Insight into the Alpha-Glucosidase Inhibitory Potentials of Curcuma longa Methanolic Extracts and Phytochemicals: An In Vitro and In Silico Study †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Methanolic Solvent Extraction of Dried Pulverized Turmeric Rhizome
2.2.2. Qualitative and Quantitative Phytochemical Analysis
2.2.3. Alpha-Glucosidase Inhibition
2.2.4. Phytochemicals and Bioactive Metabolite from Curcuma longa (Turmeric)
2.2.5. Preparation and Refinement of the Alpha-Glucosidase Crystal Structure for Molecular Docking
2.2.6. Molecular Docking for Probable Inhibitors of Alpha-Glucosidase and ADMET Analysis
3. Results
3.1. Methanolic Extract Yield and Its Phytochemicals Constituents
3.2. In Vitro Inhibition of Alpha-Glucosidase by the Methanolic Extract of Turmeric Rhizome
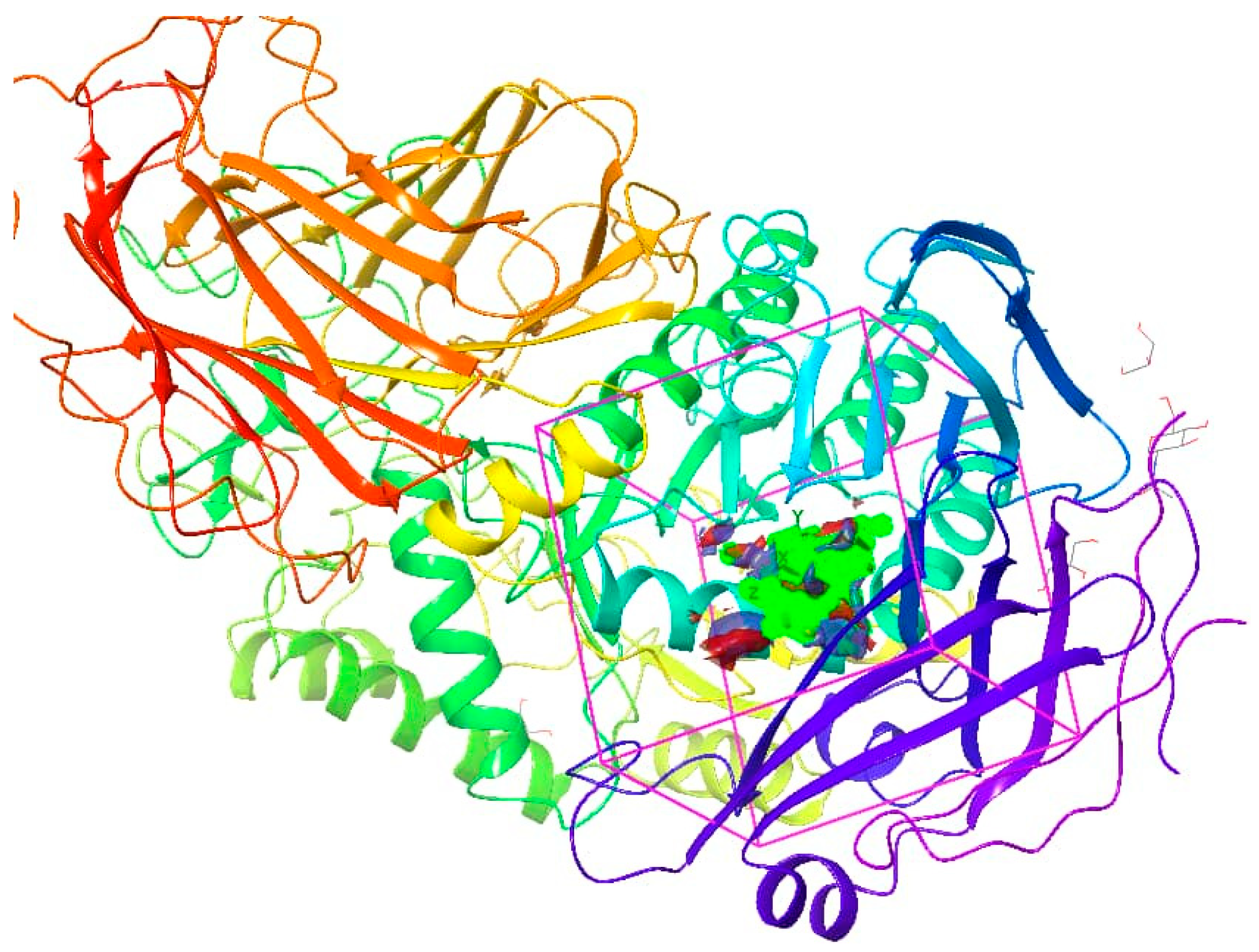
3.3. Molecular Docking of Curcuma longa Phytochemical Library to the Binding Pocket of Alpha-Glucosidase
3.4. Examining the ADMET Properties of Acarbose and Guaiacol (Top-Scoring Compound from Turmeric against Alpha-Glucosidase)
4. Discussion
5. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chukwuma, I.F.; Nworah, F.N.; Apeh, V.O.; Omeje, K.O.; Nweze, E.J.; Asogwa, C.D.; Ezeorba, T.P.C. Phytochemical Characterization, Functional Nutrition, and Antidiabetic Potentials of Leptadenia Hastata (Pers) Decne Leaves: In Silico and In Vitro Studies. Bioinform. Biol. Insights 2022, 16, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Lopez, A.D. Measuring the Global Burden of Disease. N. Engl. J. Med. 2013, 369, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Mambiya, M.; Shang, M.; Wang, Y.; Li, Q.; Liu, S.; Yang, L.; Zhang, Q.; Zhang, K.; Liu, M.; Nie, F.; et al. The Play of Genes and Non-Genetic Factors on Type 2 Diabetes. Front. Public Heal. 2019, 7, 447628. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Hwang, J.S. Genetic Aspects of Type 1 Diabetes. Ann. Pediatr. Endocrinol. Metab. 2019, 24, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Sami, W.; Ansari, T.; Butt, N.S.; Hamid, M.R.A. Effect of Diet on Type 2 Diabetes Mellitus: A Review. Int. J. Health Sci. 2017, 11, 65. [Google Scholar]
- Raman, P.G. Environmental Factors in Causation of Diabetes Mellitus. In Environ. Health Risk Hazard. Factors Living Species; Larramendy, M.L., Soloneski, S., Eds.; InTechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Rahn, S.; Zimmermann, V.; Viol, F.; Knaack, H.; Stemmer, K.; Peters, L.; Lenk, L.; Ungefroren, H.; Saur, D.; Schäfer, H.; et al. Diabetes as Risk Factor for Pancreatic Cancer: Hyperglycemia Promotes Epithelial-Mesenchymal-Transition and Stem Cell Properties in Pancreatic Ductal Epithelial Cells. Cancer Lett. 2018, 415, 129–150. [Google Scholar] [CrossRef] [PubMed]
- Abudawood, M. Diabetes and Cancer: A Comprehensive Review. J. Res. Med. Sci. 2019, 24, 94. [Google Scholar] [CrossRef] [PubMed]
- Clemmensen, C.; Müller, T.D.; Finan, B.; Tschöp, M.H.; Dimarchi, R. Current and Emerging Treatment Options in Diabetes Care. Handb. Exp. Pharmacol. 2015, 233, 437–459. [Google Scholar]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A Review of Alpha-Glucosidase Inhibitors from Plants as Potential Candidates for the Treatment of Type-2 Diabetes. Phytochem. Rev. 2022, 21, 1049. [Google Scholar] [CrossRef]
- Alam, S.; Sarker, M.M.R.; Sultana, T.N.; Chowdhury, M.N.R.; Rashid, M.A.; Chaity, N.I.; Zhao, C.; Xiao, J.; Hafez, E.E.; Khan, S.A.; et al. Antidiabetic Phytochemicals from Medicinal Plants: Prospective Candidates for New Drug Discovery and Development. Front. Endocrinol. 2022, 13, 1. [Google Scholar] [CrossRef]
- Matoori, S. Diabetes and Its Complications. ACS Pharmacol. Transl. Sci. 2022, 5, 513–515. [Google Scholar] [CrossRef] [PubMed]
- Hossain, U.; Das, A.K.; Ghosh, S.; Sil, P.C. An Overview on the Role of Bioactive α-Glucosidase Inhibitors in Ameliorating Diabetic Complications. Food Chem. Toxicol. 2020, 145, 111738. [Google Scholar] [CrossRef] [PubMed]
- Ezeorba, T.P.C.; Chukwudozie, K.I.; Ezema, C.A.; Anaduaka, E.G.; Nweze, E.J.; Okeke, E.S. Potentials for Health and Therapeutic Benefits of Garlic Essential Oils: Recent Findings and Future Prospects. Pharmacol. Res. Mod. Chin. Med. 2022, 3, 100075. [Google Scholar] [CrossRef]
- Ezeorba, T.P.C.; Uchendu, N.O.; Nweze, E.J.; Okoroafor, C.K.; Ogbu, P.O.; Okpara, M.C.; Asomadu, R.O.; Joshua, P.E.; Ng, R.O.A. A Probable Anti-COVID Phytochemical (1,7-Bis-(4-Hydroxyphenyl)-1-Heptene-3,5-Dione) Screened Computationally from the Rhizome of Curcuma longa. Med. Sci. Forum 2021, 7, 6. [Google Scholar] [CrossRef]
- Rajagopal, K.; Varakumar, P.; Baliwada, A.; Byran, G. Activity of Phytochemical Constituents of Curcuma longa (Turmeric) and Andrographis Paniculata against Coronavirus (COVID-19): An In Silico Approach. Futur. J. Pharm. Sci. 2020, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, A.R.; Haque, M. Preparation of Medicinal Plants: Basic Extraction and Fractionation Procedures for Experimental Purposes. J. Pharm. Bioallied Sci. 2020, 12, 1. [Google Scholar] [CrossRef]
- Sharma, A.; Ray, A.; Singhal, R.S. Co-Extraction of Turmeric (Curcuma longa L.) and Dried Coconut Shreds by Supercritical Fluid Extraction (SFE): Chemical and Bioactivity Profile. J. Clean. Prod. 2023, 382, 135313. [Google Scholar] [CrossRef]
- Gul, P.; Bakht, J. Antimicrobial Activity of Turmeric Extract and Its Potential Use in Food Industry. J. Food Sci. Technol. 2015, 52, 2272–2279. [Google Scholar] [CrossRef]
- Rajesh, H.; Rao, S.N.; Megha, R.N.; Prathima, K.S.; Rejeesh, E.P.; Chandrashekar, R. Phytochemical Analysis of Methanolic Extract of Curcuma longa Linn Rhizome. Int. J. Univers. Pharm. Bio. Sci. 2013, 2, 285–297. [Google Scholar]
- Velurajan, S.; Balamurugan, V. Phytochemical Comparison of Alcoholic Extract of Fresh and Dry Curcuma longa. IJARIIE 2019, 5, 419–428. [Google Scholar]
- Meena, N.K.; Raben, N. Pompe Disease: New Developments in an Old Lysosomal Storage Disorder. Biomolecules 2020, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhou, W.; Seshan, K.; Li, Y. Green and Efficient Synthesis Route of Catechol from Guaiacol. J. Mol. Catal. A Chem. 2013, 368–369, 61–65. [Google Scholar] [CrossRef]
- Knezevic, S.; Ghafoor, A.; Mehri, S.; Barazi, A.; Dziura, M.; Trant, J.F.; Dieni, C.A. Catechin and Other Catechol-Containing Secondary Metabolites: Bacterial Biotransformation and Regulation of Carbohydrate Metabolism. PharmaNutrition 2021, 17, 100273. [Google Scholar] [CrossRef]


| S/N | Phytochemical Classes | Qualitative Analysis | Quantitative Analysis |
|---|---|---|---|
| 1. | Phenols | +++ | 790.32 ± 129.20 |
| 2. | Flavonoids | ++ | 171.08 ± 0.04 |
| 3. | Alkaloid | +++ | 494.99 ± 1.27 |
| 4. | Tannin | + | 9.52 ± 6.59 |
| 5. | Reducing Sugar | − | 23.40 ± 7.74 |
| 6. | Glycosides | − | 0.00 ± 0.00 |
| 7. | Steroids | + | 3.40 ± 0.00 |
| 8. | Terpenoid | +++ | 131.99 ± 6.59 |
| Concentration (µg/mL) | % Inhibition ± SEM |
|---|---|
| 10 | 19.14 ± 0.46 |
| 20 | 22.29 ± 0.09 |
| 30 | 37.01 ± 0.28 |
| 40 | 31.23 ± 0.16 |
| 1C50-20.9184 | |
| Site Number | Site Score | D-Score | Volume |
|---|---|---|---|
| Site 1 | 1.031 | 1.031 | 144.746 |
| Site 2 | 0.937 | 0.886 | 166.698 |
| Site 3 | 0.853 | 0.796 | 192.08 |
| Site 4 | 0.692 | 0.643 | 149.548 |
| S/N | Phytochemical Compounds | Entry ID | Canonical SMILE | Docking Score |
|---|---|---|---|---|
| 1. | Guaiacol | CID 460 | COC1=CC=CC=C1O | −5.266 |
| 2. | P-Tolyl-Methyl | CID 110953 | CC1=CC=CC=C1C(C)O | −3.939 |
| 3. | Limonene | CID 22311 | CC1=CCC(CC1)C(=C)C | −3.702 |
| 4. | Quercetine | CID 5280343 | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O)O | −3.256 |
| 5. | Azulene | CID 9231 | C1=CC=C2C=CC=C2C=C1 | −3.215 |
| 6. | * Acarbose | CID 41774 | CC1C(C(C(C(O1)OC2C(OC(C(C2O)O)OC3C(OC(C(C3O)O)O)CO)CO)O)O)NC4C=C(C(C(C4O)O)O)CO | −9.522 |
| Parameters | Guaiacol | Acarbose (Standard Drug) |
|---|---|---|
| Physiochemical Properties | ||
| Mol. Weight (g/mol) | 124.14 | 645.60 |
| Num. Rotatable bond | 1 | 9 |
| Num. H—bond acceptors | 2 | 19 |
| Num. H—bond donor | 1 | 14 |
| Molar refractivity | 34.96 | 136.69 |
| TPSA (A2) | 29.46 | 321.17 |
| Drug-Likeness | ||
| Lipinski violations | 0 | 3 |
| Verber violations | No | Yes |
| Bioavailabilty score | 0.55 | 0.17 |
| Pharmacokinetics | ||
| GI absorption | High | Low |
| BBB permeant | Yes | No |
| P-gp substrate | No | Yes |
| Log Kp (cm/s) (skin permeation) | −6.12 | −16.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okechukwu, A.-J.M.; Okeke, E.S.; Eze, K.N.; Ezeorba, W.F.C.; Ezeorba, T.P.C. Insight into the Alpha-Glucosidase Inhibitory Potentials of Curcuma longa Methanolic Extracts and Phytochemicals: An In Vitro and In Silico Study. Biol. Life Sci. Forum 2023, 26, 92. https://doi.org/10.3390/Foods2023-15514
Okechukwu A-JM, Okeke ES, Eze KN, Ezeorba WFC, Ezeorba TPC. Insight into the Alpha-Glucosidase Inhibitory Potentials of Curcuma longa Methanolic Extracts and Phytochemicals: An In Vitro and In Silico Study. Biology and Life Sciences Forum. 2023; 26(1):92. https://doi.org/10.3390/Foods2023-15514
Chicago/Turabian StyleOkechukwu, Ada-Jesus Mercy, Emmanuel Sunday Okeke, Kingsley Nnaechetam Eze, Wisdom Favour Chinedu Ezeorba, and Timothy Prince Chidike Ezeorba. 2023. "Insight into the Alpha-Glucosidase Inhibitory Potentials of Curcuma longa Methanolic Extracts and Phytochemicals: An In Vitro and In Silico Study" Biology and Life Sciences Forum 26, no. 1: 92. https://doi.org/10.3390/Foods2023-15514
APA StyleOkechukwu, A.-J. M., Okeke, E. S., Eze, K. N., Ezeorba, W. F. C., & Ezeorba, T. P. C. (2023). Insight into the Alpha-Glucosidase Inhibitory Potentials of Curcuma longa Methanolic Extracts and Phytochemicals: An In Vitro and In Silico Study. Biology and Life Sciences Forum, 26(1), 92. https://doi.org/10.3390/Foods2023-15514







