mRNA-Based Biomarker Identification for Targeted Therapy Development in Pancreatic Cancer †
Abstract
1. Introduction
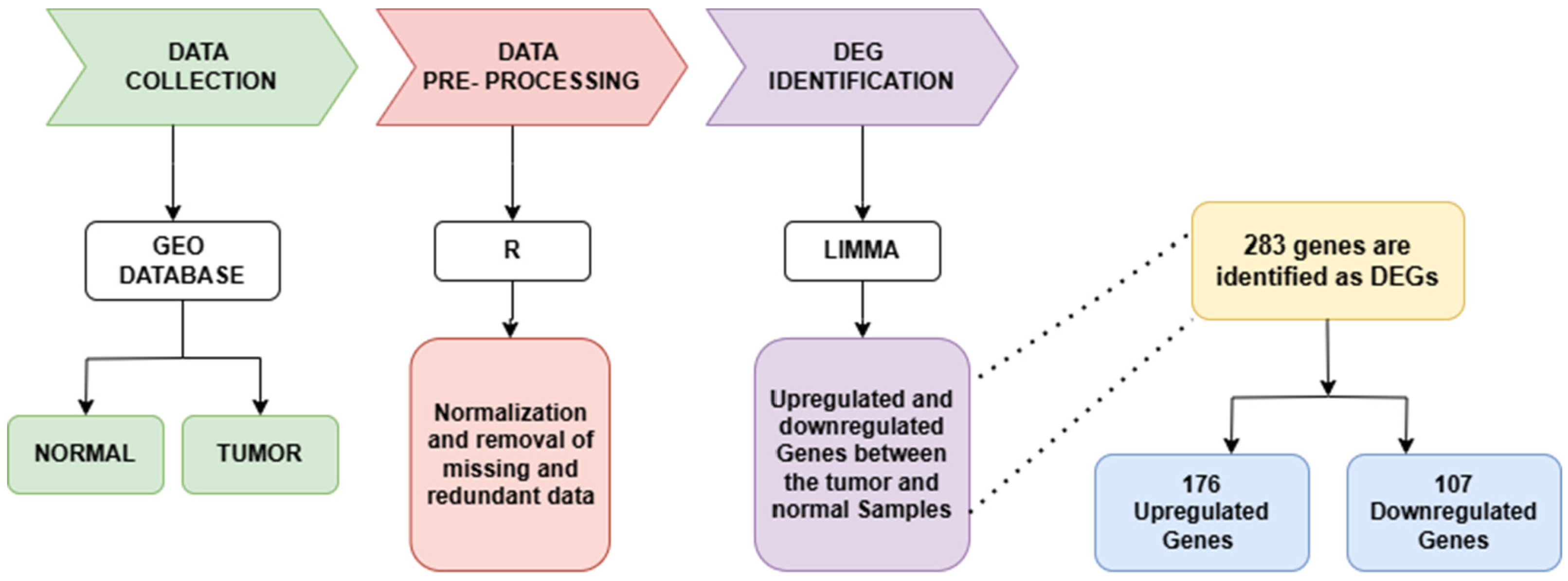
2. Materials and Methods
2.1. Database Screening and Preprocessing
2.2. Differential Gene Expression Analysis
3. Results
3.1. DEG Identification
3.2. Identification of Key Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Lyssiotis, C.A.; di Magliano, M.P.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lok, V.; Ngai, C.H.; Zhang, L.; Yuan, J.; Lao, X.Q.; Ng, K.; Chong, C.; Zheng, Z.J.; Wong, M.C. Worldwide burden of, risk factors for, and trends in pancreatic cancer. Gastroenterology 2021, 160, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, G.; Tang, T.Y.; Gao, X.; Liang, T.B. Personalized pancreatic cancer therapy: From the perspective of mRNA vaccine. Mil. Med. Res. 2022, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, H.; Taher, L.; Denz, A.; Grützmann, R.; Pilarsky, C.; Weber, G.F. Identification of prognostic biomarkers by combined mRNA and miRNA expression microarray analysis in pancreatic cancer. Transl. Oncol. 2018, 11, 700–714. [Google Scholar] [CrossRef]
- Stefanovic, S.; Wirtz, R.; Deutsch, T.M.; Hartkopf, A.; Sinn, P.; Varga, Z.; Sobottka, B.; Sotiris, L.; Taran, F.A.; Domschke, C.; et al. Tumor biomarker conversion between primary and metastatic breast cancer: mRNA assessment and its concordance with immunohistochemistry. Oncotarget 2017, 8, 51416. [Google Scholar] [CrossRef]
- He, J.; Wu, F.; Han, Z.; Hu, M.; Lin, W.; Li, Y.; Cao, M. Biomarkers (mRNAs and Non-Coding RNAs) for the diagnosis and prognosis of colorectal cancer–from the body fluid to tissue level. Front. Oncol. 2021, 11, 632834. [Google Scholar] [CrossRef]
- Xing, L.; Lv, L.; Ren, J.; Yu, H.; Zhao, X.; Kong, X.; Xiang, H.; Tao, X.; Dong, D. Advances in targeted therapy for pancreatic cancer. Biomed. Pharmacother. 2023, 168, 115717. [Google Scholar] [CrossRef]
- Tan, A.C.; Tan, D.S. Targeted therapies for lung cancer patients with oncogenic driver molecular alterations. J. Clin. Oncol. 2022, 40, 611–625. [Google Scholar] [CrossRef]
- Zhu, K.; Wu, Y.; He, P.; Fan, Y.; Zhong, X.; Zheng, H.; Luo, T. PI3K/AKT/mTOR-targeted therapy for breast cancer. Cells 2022, 11, 2508. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Agapito, G.; Milano, M.; Cannataro, M. A statistical network pre-processing method to improve relevance and significance of gene lists in microarray gene expression studies. BMC Bioinform. 2022, 23, 393. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar]
- Baus-Loncar, M.; Giraud, A.S. Trefoil factors: Multiple regulatory pathways for trefoil factor (TFF) genes. Cell. Mol. Life Sci. CMLS 2005, 62, 2921–2931. [Google Scholar]
- Fu, T.; Liu, J.X.; Xie, J.; Gao, Z.; Yang, Z. LAMC2 as a prognostic biomarker in human cancer: A systematic review and meta-analysis. BMJ Open 2022, 12, e063682. [Google Scholar] [CrossRef]
- O’Neill, R.S.; Stoita, A. Biomarkers in the diagnosis of pancreatic cancer: Are we closer to finding the golden ticket? World J. Gastroenterol. 2021, 27, 4045. [Google Scholar]
- Shen, G.Q.; Aleassa, E.M.; Walsh, R.M.; Morris-Stiff, G. Next-generation sequencing in pancreatic cancer. Pancreas 2019, 48, 739–748. [Google Scholar]
- Cicenas, J.; Kvederaviciute, K.; Meskinyte, I.; Meskinyte-Kausiliene, E.; Skeberdyte, A.; Cicenas, J., Jr. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers 2017, 9, 42. [Google Scholar] [CrossRef]
- Jahan, R.; Shah, A.; Kisling, S.G.; Macha, M.A.; Thayer, S.; Batra, S.K.; Kaur, S. Odyssey of trefoil factors in cancer: Diagnostic and therapeutic implications. Biochim. Biophys. Acta (BBA) Rev. Cancer 2020, 1873, 188362. [Google Scholar]
- Erice, O.; Narayanan, S.; Feliu, I.; Entrialgo-Cadierno, R.; Malinova, A.; Vicentini, C.; Guruceaga, E.; Delfino, P.; Trajkovic-Arsic, M.; Moreno, H.; et al. LAMC2 regulates key transcriptional and targetable effectors to support pancreatic cancer growth. Clin. Cancer Res. 2023, 29, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firdaus, S.; Parveen, R. mRNA-Based Biomarker Identification for Targeted Therapy Development in Pancreatic Cancer. Biol. Life Sci. Forum 2025, 43, 2. https://doi.org/10.3390/blsf2025043002
Firdaus S, Parveen R. mRNA-Based Biomarker Identification for Targeted Therapy Development in Pancreatic Cancer. Biology and Life Sciences Forum. 2025; 43(1):2. https://doi.org/10.3390/blsf2025043002
Chicago/Turabian StyleFirdaus, Saima, and Rafat Parveen. 2025. "mRNA-Based Biomarker Identification for Targeted Therapy Development in Pancreatic Cancer" Biology and Life Sciences Forum 43, no. 1: 2. https://doi.org/10.3390/blsf2025043002
APA StyleFirdaus, S., & Parveen, R. (2025). mRNA-Based Biomarker Identification for Targeted Therapy Development in Pancreatic Cancer. Biology and Life Sciences Forum, 43(1), 2. https://doi.org/10.3390/blsf2025043002





