Synergizing Crop Growth Models and Digital Phenotyping: The Design of a Cost-Effective Internet of Things-Based Sensing Network †
Abstract
:1. Introduction
2. Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pieruschka, R.; Schurr, U. Plant Phenotyping: Past, Present, and Future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Martre, P.; Buis, S.; Abichou, M.; Andrieu, B.; Baret, F. Estimation of Plant and Canopy Architectural Traits Using the Digital Plant Phenotyping Platform. Plant Physiol. 2019, 181, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Zhang, J.; Guo, D.; Chang, L.; Huang, D. Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris Ssp. Chinensis L. Sensors 2019, 19, 2448. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Magalhães, S.A.; da Silva, D.Q.; dos Santos, F.N.; Cunha, M. Computer Vision and Deep Learning as Tools for Leveraging Dynamic Phenological Classification in Vegetable Crops. Agronomy 2023, 13, 463. [Google Scholar] [CrossRef]
- Dhondt, S.; Wuyts, N.; Inzé, D. Cell to whole-plant phenotyping: The best is yet to come. Trends Plant Sci. 2013, 8, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Campbell, Z.; Acosta-Gamboa, L.; Nepal, N.; Lorence, A. Engineering plants for tomorrow: How high-throughput phenotyping is contributing to the development of better crops. Phytochem. Rev. 2018, 17, 1329–1343. [Google Scholar] [CrossRef]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic Technologies for High-Throughput Plant Phenotyping: Contemporary Reviews and Future Perspectives. Front. Plant Sci. 2021, 12, 611940. [Google Scholar] [CrossRef]
- Coppens, F.; Wuyts, N.; Inzé, D.; Dhondt, S. Unlocking the potential of plant phenotyping data through integration and data-driven approaches. Curr. Opin. Syst. Biol. 2017, 4, 58–63. [Google Scholar] [CrossRef]
- Saint Cast, C.; Lobet, G.; Cabrera-Bosquet, L.; Couvreur, V.; Pradal, C.; Tardieu, F.; Draye, X. Connecting plant phenotyping and modelling communities: Lessons from science mapping and operational perspectives. Silico Plants 2022, 4, diac005. [Google Scholar] [CrossRef]
- Nabwire, S.; Suh, H.-K.; Kim, M.S.; Baek, I.; Cho, B.-K. Review: Application of Artificial Intelligence in Phenomics. Sensors 2021, 21, 4363. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, P.; Nicastro, N.; Pane, C. Digital applications and artificial intelligence in agriculture toward next-generation plant phenotyping. Crop Pasture Sci. 2022, 74, 597–614. [Google Scholar] [CrossRef]
- Wallach, D.; Makowski, D.; Jones, J.W.; Brun, F. Working with Dynamic Crop Models: Methods, Tools and Examples for Agriculture and Environment, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar] [CrossRef]
- Parent, B.; Tardieu, F. Can current crop models be used in the phenotyping era for predicting the genetic variability of yield of plants subjected to drought or high temperature? J. Exp. Bot. 2014, 65, 6179–6189. [Google Scholar] [CrossRef] [PubMed]
- Kasampalis, D.A.; Alexandridis, T.K.; Deva, C.; Challinor, A.; Moshou, D.; Zalidis, G. Contribution of Remote Sensing on Crop Models: A Review. J. Imaging 2018, 4, 52. [Google Scholar] [CrossRef]
- Leng, G.; Hall, J.W. Predicting spatial and temporal variability in crop yields: An inter-comparison of machine learning, regression and process-based models. Environ. Res. Lett. 2020, 15, 044027. [Google Scholar] [CrossRef] [PubMed]
- Corrales, D.C.; Schoving, C.; Raynal, H.; Debaeke, P.; Journet, E.-P.; Constantin, J. A surrogate model based on feature selection techniques and regression learners to improve soybean yield prediction in southern France. Comput. Electron. Agric. 2022, 192, 106578. [Google Scholar] [CrossRef]
- Kallenberg, M.G.; Maestrini, B.; van Bree, R.; Ravensbergen, P.; Pylianidis, C.; van Evert, F.K.; Athanasiadis, I.N. Integrating processed-based models and machine learning for crop yield prediction. arXiv 2023, arXiv:2307.13466. [Google Scholar] [CrossRef]
- Pylianidis, C.; Snow, V.; Overweg, H.; Osinga, S.; Kean, J.; Athanasiadis, I.N. Simulation-assisted machine learning for operational digital twins. Environ. Model. Softw. 2021, 148, 105274. [Google Scholar] [CrossRef]
- Nasirahmadi, A.; Hensel, O. Toward the Next Generation of Digitalization in Agriculture Based on Digital Twin Paradigm. Sensors 2022, 22, 498. [Google Scholar] [CrossRef]
- Terra, F.; Rodrigues, L.; Magalhães, S.; Santos, F.; Moura, P.; Cunha, M. PixelCropRobot, a cartesian multitask platform for microfarms automation. In Proceedings of the International Symposium of Asian Control Association on Intelligent Robotics and Industrial Automation, Goa, India, 20–22 September 2021. [Google Scholar] [CrossRef]
- Moreira, T.; Santos, F.; Santos, L.; Sarmento, J.; Terra, F.; Sousa, A. Mission supervisor for food factories robots. Lect. Notes Netw. Syst. 2023; accepted. [Google Scholar]
- Brisson, N.; Gary, C.; Justes, E.; Roche, R.; Mary, B.; Ripoche, D.; Zimmer, D.; Sierra, J.; Bertuzzi, P.; Burger, P. An overview of the crop model stics. Eur. J. Agron. 2003, 18, 309–332. [Google Scholar] [CrossRef]
- Droutsas, I.; Challinor, A.J.; Deva, C.R.; Wang, E. Integration of machine learning into process-based modelling to improve simulation of complex crop responses. Silico Plants 2022, 4, diac017. [Google Scholar] [CrossRef]
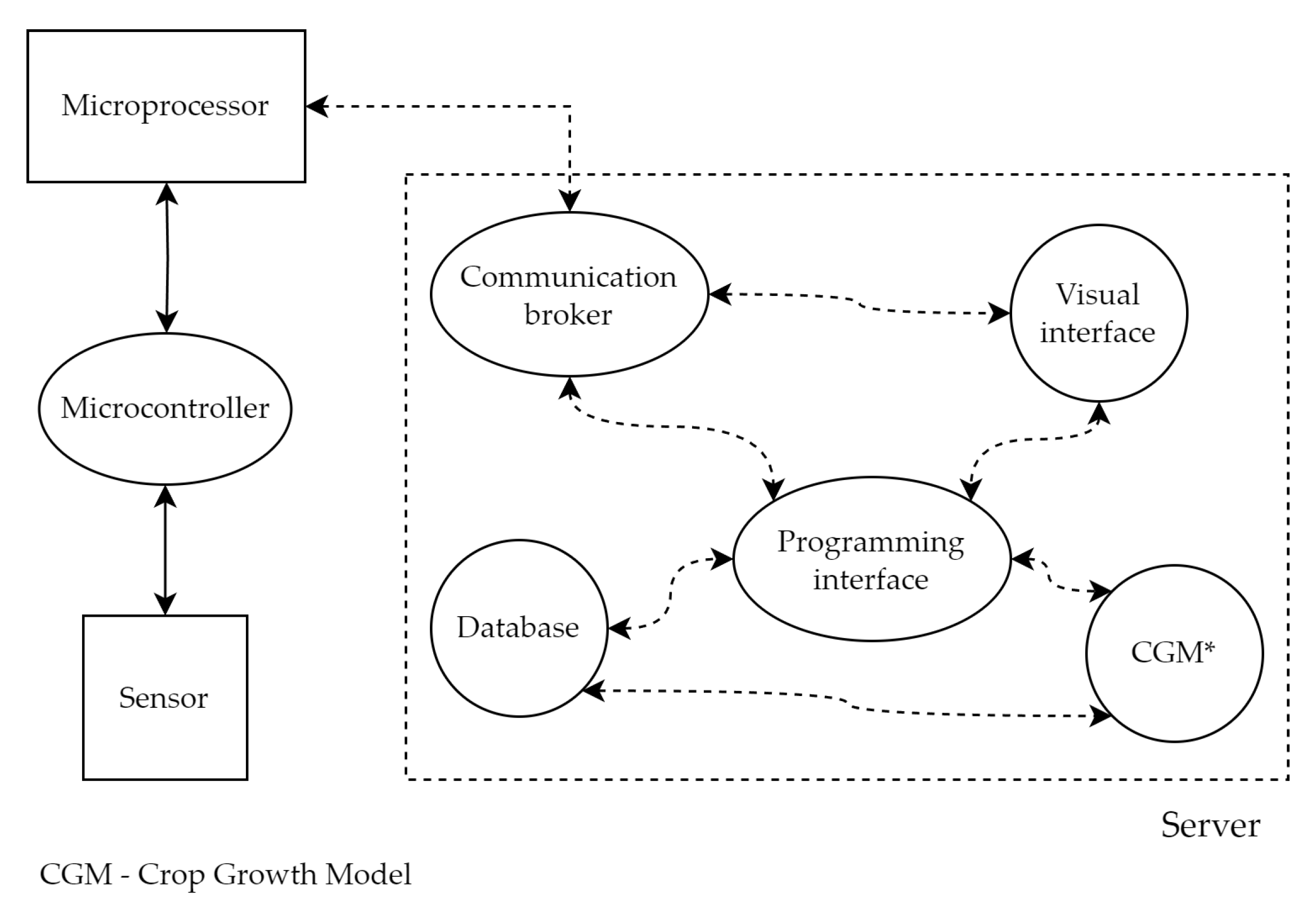


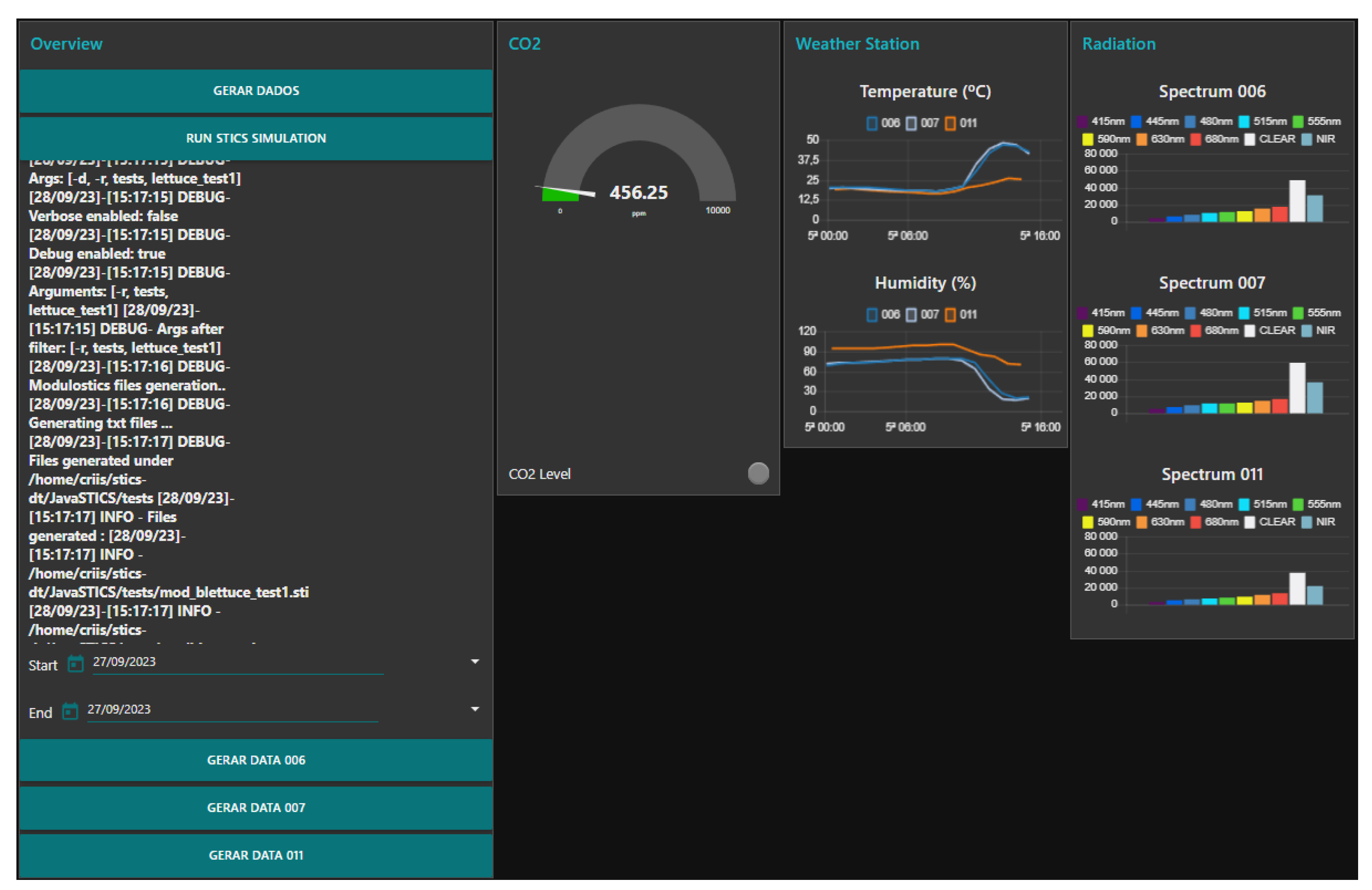
| Sensor | Quantity (n) | Daily Requests (n) | Average Size |
|---|---|---|---|
| Stationary | |||
| RPi Camera | 1 | 24 | 10 MB |
| AS7341 | 2 | 24 | 400 B |
| HTU21D | 2 | 24 | 170 B |
| SEN0159 | 1 | 24 | 120 B |
| Lysimeter | 12 | 24 | 160 B |
| SEN0308 | 12 | 24 | 170 B |
| PixelCropRobot | |||
| RPi Camera | 1 | 5 | 10 MB |
| Multispectral sensor | 1 | 5 | 400 B |
| LiDAR | 1 | 5 | 370 B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, L.; Moura, P.; Terra, F.; Carvalho, A.M.; Sarmento, J.; dos Santos, F.N.; Cunha, M. Synergizing Crop Growth Models and Digital Phenotyping: The Design of a Cost-Effective Internet of Things-Based Sensing Network. Biol. Life Sci. Forum 2023, 27, 41. https://doi.org/10.3390/IECAG2023-16276
Rodrigues L, Moura P, Terra F, Carvalho AM, Sarmento J, dos Santos FN, Cunha M. Synergizing Crop Growth Models and Digital Phenotyping: The Design of a Cost-Effective Internet of Things-Based Sensing Network. Biology and Life Sciences Forum. 2023; 27(1):41. https://doi.org/10.3390/IECAG2023-16276
Chicago/Turabian StyleRodrigues, Leandro, Pedro Moura, Francisco Terra, Alexandre Magno Carvalho, José Sarmento, Filipe Neves dos Santos, and Mário Cunha. 2023. "Synergizing Crop Growth Models and Digital Phenotyping: The Design of a Cost-Effective Internet of Things-Based Sensing Network" Biology and Life Sciences Forum 27, no. 1: 41. https://doi.org/10.3390/IECAG2023-16276
APA StyleRodrigues, L., Moura, P., Terra, F., Carvalho, A. M., Sarmento, J., dos Santos, F. N., & Cunha, M. (2023). Synergizing Crop Growth Models and Digital Phenotyping: The Design of a Cost-Effective Internet of Things-Based Sensing Network. Biology and Life Sciences Forum, 27(1), 41. https://doi.org/10.3390/IECAG2023-16276









