Abstract
Barley is one of the economically important crop species and the net form of net blotch (NFNB) caused by Pyrenophora teres f. teres has a significant impact on the quantity and quality of grain yield. Selection and inbreeding have resulted in a lack of genetic diversity in elite barley accessions. Old varieties often possess unique genetic traits that may have been lost in modern crop breeding. Therefore, the aim of the current study was to identify sources of resistance to barley NFNB in the collection of European old varieties. In this study, 431 European barley accessions were evaluated phenotypically under field conditions scoring APR to NFNB and genotypically using DArTseq. The range of adult plant resistance (APR) variability at the HA growth stage was sufficient to determine marker–trait associations (MTAs). The net form of net blotch at the HA stage was scored with a range of 1.0–4.0 according to a scale of 1–9, and GWAS identified 10 marker–trait associations (MTAs) for NFNB resistance. In the HA stage, two MTAs were identified on each chromosome 1H, 3H, 5H and 6H. Moreover, one was identified on the chromosome 7H and un. One of these MTA is localized on chromosome 6H, corresponding with findings from other studies, and could contribute to the exploration of genetic resistance of barley to NFNB. Additionally, the results of this study will be utilized to establish a Polish Gene Bank platform for precise breeding programs.
1. Introduction
Barley is important in agriculture for several reasons, and its significance extends to both human and animal consumption as well as its role in sustainable farming systems. It is used in various food products, and it is an essential component of animal feed. Moreover, barley is a primary ingredient in the production of malt, a crucial component in the brewing industry. Due to climate change, it is important that barley is frequently used as a cover crop or as part of crop rotation systems. Because it has relatively low water and fertilizer requirements compared to some other cereal crops, making it a more sustainable option in regions with limited resources or concerns about environmental impact [1]. It is ranked fourth in terms of the most cultivated crop (by area) in the world, following wheat, maize, and rice. Almost half of the world’s barley-growing area is in Europe, including Poland, where it is the second most cultivated crop after wheat.
The net form of net blotch is a common foliar disease that affects barley, a cereal grain crop. It is caused by fungal pathogens belonging to the Pyrenophora genus, specifically Pyrenophora teres. Pyrenophora teres is further classified into two forms: P. teres f. teres (Drechsler) (Ptt) and P. teres f. maculata (Smedegaard-Petersen) (Ptm). Both forms of net blotch can weaken the barley plant, reduce photosynthesis, and ultimately lead to decreased grain yield and quality if left untreated. Ptt causes net-form net blotch (NFNB), whereas Ptm causes spot-form net blotch (SFNB). In the case of NFNB, net-like patterns typically appear as elongated, rectangular lesions with a net-like appearance on the leaves of barley plants. These lesions can be brown to grayish-green in color and can coalesce as the disease progresses. It primarily affects the lower leaves of the plant. The symptoms of SFNB appear as small, round to irregularly shaped lesions on the leaves of barley. Unlike the net-like lesions of the more common form, this type of NFNB creates discrete, darker spots with a defined margin [2,3].
The potential yield loss due to diseases is influenced by multiple factors. Maximized yield losses are often observed when these diseases manifest themselves before the heading stage of plant development. This critical period serves as a crucial juncture for the timely identification of early activated pathogen-associated molecular patterns. These patterns hold the potential to elicit non-specific defense cascades within the plant, thereby highlighting the significance of early detection during this developmental phase.
Conversely, resistance mechanisms that come into play during late growth stages, such as the milky-waxy stage, become essential for identifying key resistance proteins. To summarize, effective disease management during the growth period relies heavily on timely detection and precise identification of pathogens. Proactive measures at this stage significantly contribute to successful disease control strategies. Moreover, these data can guide the selection of future varieties and the integration of these genes into breeding programs [4].
Modern barley breeding programs often prioritize the development of varieties with improved disease resistance, including resistance to NFNB. The Polish Gene Bank, in conjunction with other gene banks, plays a two-fold role: preserving plant genetic resources and acting as a reservoir of new genetic variations. As a result, they serve as invaluable reservoirs of genetic material crucial for essential traits in breeding programs [5]. Without meticulously curated genetic collections, the potential of the genetic material stored in gene banks remains untapped [6]. Therefore, the long-term strategies of conventional gene banks should shift towards becoming comprehensive biological resource centers, offering access to the wealth of metadata associated with their holdings [7,8,9]. Old varieties often possess unique genetic traits and the process of incorporating new alleles into elite cultivars is more straightforward and efficient when sourcing from old cultivars and landraces, as opposed to wild relatives [10]. Given these factors, it is imperative to extensively explore old barley cultivars and landraces gathered from European countries in the quest for novel genes [11,12,13,14].
Recent progress in next-generation sequencing has empowered plant scientists to produce numerous single nucleotide polymorphism (SNP) markers and create precise genetic maps. The availability of cost-effective sequencing platforms has allowed researchers to conduct genome-wide association studies, enhancing their ability to map quantitative trait loci (QTL) related to agronomic traits and disease resistance.
Multiple studies mapping sources of NFNB resistance have been conducted in barley using linkage mapping [15,16,17,18,19,20] and association mapping approaches [20,21,22,23,24,25]. Numerous quantitative trait loci (QTL) have been documented to contribute to the resistance against the net form of net blotch, demonstrating minor effects during both the seedling and adult plant stages. These QTLs have been precisely identified and positioned on chromosomes 1H, 3H, 4H, 5H, and 7H. Additionally, various previous studies have specifically documented QTL on chromosome 6H. Despite its importance, the broad expanse of the identified genomic region makes it less favorable for direct integration into breeding programs targeting NFNB.
The objective of this study was to establish associations between genetic loci and adult plant resistance (APR) against the net form of net blotch (NFNB) at both the heading and seed’s milky-waxy plant development stages. To accomplish this, we conducted a GWAS analysis using DArTseq-derived markers and phenotypic data for 431 barley accessions segregating for these specific disease-resistance traits.
2. Materials and Methods
The plant material used in the preset study was characterized in terms of adult powdery mildew and rusts resistance and agronomic traits under field conditions in the period of 2018–2019 as was described by authors in the previous study [8,9].
2.1. Plant Material
In summary, a collection of 431 barley accessions, including landraces and old cultivars, stored at the Polish Gene Bank (National Centre for Plant Genetic Resources: NCPGR) were phenotyped and evaluated using DArTseq: 137 POL, 67 DEU, 38 SWE, 35 CSK, 34 FRA, 27 GBR, 25 DNK, 21 NLD, 12 AUT, 8 SUN, 6 NOR, 4 FIN, 3 IRL, 3 CAN, 2 USA, 2 HUN and 1 each from UKR, TUR, PRK, NZL, JPN, BEL and one of unknown origin [9].
2.2. Field Experiment and Phenotypic Evaluation
In 2019, field experiments were meticulously carried out at the Plant Breeding and Acclimatization Institute-National Research Institute (PBAI-NRI) in Radzikow, situated near Warsaw, Poland. Notably, no specific permissions were deemed necessary. The experimental setup comprised two replications, each consisting of two rows with a row length of 2.0 m. The planting configuration included a plant spacing of 4.0 cm and a row spacing of 20.0 cm [8,9].
Net form of net blotch (NFNB) was scored according to IPGRI descriptors (https://cropgenebank.sgrp.cgiar.org/index.php/learning-space-mainmenu-454/manuals-and-handbooks-mainmenu-533/descriptors-mainmenu-547 (accessed on 29 October 2021), using a 1–9 scale, where 1 means no symptoms of the disease (immune reaction). Measurements were undertaken during the heading stage (HA), precisely when half of the heads had emerged for 50% of the plants within a plot (identified as Z55 on the Zadoks growth scale). Subsequently, the assessment was repeated two weeks later, precisely during the early milky-waxy seed maturity stage (MW; designated as Z75 on the Zadoks growth scale).
2.3. Statistical Analysis
Due to the limited number of seeds accessible for each accession in the NCPGR, the experiment could only be conducted in a single environment and during one specific year. To ensure comprehensive and reliable results, it is advisable to replicate the experiment and perform phenotypic assessments across multiple environments. Basic analysis of phenotypic data was performed using the Statistica software and Excel 2019. This was used to obtain the range, mean, standard deviation (SD), coefficient of variation (CV) and frequency distribution of barley accessions for NFNB resistance.
2.4. Genotyping and Data Filtering Process
A total of 454 accessions were genotyped using Diversity Arrays Technology (DArT) were (431 evaluated under field conditions and 25 additional, used as a controls). SNP calls were made against IBSC Barley Morex v2 assembly [26]. The Barley GBS 1.0 platform DArT genotyping service returned 28,530 in silico DArT-seq markers. DArT were was handled in the same manner as described previously [27,28]. That is, we used the dartR v1.1.11 package [29] in the R programming language. SNPs and genotypes were removed if SNP markers contained >5% missing data and genotypes contained >10% missing data, respectively. SNPs with a reproducibility score (RepAvg) <100% were removed. Non-informative monomorphic SNPs were removed, as were rare SNPs with a minor allele frequency of <1%. After filtering, 453 (1 individual was removed due to having >10% missing calls) and 10,153 SNPs were retained for further analysis.
2.5. Genome-Wide Association Studies (GWAS)
The GWAS analysis followed the methodology described by the authors [8,9,14,27,28]. We utilized the GAPIT v2018.08.18 R package for the analysis. Our study employed the recently developed Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway (BLINK) model, known for minimizing false positives, enhancing true positive identification, and its capacity to handle large datasets [28]. Markers’ physical genome positions were obtained from the DArTseq SNP genotype file. Considering that GAPIT requires complete data, only markers with a physical position on one of the chromosomes and zero missing data were included in the GWAS analysis. GWAS for NFNB was performed to assess disease resistance scoring at the heading and milky-waxy seed stages. Additionally, Manhattan plots were generated to visualize the distribution of SNPs across the chromosomes.
3. Results and Discussion
A collection of 431 accessions was evaluated under field conditions for net form of net blotch (NFNB) resistance at the heading (HA) and milky-waxy (MW) stages. Range of adult plant resistance (APR) variability at plants HA and MW growth stages were sufficient to determine marker–trait associations (MTAs). NFNB at the HA stage was scored with a range of 1.0–4.0 according to a scale of 1–9 with standard deviation (SD) 0.54 and coefficient variation 0.29%. Phenotypic variations in disease severity at the HA stage are presented in Figure 1.
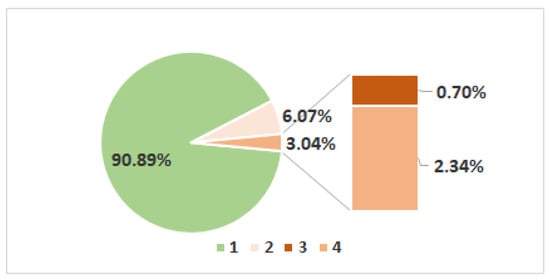
Figure 1.
The frequency distribution histogram presents the percentage of old varieties scored for the net form of net blotch (NFNB) at heading stage (HA) using 1–9 scale (1 = no symptoms of the disease).
Based on the number of accessions in the Polish Gene bank database, accessions with resistance for NFNB scored at 4.0 originated from DEU/DNK (43672, 19I00614, 43727, 43727), POL (19I00609, 42388, 19I00601, 43319), FRA (43760), GBR (43689), IRL (40388), AUT (43778), SUN (43778), TUR (43779) (https://wyszukiwarka.ihar.edu.pl/en (accessed on 8 September 2023)). At the MW stage, the severity of the disease was lower than at the HA stage, and only a few genotypes had symptoms of the disease scored at 3.
At the HA stage, GWAS identified 10 marker–trait associations (MTAs) for NFNB resistance (Table 1). Specifically, two MTAs were identified on each chromosome 1H, 3H, 5H, 6H (Table 1 and Figure 2). Moreover, one was identified on the chromosome 7H and un.

Table 1.
Significant marker–trait associations (MTAs) associated with the net form of net blotch (NFNB) at heading (HA) stage.

Figure 2.
Single nucleotide polymorphism (SNP) significantly associated with net form of net blotch (NFNB) resistance in barley identified by genome-wide association study (GWAS) with BLINK model. Manhattan plot and quantile–quantile plot for heading stage (A1,A2); Manhattan plot and quantile–quantile plot for milky-waxy stage (B1,B2).
It is important that the two MTAs identified at the HA stage, 3432352-13-G/T and 4414028-24-G/C, are adjacent on chromosome 6HAt the MW stage, one, two, one, and three significant trait-associated markers (MTAs) were identified on chromosomes 1H, 2H, 5H, and 7H, respectively (Table 2, Figure 2). Since the severity of the disease was low at the MW stage, specific MTAs can only provide limited support for the interpretation of barley genetic resistance determination for NFNB.

Table 2.
Significant marker–trait associations (MTAs) associated with the net form of net blotch (NFNB) at the milky-waxy (MW) stage.
These results correspond to those of the previous study, and confirm that chromosome 6H has been pinpointed as a hotspot for Ptt resistance loci in numerous QTL mapping studies [2,7,15,16,17,19,30,31]. Most of these investigations have highlighted an extensive section of chromosome 6H associated with resistance to NFNB. Unlike this particular study, many of these research efforts have employed smaller mapping populations [30]. It is worth noting that smaller mapping populations lack the statistical robustness needed to precisely define QTL boundaries with high confidence. Therefore, the regions ((,2.00 cM, 22 Mb)) on chromosome 6H and ((,1.00 cM, 0.11 Mb)) on chromosome 3H are considered high-confidence regions of interest for further investigations, including validation, fine mapping, and eventual cloning [23,25]. A BLAST search unveiled multiple predicted genes in these regions, some of which have well-established annotations related to disease resistance functions. Notably, the region on chromosome 3H contains two predicted genes from the NBS-LRR gene family.
Based on the conducted study, it is worth mentioning that the region on 3H seems to be particularly relevant to the six-row germplasm, as it was identified in the six-row panel, while the region on 6H appears to be more specialized for the two-row germplasm. Importantly, these markers can be employed in marker-assisted selection processes without the risk of significant linkage drag, as they target relatively small regions. The initial step in this direction would involve validating these markers in other populations. Plant material used in recent study was used to determine resistance to powdery mildew, barley brown rust and stem rust at the same stages as HA and MW.
4. Conclusions
Gene banks play a dual role, actively facilitating the preservation of plant genetic resources while simultaneously serving as invaluable repositories for accessing new genetic alleles. This study enhances our understanding of the genomic regions linked to barley’s APR (Adult Plant Resistance) against NFNB (net form of net blotch). It reaffirms the efficiency of GWAS (Genome-Wide Association Studies) with DArT (Diversity Arrays Technology) data in identifying markers associated with these traits. This opens up the possibility of establishing a gene bank platform that includes comprehensive trait descriptions, making it well-suited for utilization in breeding programs and research. Furthermore, the study confirms the presence of closely related markers on chromosomes 1H and 6H at heading stage and at heading stage additional markers on chromosomes 1H and 7H. Lastly, the inclusion of evaluated landraces and old cultivars brings added value, as these resources can play a pivotal role in preserving agrobiodiversity through a range of diverse strategies.
Author Contributions
Conceptualization J.H.C. and E.C. Methodology: E.C. and J.H.C. Investigation: E.C. and J.H.C. Phenotypic assessment and statistical analysis of data, visualization: E.C. Project administration was overseen by E.C. and J.H.C. was responsible for managing the project’s resources. Both authors, E.C. and J.H.C., collaborated on the initial draft, review, and editing of the manuscript. Furthermore, all authors have thoroughly reviewed and provided their consent for the publication of the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study is a part of the AGROBANK project “Creation of bioinformatic management system about national genetic resources of useful plants and development of social and economic resources of Poland throughout the protection and use of them in the process of providing agricultural consulting services” (1/394826/10/NCBR/2018) https://agrobank.cdr.gov.pl/index.php (accessed on 8 September 2023) financed by the National Center for Research and Development under the strategic research program GOSPOSTRATEG “Social And Economic Development Of Poland In The Context Of Globalizing Markets”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available in the tables. The person to contact is Elzbieta Czembor, IHAR-PIB Radzikow, 05-870 Blonie, Poland.
Acknowledgments
We thank the Polish Genebank (National Centre for Plant Genetic Resources—KCRZG at the Plant Breeding and Acclimatization Institute—National Research Institute, Radzikow, Poland) for providing seed samples; Our sincere appreciation goes to Radoslaw Suchecki from CSIRO Agriculture and Food, Urrbrae, SA 5064, Australia, for his contributions, and to Nathan S. Watson-Haigh from the South Australian Genomics Centre, SAHMRI, North Terrace, Adelaide SA 5000, Australia, for his invaluable support in the field of bioinformatics analysis.
Conflicts of Interest
The authors declare no conflict of interest. Additionally, they disclose any personal situations or interests that might potentially affect the impartial representation or interpretation of the research findings. It is also confirmed that the funders had no involvement in shaping the study’s design, data collection, analysis, or interpretation.
References
- Kumar, A.; Verma, R.P.S.; Singh, A.; Kumar Sharma, H.; Devi, G. Barley Landraces: Ecological Heritage for Edaphic Stress Adaptations and Sustainable Production. Environ. Sustain. Indic. 2020, 6, 100035. [Google Scholar] [CrossRef]
- Clare, S.J.; Wyatt, N.A.; Brueggeman, R.S.; Friesen, T.L. Research Advances in the Pyrenophora Teres–Barley Interaction. Mol. Plant Pathol. 2020, 21, 272–288. [Google Scholar] [CrossRef]
- Tini, F.; Covarelli, L.; Ricci, G.; Balducci, E.; Orfei, M.; Beccari, G. Management of Pyrenophora Teres f. Teres, the Causal Agent of Net Form Net Blotch of Barley, in A Two-Year Field Experiment in Central Italy. Pathogens 2022, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Schnurbusch, T. Heading Date Is Not Flowering Time in Spring Barley. Front. Plant Sci. 2017, 8, 896. [Google Scholar] [CrossRef] [PubMed]
- Weise, S.; Lohwasser, U.; Oppermann, M. Document or Lose It—On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- González, M.Y.; Philipp, N.; Schulthess, A.W.; Weise, S.; Zhao, Y.; Börner, A.; Oppermann, M.; Graner, A.; Reif, J.C. Unlocking Historical Phenotypic Data from an Ex Situ Collection to Enhance the Informed Utilization of Genetic Resources of Barley (Hordeum Sp.). Theor. Appl. Genet. 2018, 131, 2009–2019. [Google Scholar] [CrossRef]
- Amezrou, R.; Gyawali, S.; Belqadi, L.; Chao, S.; Arbaoui, M.; Mamidi, S.; Rehman, S.; Sreedasyam, A.; Verma, R.P.S. Molecular and Phenotypic Diversity of ICARDA Spring Barley (Hordeum vulgare L.) Collection. Genet. Resour. Crop Evol. 2018, 65, 255–269. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E.; Suchecki, R.; Watson-haigh, N.S. Genome-Wide Association Study for Powdery Mildew and Rusts Adult Plant Resistance in European Spring Barley from Polish Gene Bank. Agronomy 2022, 12, 7. [Google Scholar] [CrossRef]
- Czembor, J.H. Genome-Wide Association Study of Agronomic Traits in European Spring Barley from Polish Gene Bank. Agronomy 2022, 12, 2135. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.; Stephen Baenziger, P.; Börner, A. GWAS: Fast-Forwarding Gene Identification and Characterization in Temperate Cereals: Lessons from Barley—A Review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Singh, D.; Ziems, L.A.; Dracatos, P.M.; Pourkheirandish, M.; Tshewang, S.; Czembor, P.; German, S.; Fowler, R.A.; Snyman, L.; Platz, G.J.; et al. Genome-Wide Association Studies Provide Insights on Genetic Architecture of Resistance to Leaf Rust in a Worldwide Barley Collection. Mol. Breed. 2018, 38, 43. [Google Scholar] [CrossRef]
- Monteagudo, A.; Casas, A.M.; Cantalapiedra, C.P.; Contreras-Moreira, B.; Gracia, M.P.; Igartua, E. Harnessing Novel Diversity from Landraces to Improve an Elite Barley Variety. Front. Plant Sci. 2019, 10, 434. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.A.; Mores, A.; Ficco, D.B.M.; Laidò, G.; Mastrangelo, A.M.; Borrelli, G.M. Importance of Landraces in Cereal Breeding for Stress Tolerance. Plants 2021, 10, 1267. [Google Scholar] [CrossRef] [PubMed]
- Riaz, A.; Kanwal, F.; Börner, A.; Pillen, K.; Dai, F.; Alqudah, A.M. Advances in Genomics-Based Breeding of Barley: Molecular Tools and Genomic Databases. Agronomy 2021, 11, 894. [Google Scholar] [CrossRef]
- Friesen, T.L.; Faris, J.D.; Lai, Z.; Steffenson, B.J. Identification and Chromosomal Location of Major Genes for Resistance to Pyrenophora Teres in a Doubled-Haploid Barley Population. Genome 2006, 49, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Grewal, T.S.; Rossnagel, B.G.; Scoles, G.J. Mapping of Quantitative Trait Loci Associated with Resistance to Net Blotch [Pyreuophora Teres] in Barley. Can. J. Plant Pathol. Can. Phytopathol. 2007, 29, 219. [Google Scholar]
- Grewal, T.S.; Rossnagel, B.G.; Pozniak, C.J.; Scoles, G.J. Mapping Quantitative Trait Loci Associated with Barley Net Blotch Resistance. Theor. Appl. Genet. 2008, 116, 529–539. [Google Scholar] [CrossRef]
- Pierre, S.S.; Gustus, C.; Steffenson, B.; Dill-Macky, R.; Smith, K.P. Mapping Net Form Net Blotch and Septoria Speckled Leaf Blotch Resistance Loci in Barley. Phytopathology 2010, 100, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Lapitan, N.L.V.; Steffenson, B. QTL Mapping of Net Blotch Resistance Genes in a Doubled-Haploid Population of Six-Rowed Barley. Euphytica 2004, 137, 291–296. [Google Scholar] [CrossRef]
- Rozanova, I.V.; Lashina, N.M.; Mustafin, Z.S.; Gorobets, S.A.; Efimov, V.M.; Afanasenko, O.S.; Khlestkina, E.K. SNPs Associated with Barley Resistance to Isolates of Pyrenophora Teres f. Teres. BMC Genom. 2019, 20, 292. [Google Scholar] [CrossRef]
- Adhikari, A.; Smith, M.J.; Dill-Macky, R. Association Mapping for Net Blotch Resistance in Barley and a Study of Barley/Cereal Yellow Dwarf Virus in Minnesota. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2017. [Google Scholar]
- Amezrou, R.; Verma, R.P.S.; Chao, S.; Brueggeman, R.S.; Belqadi, L.; Arbaoui, M.; Rehman, S.; Gyawali, S. Genome-Wide Association Studies of Net Form of Net Blotch Resistance at Seedling and Adult Plant Stages in Spring Barley Collection. Mol. Breed. 2018, 38, 58. [Google Scholar] [CrossRef]
- Adhikari, A.; Steffenson, B.J.; Smith, M.J.; Dill-Macky, R. Genome-Wide Association Mapping of Seedling Net Form Net Blotch Resistance in an Ethiopian and Eritrean Barley Collection. Crop Sci. 2019, 59, 1625–1638. [Google Scholar] [CrossRef]
- Daba, S.D.; Horsley, R.; Brueggeman, R.; Chao, S.; Mohammadi, M. Genome-Wide Association Studies and Candidate Gene Identification for Leaf Scald and Net Blotch in Barley (Hordeum vulgare L.). Plant Dis. 2019, 103, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, A.; Steffenson, B.J.; Smith, K.P.; Smith, M.; Dill-Macky, R. Identification of Quantitative Trait Loci for Net Form Net Blotch Resistance in Contemporary Barley Breeding Germplasm from the USA Using Genome-Wide Association Mapping. Theor. Appl. Genet. 2020, 133, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A Chromosome Conformation Capture Ordered Sequence of the Barley Genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated-Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A Package for the next Level of Genome-Wide Association Studies with Both Individuals and Markers in the Millions. Gigascience 2018, 8, giy154. [Google Scholar] [CrossRef]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. Dartr: An r Package to Facilitate Analysis of SNP Data Generated from Reduced Representation Genome Sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef]
- Richards, J.K.; Friesen, T.L.; Brueggeman, R.S. Association Mapping Utilizing Diverse Barley Lines Reveals Net Form Net Blotch Seedling Resistance/Susceptibility Loci. Theor. Appl. Genet. 2017, 130, 915–927. [Google Scholar] [CrossRef]
- Wonneberger, R.; Ficke, A.; Lillemo, M. Identification of Quantitative Trait Loci Associated with Resistance to Net Form Net Blotch in a Collection of Nordic Barley Germplasm. Theor. Appl. Genet. 2017, 130, 2025–2043. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).