The Acclimation Mechanisms of Chlamydomonas reinhardtii against Nitrosative Stress: A Role of NADPH Oxidase (RBOL2) in the Regulation of Nitric Oxide-Mediated ER Stress and Glutathione Redox State †
Abstract
:1. Introduction
2. Experiments
2.1. Algal Culture and Treatments
2.2. Detection of NO Production
2.3. Enzyme Assay
2.4. Determination of GSH and GSSG
2.5. RNA Isolation, cDNA Synthesis and mRNA Quantification via Real-Time Quantitative PCR
2.6. Statistics
3. Results
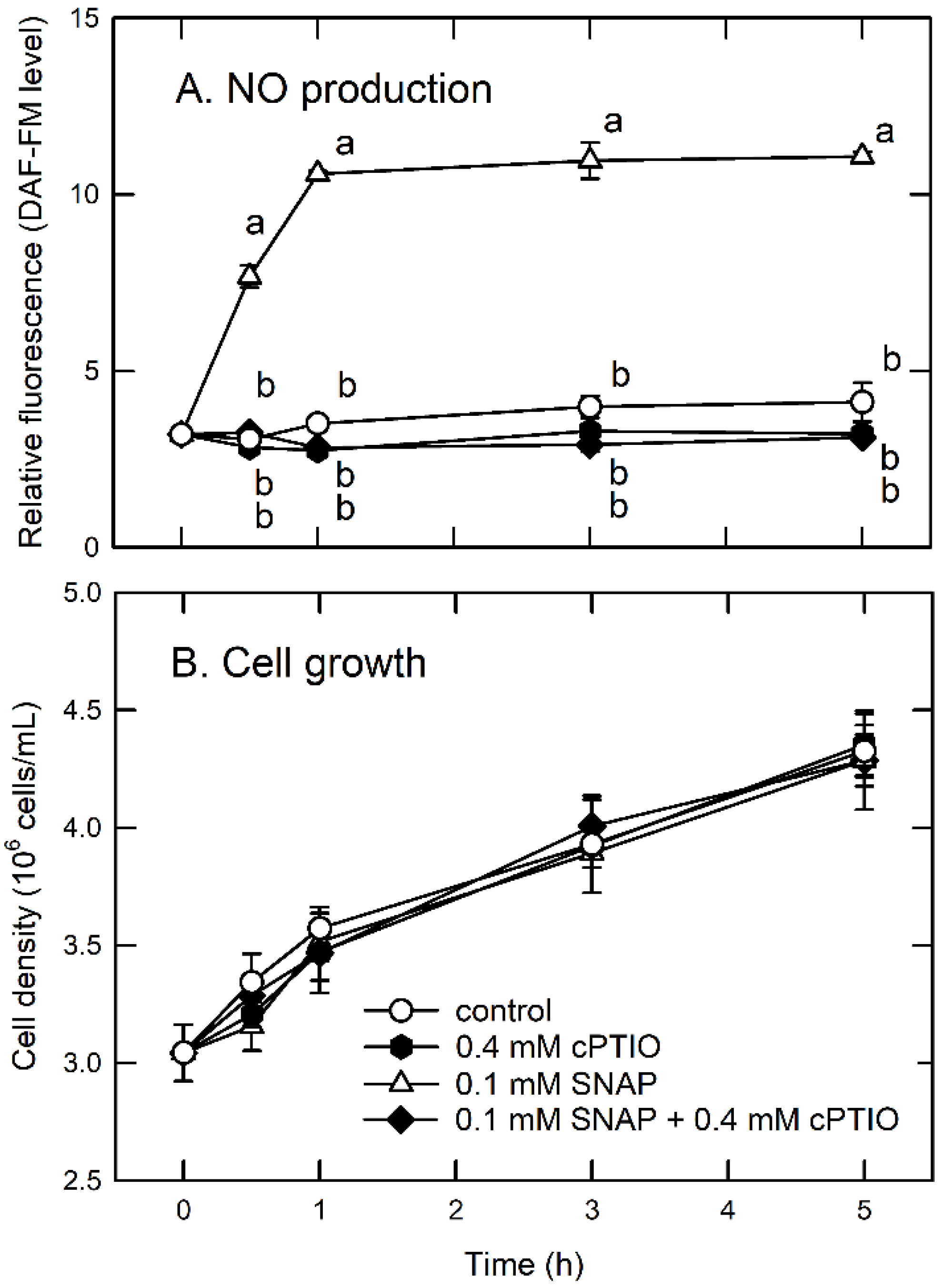
3.1. Survival under Sub-Lethal NO Treatment
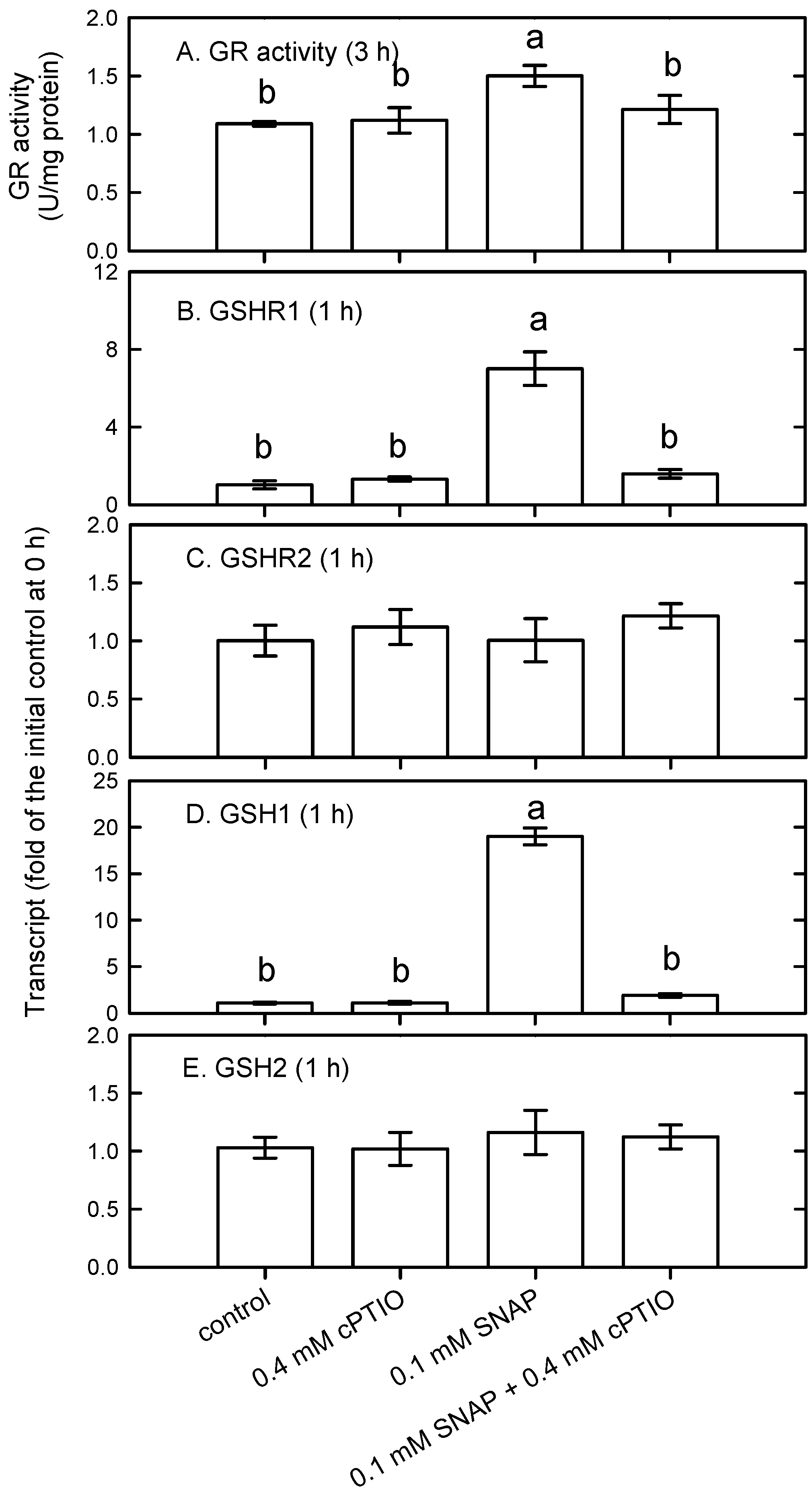
3.2. Modulation of Glutathione Availability and Redox State by NO
3.3. Negative Regulation of UPR by NO
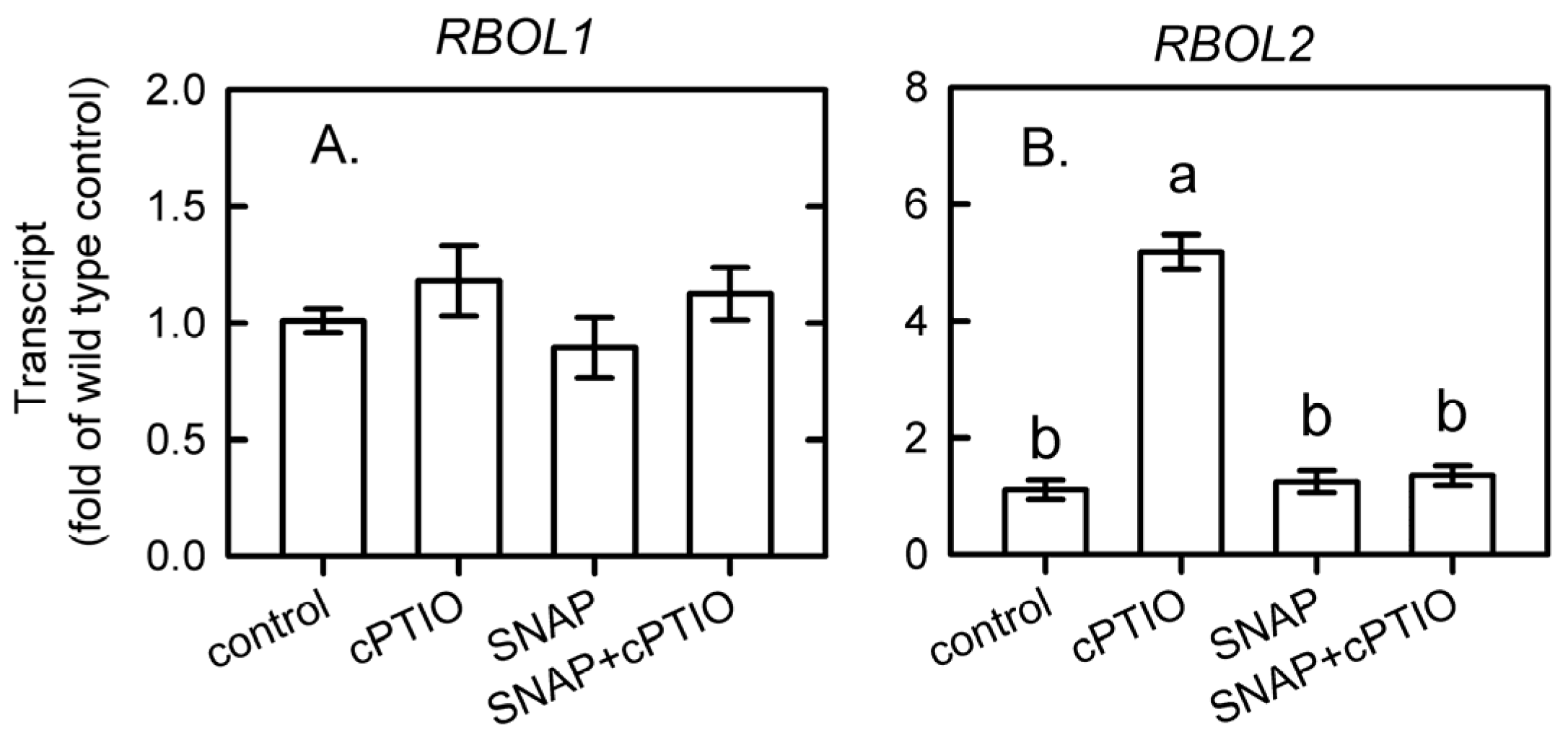
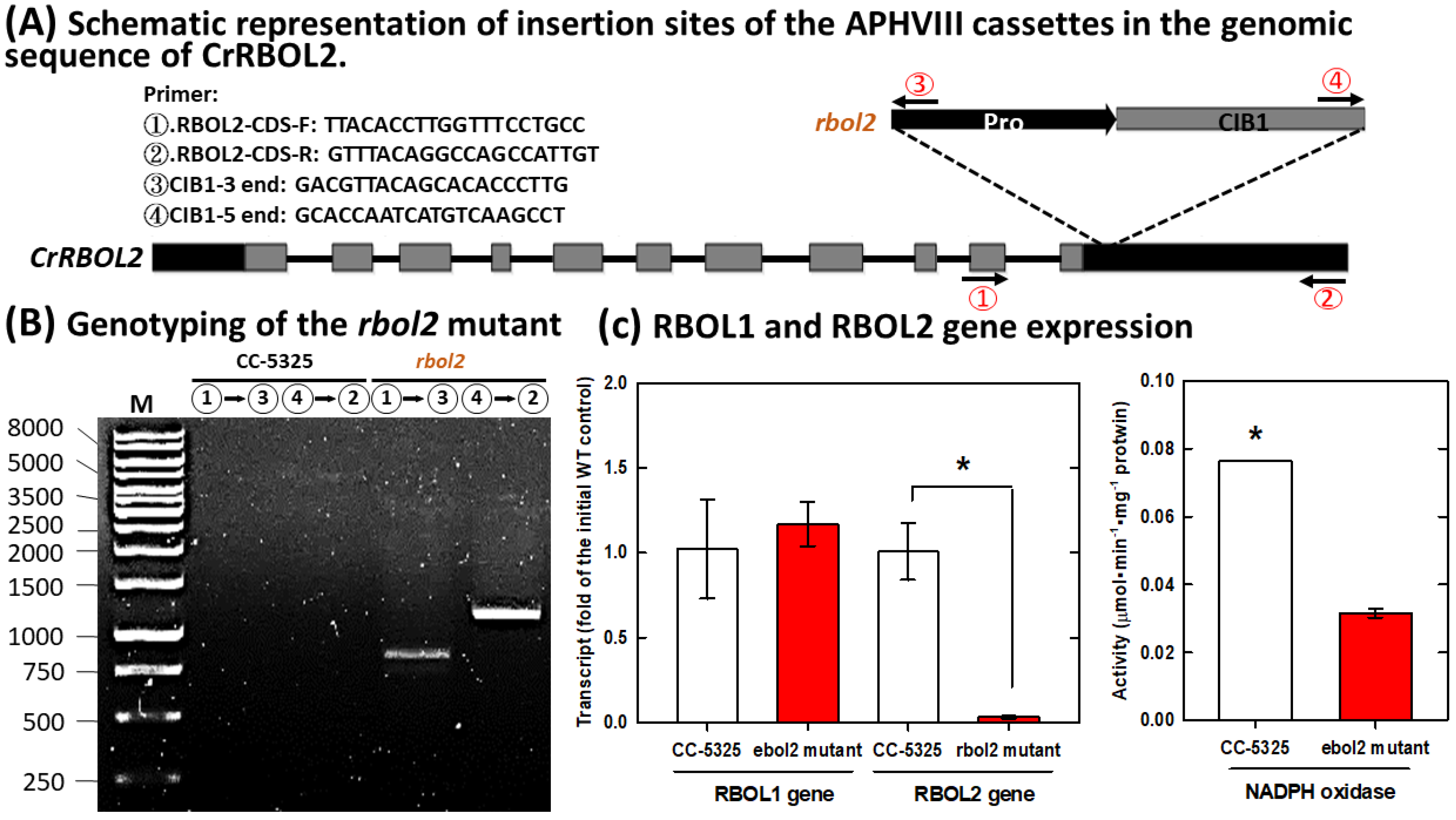
3.4. Involvement of NADPH Oxidase in the NO Modulation of Glutathione and UPR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| BiP1 | ER-resident molecular chaperone |
| CAL2 | chaperone Calreticulin 2 |
| cPTIO | 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide |
| ERO1 | disulfide-forming protein, endoplasmic reticulum oxidoreductin 1 |
| GSNO | S-nitrosoglutathione |
| IRE1 | inositol-requiring enzyme 1 |
| NO | nitric oxide |
| SNAP | S-nitroso-N-acetylpenicillamine |
| SAR1 | membrane-trafficking protein Sar-type small GTPase 1 |
| SEC61G | protein transport protein Sec61 gamma |
Appendix A
| Gene | Forward Primer Sequence (5′ → 3′) | Reverse Primer Sequence (5′ → 3′) |
|---|---|---|
| UBC | CATTAGAGGCGGGCAAA | TATCGTCATCGTGGTTGTGTAT |
| GSH1 | ACCACCTACCACCATCTTC | GTATATGAGCCCCACCCACA |
| GSH2 | CATATATTGCAGTACATACGTGG | GCACCTACTCACCTACTGTCA |
| GSHR1 | >GCCATCAAGGTGGATGAGTT | ATAGTCGGGCTTGGTCAGC |
| GSHR2 | TCCTTTCGAGGGAGAGC | GTCATTCTATACACGCCTTCCTA |
| IRE1 | CTTGGCAGTTTGCCAGTACA | AGTCCAGGTAGGACCCGAGT |
| BiP1 | AGTGAGCCCGTCTTTTAGAACTT | TCTCCTCTGTACCACCGTTTTTA |
| CAL2 | ACCCTGACTACGTCCACGAC | GTCCTCAGCGAACTTCTTGG |
| SAR1 | CGAGGAGATTCAATTGGGCG | CGGTGGGAATGTCGATCTTG |
| SEC61G | GTCAAGCCTGTGAAGGATTTTGG | GAATGAAGAGGAGCTTCACAAAG |
| PDI6 | GGTGTGGCTGGTTGAGTTCT | CTCTTTGGCGTCCTCACAGT |
| ERO1 | TGTCAACCTGCTCATCAACC | CTGCTGCTGCTACTGCTGTC |
| RB60 | CCAAGCGCTTTAAGAAGGTG | GTAGGGAAGCCCTTGACCTC |
| RBOL1 | CGATCAAGGGAGCCGAA | GCTATTTGCAGGTGCACTCA |
| RBOL2 | GCTTTCACATTGCCACGA | TACCTCCAGATGTACCTCCCTA |
References
- Durner, J.; Gow, A.J.; Stamler, J.S.; Glazebrook, J. Ancient origins of nitric oxide signaling in biological systems. Proc. Natl. Acad. Sci. USA 1999, 96, 14206–14207. [Google Scholar] [CrossRef] [Green Version]
- Uchida, A.; Jagendorf, A.T.; Hibino, T.; Takabe, T.; Takabe, T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002, 163, 515–523. [Google Scholar] [CrossRef]
- Qiao, W.; Fan, L.-M. Nitric oxide signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2008, 50, 1238–1246. [Google Scholar] [CrossRef]
- Yang, L.; Tian, D.; Todd, C.D.; Luo, Y.; Hu, X. Comparative proteome analyses reveal that nitric oxide is an important signal molecule in the response of rice to aluminum toxicity. J. Proteome Res. 2013, 12, 1316–1330. [Google Scholar] [CrossRef]
- Clark, D.; Durner, J.; Navarre, D.A.; Klessig, D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 2002, 13, 1380–1384. [Google Scholar] [CrossRef] [Green Version]
- Murgia, I.; De Pinto, M.C.; Delledonne, M.; Soave, C.; De Gara, L. Comparative effects of various nitric oxide donors on ferritin regulation, programmed cell death, and cell redox state in plant cells. J. Plant Physiol. 2004, 161, 777–783. [Google Scholar] [CrossRef]
- Arasimowicz-Jelonek, M.; FloryszakWieczorek, J.; Kosmala, A. Are nitric oxide donors a valuable tool to study the functional role of nitric oxide in plant metabolism. Plant Biol. 2011, 13, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Martí, M.C.; Florez-Sarasa, I.; Camejo, D.; Pallol, B.; Ortiz, A.; Ribas-Carbó, M.; Jiménez, A.; Sevilla, F. Response of mitochondrial antioxidant system and respiratory pathways to reactive nitrogen species in pea leaves. Physiol. Plant. 2013, 147, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J. Chlamydomonas Reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 1995, 29, 209–230. [Google Scholar] [CrossRef] [PubMed]
- de Montaigu, A.; Sanz-Luque, E.; Galván, A.; Fernández, E. A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 2010, 22, 1532–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Luque, E.; Ocaña-Calahorro, F.; Llamas, A.; Galvan, A.; Fernandez, E. Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J. Exp. Bot. 2013, 64, 3373–3383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calatrava, V.; Chamizo-Ampudia, A.; Sanz-Luque, E.; Ocaña-Calahorro, F.; Llamas, A.; Fernandez, E.; Galvan, A. How Chlamydomonas handles nitrate and the nitric oxide cycle. J. Exp. Bot. 2017, 68, 2593–2602. [Google Scholar] [CrossRef] [Green Version]
- Yordanova, Z.P.; Iakimova, E.T.; Cristescu, S.M.; Harren, F.J.; Kapchina-Toteva, V.M.; Woltering, E.J. Involvement of ethylene and nitric oxide in cell death in mastoparan-treated unicellular alga Chlamydomonas reinhardtii. Cell Biol. Int. 2010, 34, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.L.; Hsu, Y.T.; Kang, C.Y.; Lee, T.M. Nitric oxide down-regulation of carotenoid synthesis and PSII activity in relation to very high light-induced singlet oxygen production and oxidative stress in Chlamydomonas reinhardtii. Plant Cell Physiol. 2013, 54, 1296–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, E.Y.H.; Chang, H.L.; Lin, S.T.; Lee, T.M. High light-induced nitric oxide production induces autophagy and cell death in Chlamydomonas reinhardtii. Front. Plant Sci. 2020, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.P.; Mehta, S.K.; Liu, Z.P.; Yang, Z.M. Copper-induced proline synthesis is associated with nitric oxide generation in Chlamydomonas reinhardtii. Plant Cell Physiol. 2008, 49, 411–419. [Google Scholar] [CrossRef]
- Zalutskaya, Z.; Ostroukhova, M.; Filina, V.; Ermilova, E. Nitric oxide upregulates expression of alternative oxidase 1 in Chlamydomonas reinhardtii. J. Plant Physiol. 2017, 219, 123–127. [Google Scholar] [CrossRef]
- Vitale, A.; Ceriotti, A.; Denecke, J. The role of the endoplasmic reticulum in protein synthesis, modification and intracellular transport. J. Exp. Bot. 1993, 44, 1417–1444. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, Y.; Choi, B.Y.; Kim, H.; Shin, S.; Kim, Y.; Jang, S.; Song, W.Y.; Cho, C.H.; Yoon, H.S.; Kohno, K.; et al. Identification and functional study of the endoplasmic reticulum stress sensor IRE1 in Chlamydomonas reinhardtii. Plant J. 2018, 94, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, Y.; Shin, S.; Choi, B.Y.; Kim, H.; Jang, S.; Kajikawa, M.; Yamano, T.; Kong, F.; Légeret, B.; Fukuzawa, H.; et al. The bZIP1 transcription factor regulates lipid remodeling and contributes to ER stress management in Chlamydomonas reinhardtii. Plant Cell 2019, 31, 1127–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyadomari, S.; Takeda, K.; Takiguchi, M.; Gotoh, T.; Matsumoto, M.; Wada, I.; Mori, M. Nitric oxide-induced apoptosis in pancreatic β cells is mediated by the endoplasmic reticulum stress pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 10845–10850. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T. Accumulation of misfolded protein through nitrosative stress linked to neurodegenerative disorders. Antioxid. Redox Signal. 2007, 9, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Jiang, T.; Lian, C.; Wang, H.; Zheng, Q.; Ma, H. QKI deficiency promotes FoxO1 mediated nitrosative stress and endoplasmic reticulum stress contributing to increased vulnerability to ischemic injury in diabetic heart. J. Mol. Cell. Cardiol. 2014, 75, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Ozgur, R.; Uzilday, B.; Iwata, Y.; Koizumi, N.; Turkan, I. Interplay between the unfolded protein response and reactive oxygen species: A dynamic duo. J. Exp. Bot. 2018, 69, 3333–3345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelos, E.; Brandizzi, F. NADPH Oxidase activity is required for ER stress survival in plants. Plant J. 2018, 96, 1106–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, E.H. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use; Academic Press: San Diego, CA, USA, 1989. [Google Scholar]
- Kojima, H.; Nakatsubo, N.; Kikuchi, K.; Kawahara, S.; Kirino, Y.; Nagoshi, H.; Hirata, Y.; Nagano, T. Detection and imaging of nitric oxide with novel fluorescent indicators: Diaminofluoresceins. Anal. Chem. 1998, 70, 2446–2453. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.; Hebelstrup, K.H.; Gupta, K.J. Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 2013, 5, pls052. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Lee, T.M. Nitric oxide up-regulates the expression of methionine sulfoxide reductase genes in the intertidal macroalga Ulva fasciata for high light acclimation. Plant Cell Physiol. 2012, 53, 445–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamattina, L.; Polacco, J.C. Nitric Oxide in Plant Growth, Development and Stress Physiology; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 2008, 59, 21–39. [Google Scholar] [CrossRef]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; Allah, E.F.A.; Hu, X.; Jia, A. The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef]
- de Mia, M.; Lemaire, S.D.; Choquet, Y.; Wollmana, F. Nitric oxide remodels the photosynthetic apparatus upon S- starvation in Chlamydomonas reinhardtii. Plant Physiol. 2019, 179, 718–731. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Derrien, B.; Gautier, A.; Houille-Vernes, L.; Boulouis, A.; Saint-Marcoux, D.; Malnoë, A.; Rappaport, F.; de Vitry, C.; Vallon, O.; et al. Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 353–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.H.; Rao, M.Y.; Lu, H.W.; Chiou, C.W.; Lin, S.T.; Chao, H.W.; Zheng, Z.L.; Chen, H.C.; Lee, T.M. A role for glutathione reductase and glutathione in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Physiol. Plant 2018, 16, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Martín, M.; Pérez-Pérez, M.E.; Lemaire, S.D.; Crespo, J.L. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 2014, 166, 997–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Meyer, H.H.; Wang, Y.; Warren, G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and UFD1-Np14. EMBO J. 2002, 21, 5645. [Google Scholar] [CrossRef]
- Taxis, C.; Hitt, R.; Park, S.H.; Deak, P.M.; Kostova, Z.; Wolf, D.H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 2003, 278, 35903–35913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.H.; Wang, P.Q.; Zhang, P.P.; Nie, X.M.; Li, B.B.; Tai, L.; Liu, W.T.; Li, W.Q.; Chen, K.M. NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 2002, 9, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, E.Y.; Chien, Y.-L.; Dai, W.-C.; Huang, M.J.-H.; Lee, T.-M. The Acclimation Mechanisms of Chlamydomonas reinhardtii against Nitrosative Stress: A Role of NADPH Oxidase (RBOL2) in the Regulation of Nitric Oxide-Mediated ER Stress and Glutathione Redox State. Biol. Life Sci. Forum 2021, 4, 33. https://doi.org/10.3390/IECPS2020-08609
Kuo EY, Chien Y-L, Dai W-C, Huang MJ-H, Lee T-M. The Acclimation Mechanisms of Chlamydomonas reinhardtii against Nitrosative Stress: A Role of NADPH Oxidase (RBOL2) in the Regulation of Nitric Oxide-Mediated ER Stress and Glutathione Redox State. Biology and Life Sciences Forum. 2021; 4(1):33. https://doi.org/10.3390/IECPS2020-08609
Chicago/Turabian StyleKuo, Eva YuHua, Yi-Lin Chien, Wen-Chyi Dai, Michael Jian-Hao Huang, and Tse-Min Lee. 2021. "The Acclimation Mechanisms of Chlamydomonas reinhardtii against Nitrosative Stress: A Role of NADPH Oxidase (RBOL2) in the Regulation of Nitric Oxide-Mediated ER Stress and Glutathione Redox State" Biology and Life Sciences Forum 4, no. 1: 33. https://doi.org/10.3390/IECPS2020-08609
APA StyleKuo, E. Y., Chien, Y.-L., Dai, W.-C., Huang, M. J.-H., & Lee, T.-M. (2021). The Acclimation Mechanisms of Chlamydomonas reinhardtii against Nitrosative Stress: A Role of NADPH Oxidase (RBOL2) in the Regulation of Nitric Oxide-Mediated ER Stress and Glutathione Redox State. Biology and Life Sciences Forum, 4(1), 33. https://doi.org/10.3390/IECPS2020-08609






