A Cytogenomic Analysis Reveals a New Fusarium fujikuroi Species Associated with Lemongrass (Cymbopogon citratus) †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Infected Plant Material and Isolation
2.2. Cultural and Morphological Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Cytogenomic Analysis
3. Results
3.1. Symptomatological Characteristics
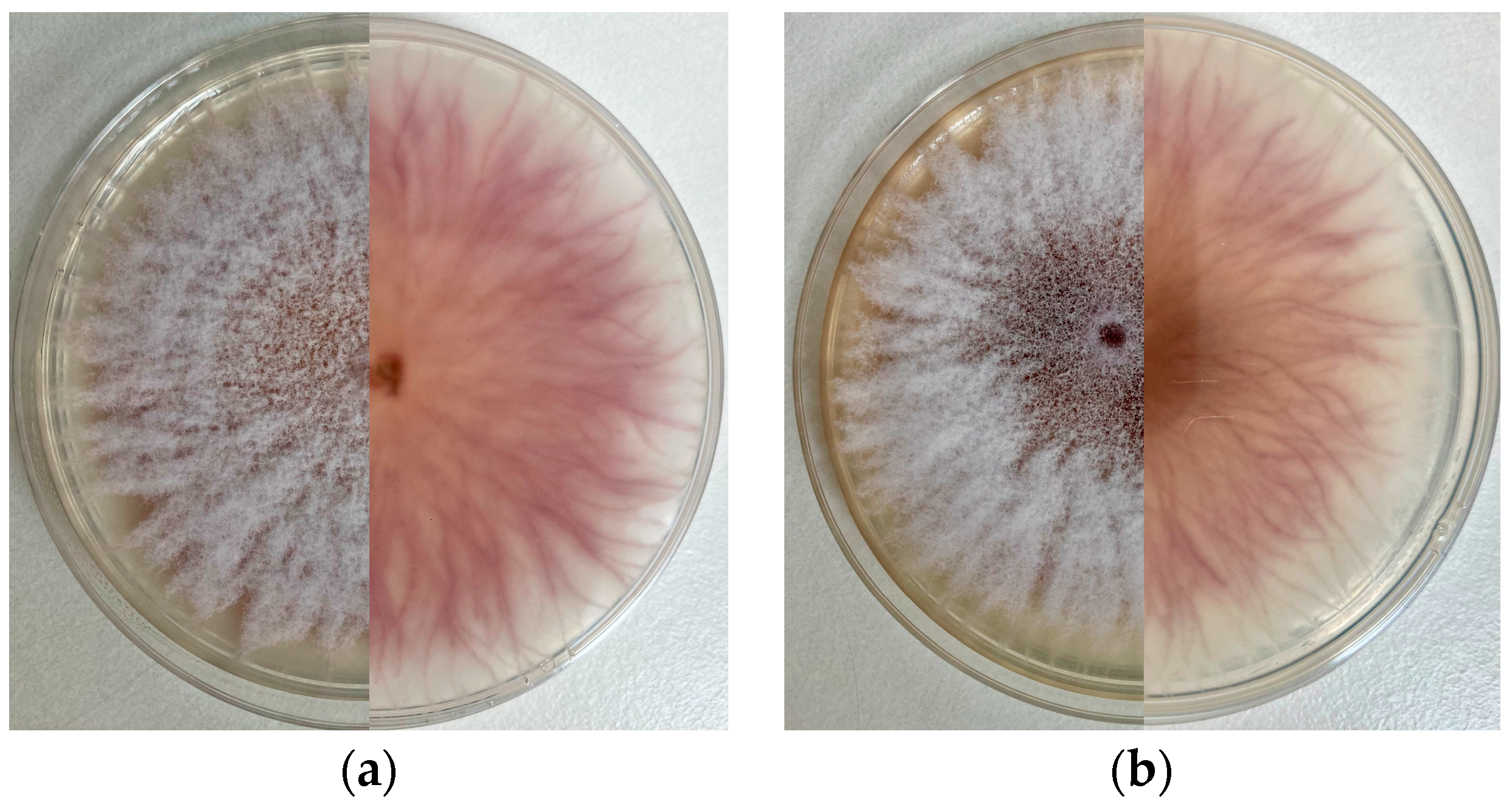
3.2. Cultural Characteristics
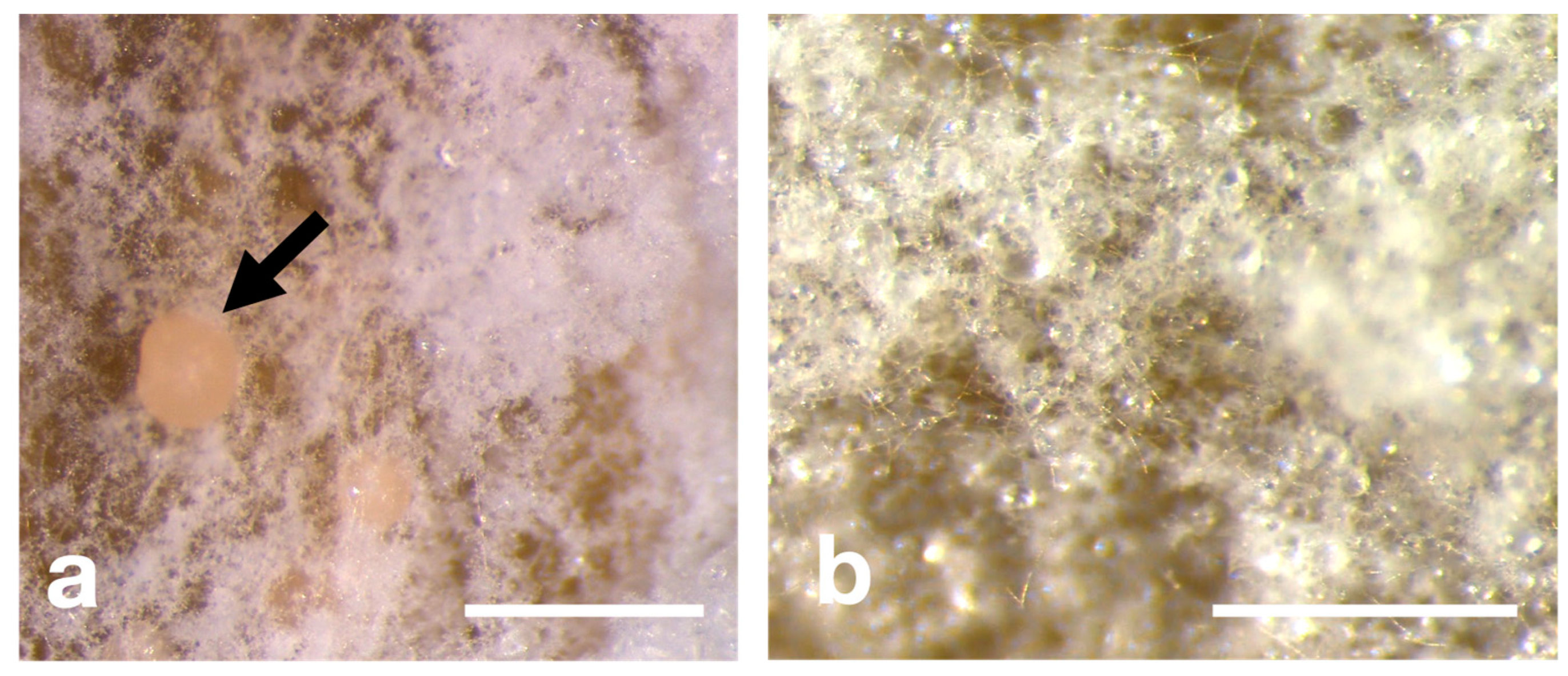
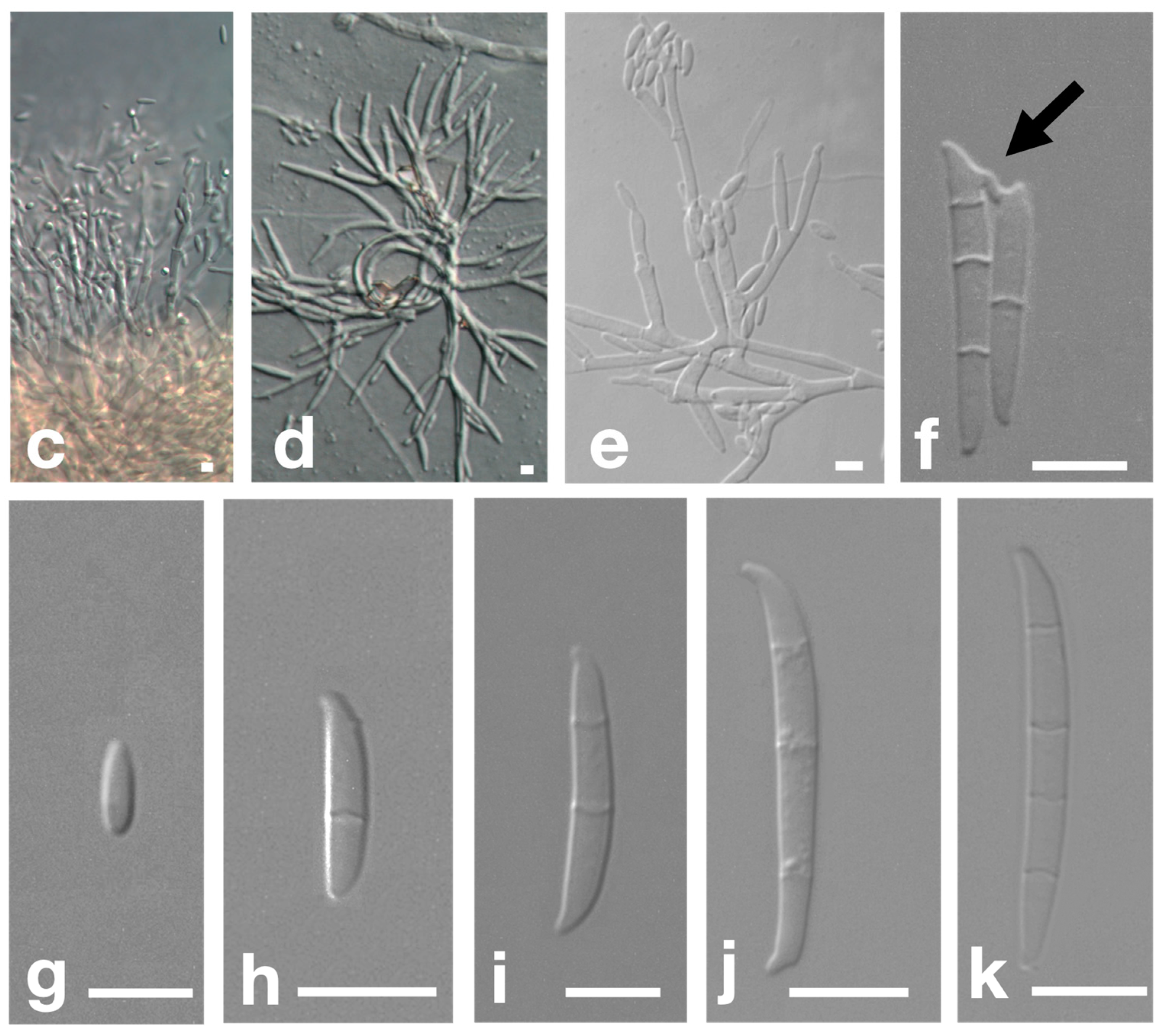
3.3. Morphological Characteristics
3.4. Genetic Characteristics
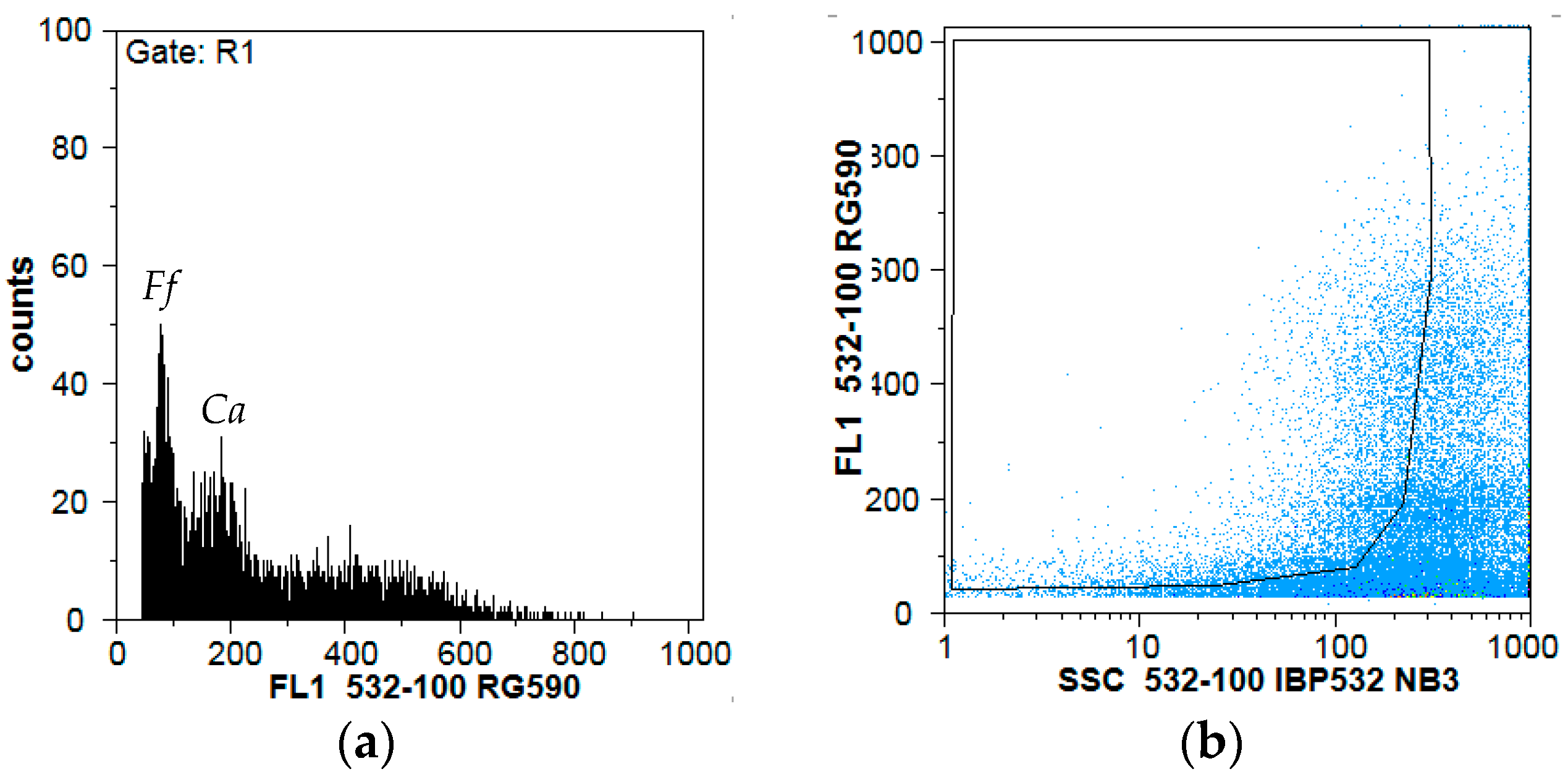
3.5. Cytogemonic Characteristics
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, E.M.; Münsterkötter, M.; Proctor, R.H.; Brown, D.W.; Sharon, A.; Idan, Y.; Oren-Young, L.; Sieber, C.M.; Novák, O.; Pěnčík, A.; et al. Comparative “Omics” of the Fusarium fujikuroi Species Complex Highlights Differences in Genetic Potential and Metabolite Synthesis. Genome Biol. Evol. 2016, 8, 3574–3599. [Google Scholar] [CrossRef]
- Lawal, O.A.; Ogundajo, A.L.; Avoseh, N.O.; Ogunwande, I.A. Cymbopogon citratus. In Medicinal Spices and Vegetables from Africa; Elsevier: Amsterdam, The Netherlands, 2017; pp. 397–423. [Google Scholar]
- Majewska, E.; Kozlowska, M.; Gruszczynska-Sekowska, E.; Kowalska, D.; Tarnowska, K. Lemongrass (Cymbopogon citratus) Essential Oil: Extraction, Composition, Bioactivity and Uses for Food Preservation—A Review. Pol. J. Food Nutr. Sci. 2019, 69, 327–341. [Google Scholar] [CrossRef]
- Akhila, A. Essential Oil-Bearing Grasses: The Genus Cymbopogon; CRC Press: Boca Raton, FL, USA, 2009; ISBN 0849378583. [Google Scholar]
- Mayaud, L.; Carricajo, A.; Zhiri, A.; Aubert, G. Comparison of Bacteriostatic and Bactericidal Activity of 13 Essential Oils against Strains with Varying Sensitivity to Antibiotics. Lett. Appl. Microbiol. 2008, 47, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Aungtikun, J.; Soonwera, M.; Sittichok, S. Insecticidal Synergy of Essential Oils from Cymbopogon citratus (Stapf.), Myristica fragrans (Houtt.), and Illicium verum Hook. f. and Their Major Active Constituents. Ind. Crops Prod. 2021, 164, 113386. [Google Scholar] [CrossRef]
- Lee, J.-E.; Seo, S.-M.; Huh, M.-J.; Lee, S.-C.; Park, I.-K. Reactive Oxygen Species Mediated-Antifungal Activity of Cinnamon Bark (Cinnamomum verum) and Lemongrass (Cymbopogon citratus) Essential Oils and Their Constituents against Two Phytopathogenic Fungi. Pestic. Biochem. Physiol. 2020, 168, 104644. [Google Scholar] [CrossRef]
- Faria, J.M.S.; Cavaco, T.; Gonçalves, D.; Barbosa, P.; Teixeira, D.M.; Moiteiro, C.; Inácio, M.L. First Report on the Synergistic Interaction between Essential Oils against the Pinewood Nematode Bursaphelenchus xylophilus. Plants 2023, 12, 2438. [Google Scholar] [CrossRef]
- Kamali-Sarvestani, S.; Mostowfizadeh-Ghalamfarsa, R.; Salmaninezhad, F.; Cacciola, S.O. Fusarium and Neocosmospora Species Associated with Rot of Cactaceae and Other Succulent Plants. J. Fungi 2022, 8, 364. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; John Wiley & Sons: Hoboken, NJ, USA, 2008; ISBN 0470276460. [Google Scholar]
- Cenis, J.L. Rapid Extraction of Fungal DNA for PCR Amplification. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef]
- De Hoog, G.S.; van den Ende, A.H.G.G. Molecular Diagnostics of Clinical Strains of Filamentous Basidiomycetes: Molekulare Diagnostik Klinischer Stämme Filamentöser Basidiomyzeten. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid Flow Cytometric Analysis of the Cell Cycle in Intact Plant Tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Two New Nuclear Isolation Buffers for Plant DNA Flow Cytometry: A Test with 37 Species. Ann. Bot. 2007, 100, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Tavares, D.; Ramos, A.P.; Gonçalves, S.; Loureiro, J. Validation of Standards Suitable for Genome Size Estimation of Fungi. J. Microbiol. Methods 2017, 142, 76–78. [Google Scholar] [CrossRef]
- Guilengue, N.; Alves, S.; Talhinhas, P.; Neves-Martins, J. Genetic and Genomic Diversity in a Tarwi (Lupinus mutabilis Sweet) Germplasm Collection and Adaptability to Mediterranean Climate Conditions. Agronomy 2019, 10, 21. [Google Scholar] [CrossRef]
- Skaria, B.P.; Joy, P.P.; Mathew, S.; Mathew, G. 24—Lemongrass. In Woodhead Publishing Series in Food Science, Technology and Nutrition; Peter, K.V., Ed.; Woodhead Publishing: Sawston, UK, 2006; pp. 400–419. ISBN 978-1-84569-017-5. [Google Scholar]
- Gardner, D.E. Lemongrass Rust Caused by Puccinia nakanishikii in Hawaii. Plant Dis. 1985, 69, 1100. [Google Scholar] [CrossRef]
- Alam, M.; Chourasia, H.K.; Sattar, A.; Janardhanan, K.K. Collar Rot and Wilt: A New Disease of Java Citronella (Cymbopogon winterianus) Caused by Fusarium moniliforme Sheldon. Plant Pathol. 1994, 43, 1057–1061. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Fandohan, P.; Gbenou, J.D.; Gnonlonfin, B.; Hell, K.; Marasas, W.F.O.; Wingfield, M.J. Effect of Essential Oils on the Growth of Fusarium verticillioides and Fumonisin Contamination in Corn. J. Agric. Food Chem. 2004, 52, 6824–6829. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavaco, T.; Gomes-Domingues, C.; Gonçalves, F. A Cytogenomic Analysis Reveals a New Fusarium fujikuroi Species Associated with Lemongrass (Cymbopogon citratus). Biol. Life Sci. Forum 2023, 27, 21. https://doi.org/10.3390/IECAG2023-15755
Cavaco T, Gomes-Domingues C, Gonçalves F. A Cytogenomic Analysis Reveals a New Fusarium fujikuroi Species Associated with Lemongrass (Cymbopogon citratus). Biology and Life Sciences Forum. 2023; 27(1):21. https://doi.org/10.3390/IECAG2023-15755
Chicago/Turabian StyleCavaco, Tomás, Catarina Gomes-Domingues, and Filipe Gonçalves. 2023. "A Cytogenomic Analysis Reveals a New Fusarium fujikuroi Species Associated with Lemongrass (Cymbopogon citratus)" Biology and Life Sciences Forum 27, no. 1: 21. https://doi.org/10.3390/IECAG2023-15755
APA StyleCavaco, T., Gomes-Domingues, C., & Gonçalves, F. (2023). A Cytogenomic Analysis Reveals a New Fusarium fujikuroi Species Associated with Lemongrass (Cymbopogon citratus). Biology and Life Sciences Forum, 27(1), 21. https://doi.org/10.3390/IECAG2023-15755






