Cultivation of Diverse Type I and Type II Methanotrophs from Tropical Wetlands in India, Including Rare Taxa (Methylocucumis and Methylolobus)
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bodelier, P.L.E.; Pérez, G.; Veraart, A.J.; Krause, S.M.B. Methanotroph Ecology, Environmental Distribution and Functioning. In Methanotrophs Microbiology Fundamentals and Biotechnological Applications; Lee, E.Y., Ed.; Springer Nature: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Aselmann, I.; Crutzen, P.J. Global distribution of natural freshwater wetlands and rice paddies, their net primary productivity, seasonality and possible methane emissions. J. Atmos. Chem. 1989, 8, 307–358. [Google Scholar] [CrossRef]
- Hu, S.; Niu, Z.; Chen, Y. Global Wetland Datasets: A Review. Wetlands 2017, 37, 807–817. [Google Scholar] [CrossRef]
- Lugo, A.E.; Brown, S.; Brinson, M.M. Concepts in wetland ecology. Ecosyst. World 1990, 15, 53–85. [Google Scholar]
- Wang, Z.; Zeng, D.; Patrick, W.H. Methane emissions from natural wetlands. Environ. Monit. Assess. 1996, 42, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Mol. Biol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murell, C.J. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jørgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623. [Google Scholar] [CrossRef] [PubMed]
- Knittel, K.; Boetius, A. Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 2009, 63, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Nauhaus, K.; Treude, T.; Boetius, A.; Krüger, M. Environmental regulation of the anaerobic oxidation of methane: A comparison of ANME-I and ANME-II communities. Environ. Microbiol. 2005, 7, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543. [Google Scholar] [CrossRef] [PubMed]
- Deutzmann, J.S. Anaerobic Methane Oxidation in Freshwater Environments. In Anaerobic Utilization of Hydrocarbons, Oils, and Lipids, Handbook of Hydrocarbon and Lipid Microbiology; Boll, M., Ed.; Springer International: Berlin/Heidelberg, Germany, 2018; pp. 1–15. [Google Scholar]
- Rahalkar, M.C.; Khatri, K.; Pandit, P.; Mohite, J.; Bahulikar, R.A. Anaerobic Oxidation of Methane: A Brief Overview. In Anaerobes and Anaerobic Processes; Prakash, O., Ranade, D.R., Eds.; New India Publishing Agency: Delhi, India, 2020; pp. 169–180. [Google Scholar]
- Rahalkar, M.; Khatri, K.; Pandit, P.; Bahulikar, R.; Mohite, J. Cultivation of Important Methanotrophs from Indian Rice Fields. Front. Microbiol. 2021, 12, 669244. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Dunfield, P.F. Cultivation of methanotrophs. In Hydrocarbon and Lipid Microbiology, Springer Protocols Handbooks; McGenity, T.J., Timmis, K.N., Nogales, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–17. [Google Scholar]
- Strong, P.J.; Xie, S.; Clarke, W.P. Methane as aresource: Can the methanotrophs add value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Knief, C. Diversity and phylogeny of described aerobic methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 17–42. [Google Scholar]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J. Methylococcaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W., Ed.; John Wliey and Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Mohite, J.A.; Manvi, S.S.; Pardhi, K.; Khatri, K.; Bahulikar, R.A.; Rahalkar, M.C. Thermotolerant methanotrophs belonging to the Methylocaldum genus dominate the methanotroph communities in biogas slurry and cattle dung: A culture-based study from India. Environ. Res. 2023, 228, 115870. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Sakata, S. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Khatri, K.; Mohite, J.; Pandit, P.; Bahulikar, R. A novel Type I methanotroph Methylolobus aquaticus gen. nov. sp. nov. isolated from a tropical wetland. Antonie Van Leeuwenhoek 2020, 113, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Mohite, J.A.; Pardhi, K.; Manvi, S.S.; Kadam, Y.S.; Patil, Y.V. Insights into Methylocucumis oryzae, a Large-sized, Phylogenetically Unique Type Ia Methanotroph with Biotechnological Potential. Indian J. Microbiol. 2024, 64, 1964–1969. [Google Scholar] [CrossRef] [PubMed]
- Mohite, J.A.; Manvi, S.S.; Pardhi, K.; Bahulikar, R.A.; Deshpande, S.; Patange, S.; Joshi, M.; Kulkarni, S.; Rahalkar, M.C. Diverse type I and type II methanotrophs cultivated from an Indian freshwater wetland habitat. Int. Microbiol. 2023, 27, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Pandit, P.S.; Hoppert, M.; Rahalkar, M.C. Description of ‘Candidatus Methylocucumis oryzae’, a novel Type I methanotroph with large cells and pale pink colour, isolated from an Indian rice field. Antonie Van Leeuwenhoek 2018, 111, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Pandit, P.; Rahalkar, M.C. Renaming of ‘Candidatus Methylocucumis oryzae’ as Methylocucumis oryzae gen. nov., sp. nov., a novel Type I methanotroph isolated from India. Antonie Van Leeuwenhoek 2018, 112, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Pandit, P.S.; Rahalkar, M.C.; Dhakephalkar, P.K.; Ranade, D.R.; Pore, S.; Arora, P.; Kapse, N. Deciphering community structure of methanotrophs dwelling in rice rhizospheres of an Indian rice field using cultivation and cultivation-independent approaches. Microb. Ecol. 2016, 71, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Pandit, P.S.; Ranade, D.R.; Dhakephalkar, P.K.; Rahalkar, M.C. A pmoA-based study reveals dominance of yet uncultured Type I methanotrophs in rhizospheres of an organically fertilized rice field in India. 3 Biotech. 2016, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Pandit, P.S.; Dhakephalkar, P.K.; Pore, S.; Arora, P.; Kapse, N. Genome characteristics of a novel Type I methanotroph ‘Sn10-6’ isolated from a flooded Indian rice field. Microb. Ecol. 2016, 71, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Pandit, P. Genome-based insights into a putative novel Methylomonas species (strain Kb3), isolated from an Indian rice field. Gene Rep. 2018, 13, 9–13. [Google Scholar] [CrossRef]
- Rahalkar, M.C.; Patil, S.; Dhakephalkar, P.K.; Bahulikar, R.A. Cultivated methanotrophs associated with rhizospheres of traditional rice landraces from Western India belong to Methylocaldum and Methylocystis. 3 Biotech 2018, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- Khatri, K.; Mohite, J.A.; Pandit, P.S.; Bahulikar, R.; Rahalkar, M.C. Description of ‘Ca. Methylobacter oryzae’ KRF1, a novel species from the environmentally important Methylobacter clade 2. Antonie Van Leeuwenhoek 2020, 113, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Khatri, K.; Pandit, P.; Mohite, J. Polyphasic Characterization of Ca. Methylomicrobium oryzae: A Methanotroph Isolated from Rice Fields. Indian J. Microbiol. 2025, 65, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xing, X.H. Mixed methanotrophic consortium for applications in environmental bioengineering and biocatalysis. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M., Xing, X.H., Eds.; Springer: Cham, Switzerland, 2018; pp. 237–251. [Google Scholar]
- Lüke, C. Molecular Ecology and Biogeography of Methanotrophic Bacteria in Wetland Rice Fields; Max-Planck-Institut für Terrestrische Mikrobiologie: Marburg, Germany, 2010; p. 159. [Google Scholar]
- Lüke, C.; Frenzel, P.; Ho, A.; Fiantis, D.; Schad, P.; Schneider, B.; Schwark, L.; Utami, S.R. Macroecology of methane-oxidizing bacteria: The β-diversity of pmoA genotypes in tropical and subtropical rice paddies. Environ. Microbiol. 2014, 16, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Costello, A.M.; Lidstrom, M.E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 1999, 65, 5066–5074. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M. Aerobic Methanotrophic Bacterial Communities from Sediments of Lake Constance. Microbiology and Limnology. Ph.D. Thesis, University of Konstanz, Konstanz, Germany, 2007. [Google Scholar]
- Rahalkar, M.; Schink, B. Comparison of aerobic methanotrophic communities in littoral and profundal sediments of Lake Constance by a molecular approach. Appl. Environ. Microbiol. 2007, 73, 4389–4394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Auman, A.J.; Lidstrom, M.E. Analysis of sMMO-containing Type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 2002, 4, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Auman, A.; Stolyar, S.; Lidstrom, M.E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 2000, 66, 5259. [Google Scholar] [CrossRef] [PubMed]
- Luke, C.; Krause, S.; Cavigiolo, S.; Greppi, D.; Lupotto, E.; Frenzel, P. Biogeography of wetland rice methanotrophs. Environ. Microbiol. 2010, 12, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Horz, H.P.; Yimga, M.T.; Liesack, W. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmox, mxaf, and 16s rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 2001, 67, 4177–4185. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, P.; Dubey, S.K. DNA microarray analysis targeting pmoA gene reveals diverse community of methanotrophs in the rhizosphere of tropical rice soils. Curr. Sci. 2010, 99, 1090–1095. [Google Scholar]
- Luke, C.; Frenzel, P. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 2011, 77, 6305–6309. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Yang, S.; Liebner, S. Evaluation and update of cutoff values for methanotrophic pmoA gene sequences. Arch. Microbiol. 2016, 198, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Heyer, J.; Galchenko, V.F.; Dunfield, P.F. Molecular phylogeny of type II methane oxidizing bacteria isolated from various environments. Microbiology 2002, 148, 2831–2846. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; Lüke, C.; Cao, Z.; Frenzel, P. Ageing well: Methane oxidation and methane oxidizing bacteria along a chronosequence of 2000 years. Environ. Microbiol. Rep. 2011, 2011, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Reim, A.; Luke, C.; Krause, S.; Pratscher, J.; Frenzel, P. One millimetre makes the difference: High-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 2012, 6, 2128–2139. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; Luke, C.; Frenzel, P. Recovery of methanotrophs from disturbance: Population dynamics, evenness and functioning. ISME J. 2011, 5, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Krause, S.; Luke, C.; Frenzel, P. Succession of methanotrophs in oxygen-methane counter-gradients of flooded rice paddies. ISME J. 2010, 4, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, P.; Dubey, S.K. Diversity of methanotrophs in urea-fertilized tropical rice agroecosystem. Indian J. Microbiol. 2010, 50, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, P.; Dumont, M.G.; Bodrossy, L.; Stralis-Pavese, N.; Murrell, J.C.; Dubey, S.K. Ecological and molecular analyses of the rhizospheric methanotroph community in tropical rice soil: Effect of crop phenology and land-use history. Curr. Sci. 2009, 96, 1082–1089. [Google Scholar]
- Graef, C.; Hestnes, A.G.; Svenning, M.M.; Frenzel, P. The active methanotrophic community in a wetland from the High Arctic. Environ. Microbiol. Rep. 2011, 3, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N. Cultivating uncultured bacteria from northern wetlands: Knowledge gained and remaining gaps. Front. Microbiol. 2011, 2, 184. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N. Exploring methanotroph diversity in acidic northern wetlands: Molecular and cultivation-based studies. Microbiology 2009, 78, 655–669. [Google Scholar] [CrossRef]
- Yun, J.; Ma, A.; Li, Y.; Zhuang, G.; Wang, Y.; Zhang, H. Diversity of methanotrophs in Zoige wetland soils under both anaerobic and aerobic conditions. J. Environ. Sci. 2010, 22, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Kip, N.; Dutilh, B.E.; Pan, Y.; Bodrossy, L.; Neveling, K.; Kwint, M.P.; Jetten, M.S.; Op den Camp, H.J. Ultra-deep pyrosequencing of pmoA amplicons confirms the prevalence of Methylomonas and Methylocystis in Sphagnum mosses from a Dutch peat bog. Environ. Microbiol. Rep. 2011, 3, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.S.; Ducklow, H.W. Microbial communities can be described by metabolic structure: A general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic Peninsula. PLoS ONE 2015, 10, e0135868. [Google Scholar] [CrossRef] [PubMed]
- Belova, S.E.; Kulichevskaya, I.S.; Bodelier, P.L.; Dedysh, S.N. Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int. J. Syst. Evol. Microbiol. 2013, 63, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Frindte, K.; Maarastawi, S.A.; Lipski, A.; Hamacher, J.; Knief, C. Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int. J. Syst. Evol. Microbiol. 2017, 67, 4507–4514. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, A.; Lee, C.G.; Ogiso, T.; Ueno, C.; Dianou, D.; Demachi, T.; Katayama, A.; Asakawa, S. Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int. J. Syst. Evol. Microbiol. 2015, 65, 3527–3534. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ganguly, D.; Chakraborty, S.; Mukherjee, A.; Kumar De, T. Methane flux dynamics in relation to methanogenic and methanotrophic populations in the soil of Indian Sundarban mangroves. Mar. Ecol. 2018, 39, e12493. [Google Scholar] [CrossRef]
- Su, G.; Jiao, N.; Hu, Y.; Zheng, Q.; Zopfi, J.; Lehmann, M.F.; Guo, Z. Tidal control on aerobic methane oxidation and mitigation of methane emissions from coastal mangrove sediments. Environ. Res. 2024, 263, 120049. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, E.N.; Suleimanov, R.Z.; Ashikhmin, A.A.; Ivanova, A.A.; Dedysh, S.N. All kinds of sunny colors synthesized from methane: Genome-encoded carotenoid production by Methylomonas species. Microorganisms 2023, 11, 2865. [Google Scholar] [CrossRef] [PubMed]
- Dall’Osto, L.; Bassi, R.; Ruban, A. Photoprotective Mechanisms: Carotenoids. In Plastid Biology, Advances in Plant Biology; Theg, S.M., Wollman, F.A., Eds.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Wartiainen, I.; Hestnes, A.G.; McDonald, I.R.; Svenning, M.M. Methylocystis rosea sp. nov., a novel methanotrophic bacterium from Arctic wetland soil, Svalbard, Norway (78° N) Free. Int. J. Syst. Evol. Microbiol. 2006, 56, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Mohite, J.; Khatri, K.; Pardhi, K.; Manvi, S.S.; Jadhav, R.; Rathod, S.; Rahalkar, M.C. Exploring the potential of methanotrophs for plant growth promotion in rice agriculture. Methane 2023, 2, 361–371. [Google Scholar] [CrossRef]
- Kumar, S.R.; David, E.M.; Pavithra, G.J.; Kumar, G.S.; Subbian, E. Methane-derived microbial biostimulant reduces greenhouse gas emissions and improves rice yield. Front. Plant Sci. 2024, 15, 1432460. [Google Scholar] [CrossRef] [PubMed]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
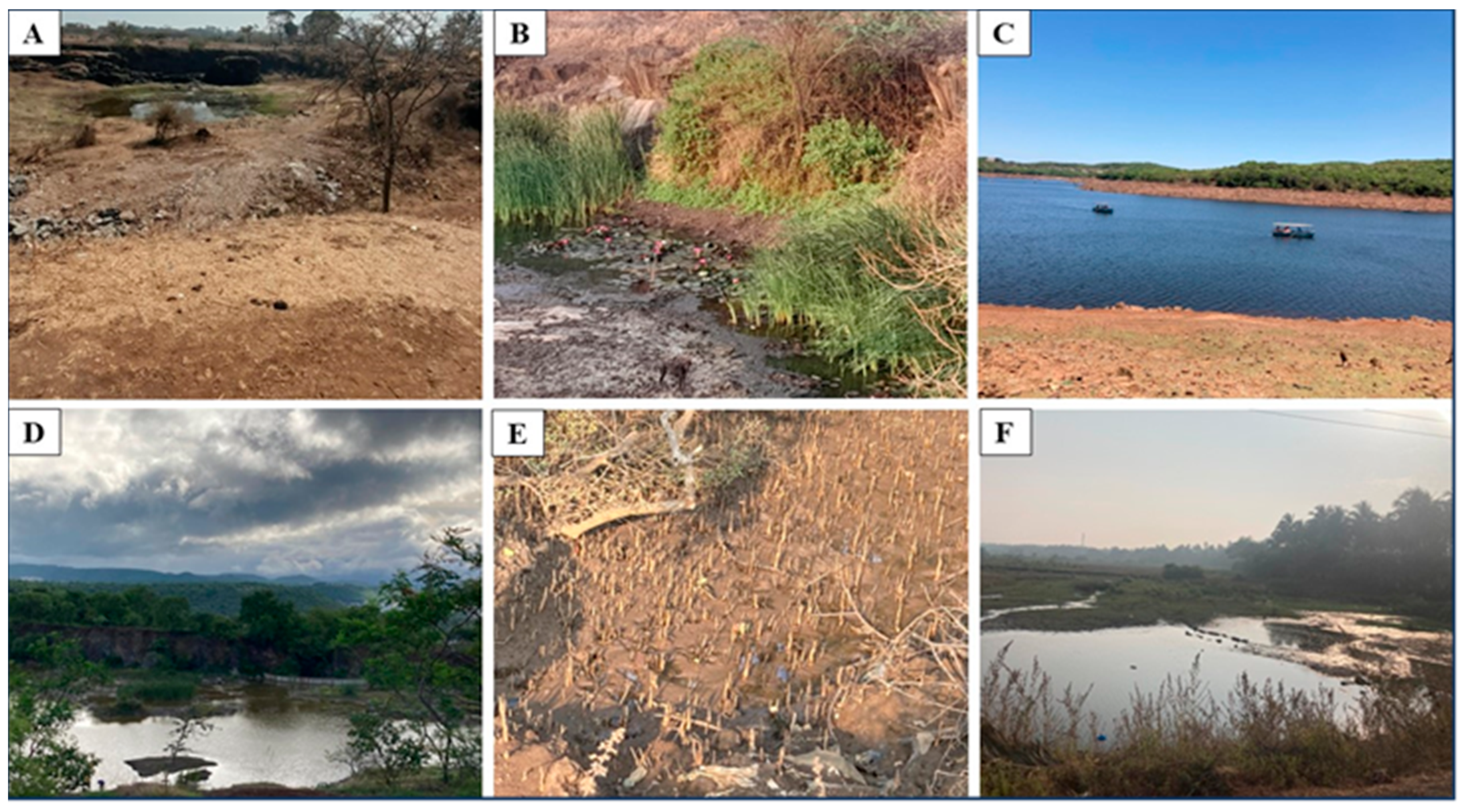

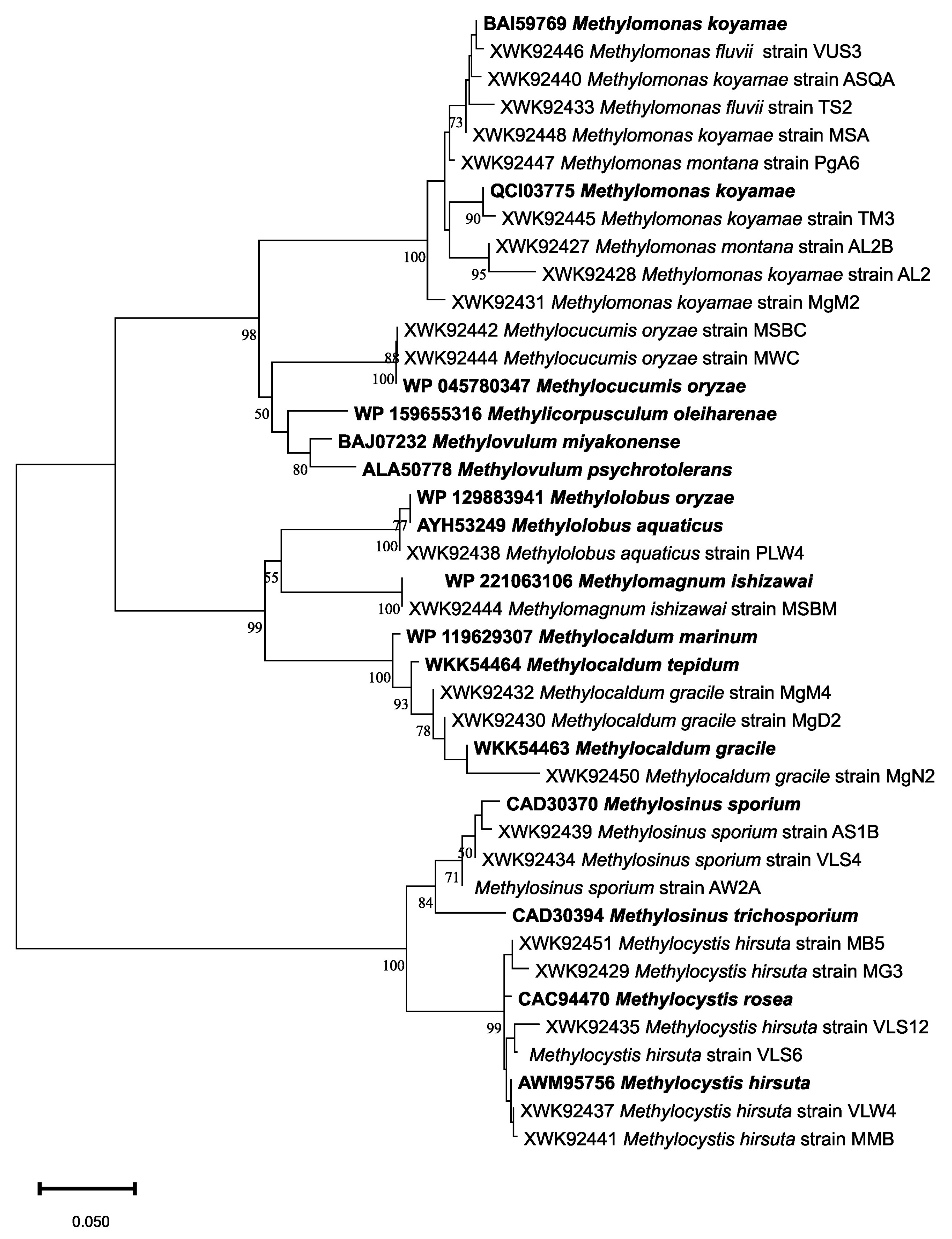

| Sampling Details | Dilution Used for Isolation of Methanotrophs | Representative Cultures | Identification Using the pmoA Gene | ||||
|---|---|---|---|---|---|---|---|
| Sampling Site and Geographical Location | Sampling Date | Strain Name | GeneBank Accession Number | Nearest Match (with Type Cultures) | % Similarity (Nucleotide) | % Similarity (Protein) | |
| Vetal Hill Pond a (Fresh water sample) 18.525474° N 73.815292° E | 15 January 2024 | 10−2 | AS1B | PQ821919 | Methylosinus sporium strain ATCC 35069 | 99.31 | 98.61 |
| 15 January 2024 | 10−2 | ASQA | PQ821920 | Methylomonas koyamae strain Fw12E-Y | 96.12 | 99.31 | |
| 15 January 2024 | 10−8 | AW2A | PQ821929 | Methylosinus sporium strain ATCC 35069 | 95.05 | 97.93 | |
| 15 January 2024 | 10−8 | AW1A | PQ821929 | Methylosinus sporium strain ATCC 35069 | 94.84 | 97.99 | |
| 15 January 2024 | 10−8 | AW2B | PQ821919 | Methylosinus sporium strain ATCC 35069 | 100 | 100 | |
| Mahatma Hill Pond a (Fresh water and mud sample) 18.4926° N 73.8013° E | 21 April 2024 | 10−2 | MSBM | PQ821923 | Methylomagnum ishizawai strain RS11D | 99.53 | 99.29 |
| 18 December 2023 | 10−3 | TM3 | PQ821925 | Methylomonas koyamae strain Fw12E-Y | 92.47 | 97.18 | |
| 21 April 2024 | 10−5 | MMB | PQ821921 | Methylocystis hirsuta strain CSC1 | 97.46 | 98.62 | |
| 21 April 2024 | 10−6 | MSA | PQ821928 | Methylomonas koyamae strain Fw12E-Y | 95.81 | 100 | |
| 21 April 2024 | 10−7 | MWC | PQ821924 | Methylocucumis oryzae strain Sn 10-6 | 98.61 | 100 | |
| 21 April 2024 | 10−8 | MSBC | PQ821922 | Methylocucumis oryzae strain Sn 10-6 | 98.66 | 100 | |
| ARI pond a (Lotus root sample) 18.5173° N 73.8475° E | 2 January 2024 | 10−2 | AL2 | PQ821908 | Methylomonas koyamae strain Fw12E-Y | 87.79 | 95.16 |
| 2 January 2024 | 10−2 | AL2B | PQ821907 | Methylomonas montana strain MW1 | 89.56 | 97.90 | |
| Paragrass BAIF pond a (Seaweed sample) 18.491005° N 74.134544° E | 9 January 2024 | 10−6 | PgA6 | PQ821927 | Methylomonas montana strain MW1 | 94.34 | 98.03 |
| Pashan Lake (Fresh water sample) 18.533752° N 73.785717° E | 8 July 2023 | 10−2 | PLW2 | PQ821916 | Methylosinus trichosporium strain OB3b | 100 | 99.31 |
| 8 July 2023 | 10−4 | PLW4 | PQ821918 | Methylolobus aquaticus strain FWC3 | 97.25 | 100 | |
| Venna Lake Sediments (Fresh water and sediment sample) 17.934° N 73.665° E | 28 December 2022 | 10−3 | VUS3 | PQ821926 | Methylomonas fluvii EbB | 95.19 | 100 |
| 28 December 2022 | 10−4 | VLS4 | PQ821914 | Methylosinus sporium ATCC 35069 | 95.06 | 98.55 | |
| 28 December 2022 | 10−4 | VLW4 | PQ821917 | Methylocystis hirsuta strain CSC1 | 97.35 | 97.93 | |
| 28 December 2022 | 10−5 | MB5 | PQ821931 | Methylocystis hirsuta strain CSC1 | 97.12 | 98.66 | |
| 28 December 2022 | 10−6 | VLS6 | PQ821915 | Methylocystis hirsuta CSC1 | 97.70 | 98.56 | |
| 28 December 2022 | 10−12 | VLS12 | PQ821915 | Methylocystis hirsuta CSC1 | 97.25 | 98.60 | |
| Tamhini river (Fresh water sediment sample) 18.134111° N 73.605728° E | 16 June 2023 | 10−2 | TS2 | PQ821913 | Methylomonas fluvii strain EbB | 95.19 | 98.53 |
| Mumbai Mangroves a (Brackish water and soil sample) 19.0374° N, 72.981° E | 21 September 2023 | 10−2 | MgM2 | PQ821911 | Methylomonas koyamae strain Fw12E-Y | 91.88 | 97.14 |
| 21 September 2023 | 10−4 | MgM4 | PQ821912 | Methylocaldum gracile strain VKM-14L | 99.18 | 100 | |
| Alibag mangroves (Brackish water and soil sample) 18.64° N 72.88° E | 23 March 2023 | 10−3 | MG3 | PQ821909 | Methylocystis hirsuta strain CSC1 | 97.50 | 99.29 |
| 23 March 2023 | 10−3 | MgN2 | PQ821930 | Methylocaldum gracile strain VKM-14L | 99.52 | 100 | |
| Diveagar mangroves (Brackish water and soil sample) 18.1920° N 72.9789° E | 29 December 2023 | 10−2 | MgD2 | PQ821910 | Methylocaldum gracile strain VKM-14L | 98.95 | 100 |
| Sampling Details | Dilution Used for Isolation of Methanotrophs | Representative Strain | Identification Using 16S rRNA Gene | |||
|---|---|---|---|---|---|---|
| Sampling Site and Geographical Location | Sampling Date | Strain Name | Gene Accession Number | Nearest Match with Type Strain | % Similarity | |
| Vetal Hill Pond (Fresh water sample) 18.525474° N 73.815292° E | 15 January 2023 | 10−8 | AW1A | PQ826297 | Methylosinus sporium strain NCIMB 11126 | 98.89 |
| 15 January 2024 | 10−8 | AW2B | PV637194 | Methylosinus trichosporium strain OB3b | 100 | |
| Venna Lake (Sediment sample) 17.934° N 73.665° E | 28 December 2022 | 10−3 | VUS3 | PQ826293 | Methylomonas fluvii strain EbB | 99.41 |
| 28 December 2022 | 10−4 | VLS4 | PQ826294 | Methylomonas sporium strain NCIMB 11126 | 98.96 | |
| Pashan Lake (Fresh water sample) 18.533752° N 73.785717° E | 8 July 2023 | 10−2 | PLW2 | PV637193 | Methylosinus trichosporium strain OB3b | 99.93 |
| 8 July 2023 | 10−4 | PLW4 | PV804843 | Methylolobus aquaticus strain FWC3 | 99.43 | |
| Tamhini river (Fresh water sediment sample) 18.134111° N 73.605728° E | 16 June 2023 | 10−2 | TS2 | PQ826293 | Methylomonas fluvii strain EbB | 99.41 |
| Mumbai Mangroves (Soil sample) 19° N 72° E | 21 September 2023 | 10−2 | MgM2 | PV630802 | Methylomonas aurea strain SURF-1 | 96.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardhi, K.; Manvi, S.; Bahulikar, R.A.; Patil, Y.; Kadam, Y.; Kadam, S.; Saraf, C.; Rahalkar, M.C. Cultivation of Diverse Type I and Type II Methanotrophs from Tropical Wetlands in India, Including Rare Taxa (Methylocucumis and Methylolobus). Methane 2025, 4, 17. https://doi.org/10.3390/methane4030017
Pardhi K, Manvi S, Bahulikar RA, Patil Y, Kadam Y, Kadam S, Saraf C, Rahalkar MC. Cultivation of Diverse Type I and Type II Methanotrophs from Tropical Wetlands in India, Including Rare Taxa (Methylocucumis and Methylolobus). Methane. 2025; 4(3):17. https://doi.org/10.3390/methane4030017
Chicago/Turabian StylePardhi, Kajal, Shubha Manvi, Rahul A. Bahulikar, Yukta Patil, Yash Kadam, Shirish Kadam, Chandani Saraf, and Monali C. Rahalkar. 2025. "Cultivation of Diverse Type I and Type II Methanotrophs from Tropical Wetlands in India, Including Rare Taxa (Methylocucumis and Methylolobus)" Methane 4, no. 3: 17. https://doi.org/10.3390/methane4030017
APA StylePardhi, K., Manvi, S., Bahulikar, R. A., Patil, Y., Kadam, Y., Kadam, S., Saraf, C., & Rahalkar, M. C. (2025). Cultivation of Diverse Type I and Type II Methanotrophs from Tropical Wetlands in India, Including Rare Taxa (Methylocucumis and Methylolobus). Methane, 4(3), 17. https://doi.org/10.3390/methane4030017







