In Silico Analysis and PCR Characterization of non-Tn4401 Transposable Elements in Pseudomonas aeruginosa †
Abstract
1. Introduction
2. Methods
2.1. In Silico Phase
Exploration of the blaKPC Genetic Environment for P. aeruginosa in the GenBank
2.2. PCR Essay for Tn4401 or NTEKPC Identification
Primer Design
3. Results and Discussion
3.1. In Silico Analysis of the Collected Reports
3.2. In Vitro Results
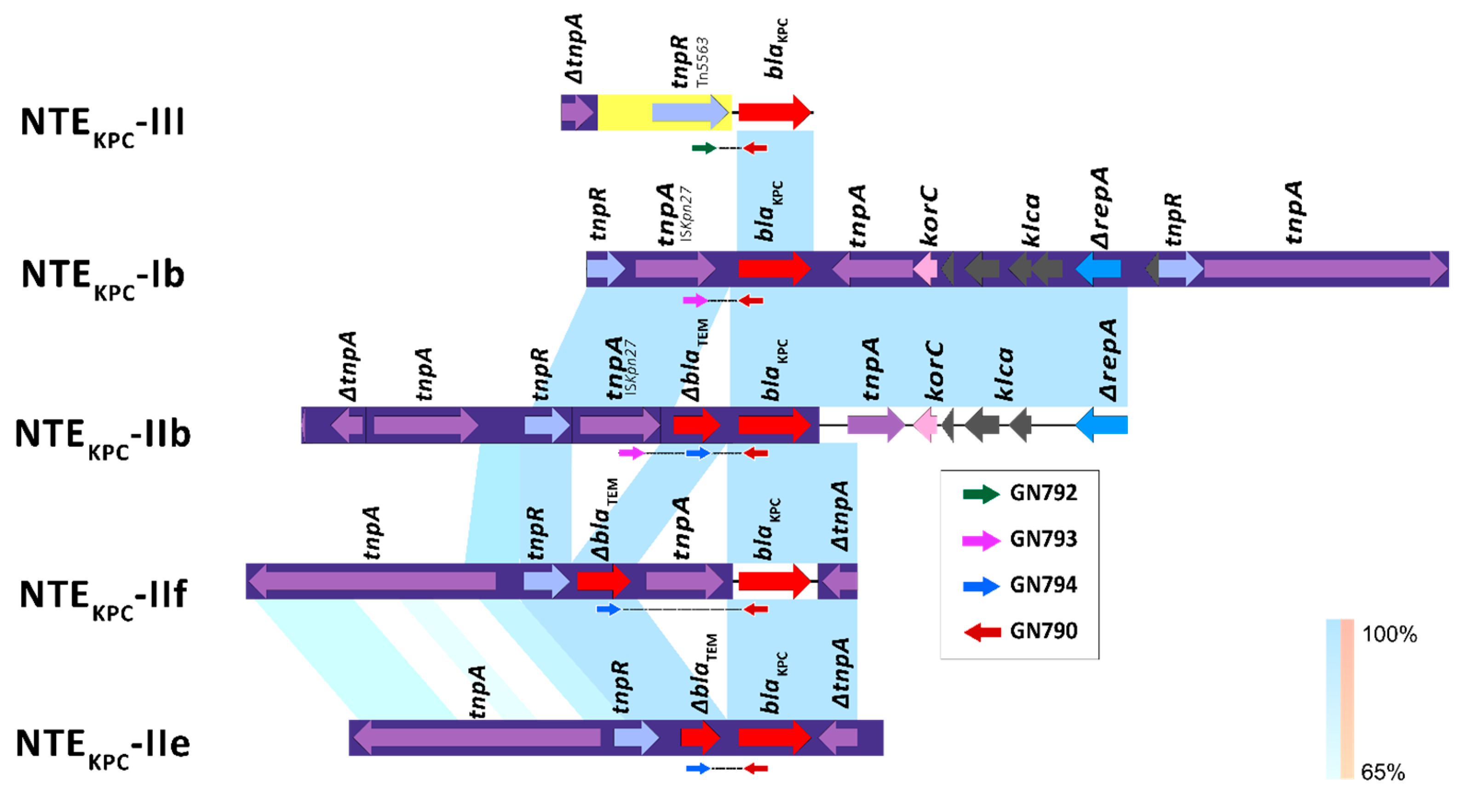
3.2.1. Tn4401 and NTEKPC Primers Design
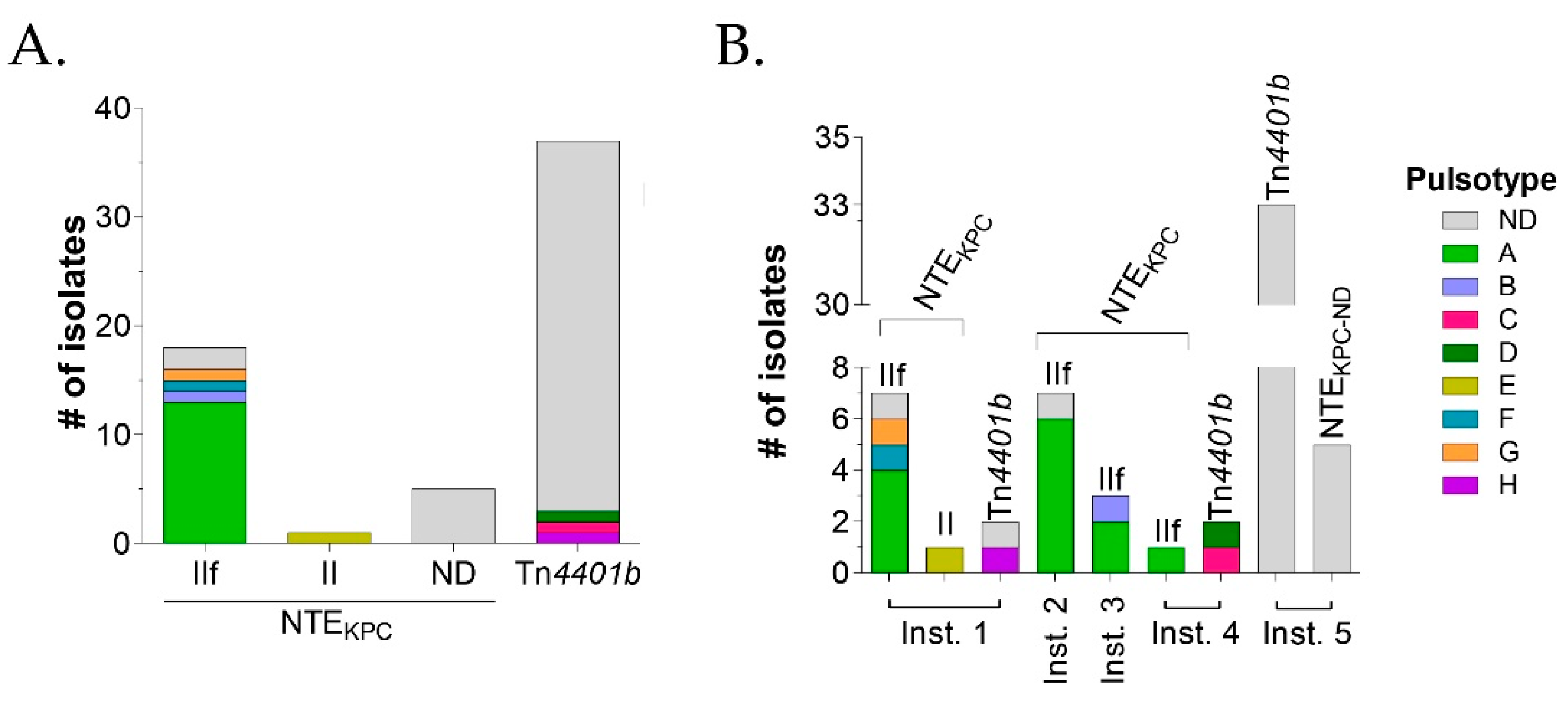
3.2.2. Characterization of the Genetic Environment Associated with blaKPC in Colombian Clinical Isolates of P. aeruginosa
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pitout, J.D.; Nordmann, P.; Poirel, L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.; Figueroa, C.; Moya-Flores, F.; Guggiana, P.; Castillo, C.; Rivas, L.; Munita, J.M.; Garcia, P.C. A multispecies outbreak of carbapenem-resistant bacteria harboring the blaKPC gene in a non-classical transposon element. BMC Microbiol. 2021, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Córdova, E.; Lespada, M.I.; Gómez, N.; Pasterán, F.; Oviedo, V.; Rodríguez-Ismael, C. Descripción clínica y epidemiológica de un brote nosocomial por Klebsiella pneumoniae productora de KPC en Buenos Aires, Argentina. Enferm. Infecc. Y Microbiol. Clínica 2012, 30, 376–379. [Google Scholar] [CrossRef]
- Vera-Leiva, A.; Barría-Loaiza, C.; Carrasco-Anabalón, S.; Lima, C.; Aguayo-Reyes, A.; Domínguez, M.; Bello-Toledo, H.; González-Rocha, G. KPC: Klebsiella pneumoniae carbapenemasa, principal carbapenemasa en enterobacterias. Rev. Chil. Infectol. 2017, 34, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Villegas, M.V.; Lolans, K.; Correa, A.; Kattan, J.N.; Lopez, J.A.; Quinn, J.P. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 2007, 51, 1553–1555. [Google Scholar] [CrossRef]
- Chen, L.; Chavda, K.D.; Al Laham, N.; Melano, R.G.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. Complete nucleotide sequence of a bla KPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 2013, 57, 5019–5025. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Chavda, K.D.; DeLeo, F.R.; Bonomo, R.A.; Kreiswirth, B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014, 22, 686–696. [Google Scholar] [CrossRef]
- Chen, L.F.; Anderson, D.J.; Paterson, D.L. Overview of the epidemiology and the threat of Klebsiella pneumoniae carbapenemases (KPC) resistance. Infect. Drug Resist. 2012, 5, 133. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Nordmann, P. Functional characterization of Tn 4401, a Tn 3-based transposon involved in bla KPC gene mobilization. Antimicrob. Agents Chemother. 2011, 55, 5370–5373. [Google Scholar] [CrossRef]
- Naas, T.; Cuzon, G.; Villegas, M.-V.; Lartigue, M.-F.; Quinn, J.P.; Nordmann, P. Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob. Agents Chemother. 2008, 52, 1257–1263. [Google Scholar] [CrossRef]
- de Lima, G.J.; Scavuzzi, A.M.L.; Beltrão, E.M.B.; Firmo, E.F.; de Oliveira, É.M.; de Oliveira, S.R.; Rezende, A.M.; de Souza Lopes, A.C. Identification of plasmid IncQ1 and NTE KPC-IId harboring bla KPC-2 in isolates from Klebsiella pneumoniae infections in patients from Recife-PE, Brazil. Rev. Soc. Bras. Med. Trop. 2020, 53, e20190526. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Tansirichaiya, S.; Rahman, M.A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Wright, M.H.; Adelskov, J.; Greene, A.C. Bacterial DNA extraction using individual enzymes and phenol/chloroform separation. J. Microbiol. Biol. Educ. 2017, 18, 18.12.60. [Google Scholar] [CrossRef]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of bla KPC-2 dissemination from non-CG258 Klebsiella pneumoniae to other Enterobacterales via IncN plasmids in an area of high endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef]
- Huang, J.; Hu, X.; Zhao, Y.; Shi, Y.; Ding, H.; Wu, R.; Zhao, Z.; Ji, J. Comparative Analysis of bla KPC Expression in Tn4401 Transposons and the Tn3-Tn4401 Chimera. Antimicrob. Agents Chemother. 2019, 63, e02434-18. [Google Scholar] [CrossRef]
- Gomez, S.A.; Pasteran, F.G.; Faccone, D.; Tijet, N.; Rapoport, M.; Lucero, C.; Lastovetska, O.; Albornoz, E.; Galas, M.; Group, K.P.C.; et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin. Microbiol. Infect. 2011, 17, 1520–1524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, B.; Sun, J.Y.; Liu, Q.Z.; Han, L.Z.; Huang, X.H.; Ni, Y.X. First report of Klebsiella oxytoca strain coproducing KPC-2 and IMP-8 carbapenemases. Antimicrob. Agents Chemother. 2011, 55, 2937–2941. [Google Scholar] [CrossRef] [PubMed]
- Treepong, P.; Kos, V.N.; Guyeux, C.; Blanc, D.S.; Bertrand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef]
- Abril, D.; Marquez-Ortiz, R.A.; Castro-Cardozo, B.; Moncayo-Ortiz, J.I.; Olarte Escobar, N.M.; Corredor Rozo, Z.L.; Reyes, N.; Tovar, C.; Sanchez, H.F.; Castellanos, J.; et al. Genome plasticity favours double chromosomal Tn4401b-blaKPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Correa, A.; Del Campo, R.; Perenguez, M.; Blanco, V.M.; Rodriguez-Banos, M.; Perez, F.; Maya, J.J.; Rojas, L.; Canton, R.; Arias, C.A.; et al. Dissemination of high-risk clones of extensively drug-resistant Pseudomonas aeruginosa in colombia. Antimicrob. Agents Chemother. 2015, 59, 2421–2425. [Google Scholar] [CrossRef] [PubMed]

| MGE | Genetic Landmark | Location | ST |
|---|---|---|---|
| NTEKPC-I n = 32 | ISKpn27 | China (n = 30) | Chromosome (n = 5) |
| Plasmid (n = 25) | |||
| Brazil (n = 2) | Plasmid (n = 2) | ||
| NTEKPC-II n = 9 | blaTEM | China (n = 5) | Plasmid (n = 5) |
| Brazil (n = 1) | Plasmid (n = 1) | ||
| Colombia (n = 3) | Plasmid (n = 3) | ||
| NTEKPC-III n = 0 | Tn5563 resolvase | Not reported | Not reported |
| NTEKPC n = 3 | ND | China (n = 1) | Plasmid (n = 1) |
| Brazil (n = 1) | Plasmid (n = 1) | ||
| France (n = 1) | Plasmid (n = 1) | ||
| Tn4401 n = 16 | ISKpn7 | France (n = 2) | Plasmid (n = 2) |
| Argentina (n = 2) | Plasmid (n = 2) | ||
| Colombia (n = 2) | Chromosome (n = 1) | ||
| Plasmid (n = 1) | |||
| USA (n = 2) | Plasmid (n = 2) | ||
| Brazil (n = 1) | Plasmid (n = 1) | ||
| Chile (n = 2) | Plasmid (n = 2) | ||
| China (n = 4) | Plasmid (n = 4) | ||
| Nepal (n = 1) | Not reported |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Castellanos, J.; Márquez-Ortiz, R.A.; Abril, D.; Hurtado, D.F.; Tíjaro, G.; Corredor-Rozo, Z.; Escobar-Pérez, J.; Vanegas, N. In Silico Analysis and PCR Characterization of non-Tn4401 Transposable Elements in Pseudomonas aeruginosa. Med. Sci. Forum 2022, 12, 33. https://doi.org/10.3390/eca2022-12738
Ruiz-Castellanos J, Márquez-Ortiz RA, Abril D, Hurtado DF, Tíjaro G, Corredor-Rozo Z, Escobar-Pérez J, Vanegas N. In Silico Analysis and PCR Characterization of non-Tn4401 Transposable Elements in Pseudomonas aeruginosa. Medical Sciences Forum. 2022; 12(1):33. https://doi.org/10.3390/eca2022-12738
Chicago/Turabian StyleRuiz-Castellanos, Julián, Ricaurte Alejandro Márquez-Ortiz, Deisy Abril, Daniela Forero Hurtado, Ginna Tíjaro, Zayda Corredor-Rozo, Javier Escobar-Pérez, and Natasha Vanegas. 2022. "In Silico Analysis and PCR Characterization of non-Tn4401 Transposable Elements in Pseudomonas aeruginosa" Medical Sciences Forum 12, no. 1: 33. https://doi.org/10.3390/eca2022-12738
APA StyleRuiz-Castellanos, J., Márquez-Ortiz, R. A., Abril, D., Hurtado, D. F., Tíjaro, G., Corredor-Rozo, Z., Escobar-Pérez, J., & Vanegas, N. (2022). In Silico Analysis and PCR Characterization of non-Tn4401 Transposable Elements in Pseudomonas aeruginosa. Medical Sciences Forum, 12(1), 33. https://doi.org/10.3390/eca2022-12738







