; Agrawal, A.; Liedl, T.; Douglas, S.; Gupta, M.K.; Smith, D.M. The art of designing DNA nanostructures with CAD software. Molecules 2021, 26, 2287. [Google Scholar] [CrossRef]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A three-dimensional editing tool for complex DNA structures. In Proceedings of the 14th International Workshop on DNA-Based Computers, DNA 14, Prague, Czech Republic, 2–9 June 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 90–101. [Google Scholar]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef]
- Levy, N.; Schabanel, N. ENSnano: A 3D modeling software for DNA nanostructures. In Proceedings of the DNA27-27th International Conference on DNA Computing and Molecular Programming, Oxford, UK, 13–16 September 2021. [Google Scholar]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef]
- Jun, H.; Zhang, F.; Shepherd, T.; Ratanalert, S.; Qi, X.; Yan, H.; Bathe, M. Autonomously designed free-form 2D DNA origami. Sci. Adv. 2019, 5, eaav0655. [Google Scholar] [CrossRef] [PubMed]
- Jun, H.; Shepherd, T.R.; Zhang, K.; Bricker, W.P.; Li, S.; Chiu, W.; Bathe, M. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 2019, 13, 2083–2093. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Limbachiya, D.; Gupta, S.K.; Joshi, F.; Pritmani, S.; Sahai, A.; Gupta, M.K. DNA pen: A tool for drawing on a molecular canvas. arXiv 2013, arXiv:1306.0369. [Google Scholar]
- Fu, D.; Narayanan, R.P.; Prasad, A.; Zhang, F.; Williams, D.; Schreck, J.S.; Yan, H.; Reif, J. Automated design of 3D DNA origami with non-rasterized 2D curvature. Sci. Adv. 2022, 8, eade4455. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Olson, W.K.; Lu, X.J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019, 47, W26–W34. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Joshi, F.; Limbachiya, D.; Gupta, M.K. 3DNA: A tool for DNA sculpting. arXiv 2014, arXiv:1405.4118. [Google Scholar]
- Kocabey, S.; Meinl, H.; MacPherson, I.S.; Cassinelli, V.; Manetto, A.; Rothenfusser, S.; Liedl, T.; Lichtenegger, F.S. Cellular uptake of tile-assembled DNA nanotubes. Nanomaterials 2014, 5, 47–60. [Google Scholar] [CrossRef]
- Sellner, S.; Kocabey, S.; Nekolla, K.; Krombach, F.; Liedl, T.; Rehberg, M. DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials 2015, 53, 453–463. [Google Scholar] [CrossRef]
- Sellner, S.; Kocabey, S.; Zhang, T.; Nekolla, K.; Hutten, S.; Krombach, F.; Liedl, T.; Rehberg, M. Dexamethasone-conjugated DNA nanotubes as anti-inflammatory agents in vivo. Biomaterials 2017, 134, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, F.; Chao, J.; Xie, M.; Liu, H.; Pan, M.; Kopperger, E.; Liu, X.; Li, Q.; Shi, J.; et al. DNA origami cryptography for secure communication. Nat. Commun. 2019, 10, 5469. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, G.D.; Mortuza, G.M.; Clay, W.; Piantanida, L.; Green, C.M.; Watson, C.; Hayden, E.J.; Andersen, T.; Kuang, W.; Graugnard, E.; et al. An alternative approach to nucleic acid memory. Nat. Commun. 2021, 12, 2371. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Bellot, G.; Voigt, N.V.; Fradkov, E.; Shih, W.M. Two design strategies for enhancement of multilayer–DNA-origami folding: Underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 2012, 3, 2587–2597. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, S.K.; Joshi, F.; Agrawal, A.; Deb, S.; Sajfutdinow, M.; Limbachiya, D.; Smith, D.M.; Gupta, M.K. 3DNA: A Tool for Sculpting Brick-Based DNA Nanostructures. SynBio 2023, 1, 226-238. https://doi.org/10.3390/synbio1030016
Gupta SK, Joshi F, Agrawal A, Deb S, Sajfutdinow M, Limbachiya D, Smith DM, Gupta MK. 3DNA: A Tool for Sculpting Brick-Based DNA Nanostructures. SynBio. 2023; 1(3):226-238. https://doi.org/10.3390/synbio1030016
Chicago/Turabian StyleGupta, Shikhar Kumar, Foram Joshi, Amay Agrawal, Sourav Deb, Martin Sajfutdinow, Dixita Limbachiya, David M. Smith, and Manish K. Gupta. 2023. "3DNA: A Tool for Sculpting Brick-Based DNA Nanostructures" SynBio 1, no. 3: 226-238. https://doi.org/10.3390/synbio1030016
APA StyleGupta, S. K., Joshi, F., Agrawal, A., Deb, S., Sajfutdinow, M., Limbachiya, D., Smith, D. M., & Gupta, M. K. (2023). 3DNA: A Tool for Sculpting Brick-Based DNA Nanostructures. SynBio, 1(3), 226-238. https://doi.org/10.3390/synbio1030016












