The specific values for the parameters were chosen such that there would be a compromise between the speed of the proposed method and its efficiency. However, a series of experiments are presented below in which various experimental parameters have been modified, in order to determine the stability of the proposed technique to possible changes in these parameters.
The machine learning methods used cover a wide range of approaches, and they have had success in many practical problems within the related literature.
The analysis of
Table 2 evaluates the error rates of various machine learning models across multiple classification datasets. Each row corresponds to a dataset, and each column represents a specific model. The values indicate the error percentages, where lower values signify better performance. The last row provides the average error rate for each model, offering an overall performance summary. Starting with the averages, the NEURALDE model exhibits the best overall performance, achieving the lowest mean error rate of 19.78%. It is followed by GENETIC, with an average error rate of 26.19%, and RBF at 30.08%. The PRUNE model achieves an average error rate of 28.39%, placing it between RBF and NEAT, which has an average error rate of 32.66%. Meanwhile, BFGS and ADAM display the highest average error rates, 33.43% and 35.88%, respectively, making NEURALDE the most efficient model among those compared. On a dataset-specific level, NEURALDE consistently delivers the lowest error rates in numerous cases. For instance, in the ALCOHOL dataset, NEURALDE achieves the best result with an error rate of 19.15%, while PRUNE follows closely with 15.75%, outperforming all other models. In the CIRCULAR dataset, NEURALDE outperforms all other models with an error rate of 4.23%, while PRUNE records a slightly higher error rate of 12.76%, still surpassing many other methods. In datasets like MAGIC and MAMMOGRAPHIC, NEURALDE again proves superior, with error rates of 11.73% and 17.52%, respectively. PRUNE, however, shows significant variability; for example, it records 30.76% in MAGIC, which is higher than GENETIC (21.75%), but still manages competitive performance in MAMMOGRAPHIC with 38.10%. There are, however, a few datasets where NEURALDE does not perform as dominantly. For example, in the HOUSEVOTES dataset, it ties with the other models at 7.48%. PRUNE performs better here, with an error rate of 5.81%, showcasing its strengths in specific datasets. Similarly, in the ZONF_S dataset, NEURALDE achieves 2.41%, while PRUNE closely follows with 3.27%. Despite this, NEURALDE’s superior consistency across datasets remains evident. Other models, such as GENETIC, show strong performance in isolated cases, such as in the SPIRAL dataset (48.66%). However, they generally fall short of NEURALDE’s consistent excellence. RBF demonstrates strong stability, with notable results in the DERMATOLOGY (62.34%) and PHONEME (23.32%) datasets, but PRUNE again outshines RBF in specific cases like DERMATOLOGY, with an error rate of 9.02%. Meanwhile, ADAM and BFGS tend to underperform, particularly in datasets like SPAMBASE, where they record high error rates of 48.05% and 18.16%, respectively. PRUNE outperforms them significantly in SPAMBASE, with an error rate of just 3.91%, illustrating its competitiveness in certain scenarios. In conclusion, the NEURALDE model stands out as the most effective model overall, achieving the lowest average error rate and excelling in the majority of datasets. PRUNE emerges as a strong contender, with competitive performance and notable success in certain datasets. While other models, such as RBF and GENETIC, perform well in specific scenarios, NEURALDE demonstrates superior consistency and effectiveness across a wide range of datasets, with PRUNE providing significant value in enhancing classification accuracy in many cases.
Table 3 presents the performance of various machine learning models on regression problems, where the values represent the error. Lower error values indicate better performance. Each row corresponds to a specific dataset, while the last row shows the average error for each model, providing an overall view of their effectiveness. Based on the average error, NEURALDE demonstrates the best performance, with the lowest overall error of 5.56. This result suggests that the model consistently performs well across most datasets. PRUNE follows as the second-best model, with an average error of 15.40, outperforming GENETIC, RBF, and other methods in several cases. GENETIC and RBF achieve average errors of 9.29 and 9.91, respectively. ADAM, NEAT, and BFGS exhibit higher average errors, at 22.47, 24.35, and 30.31, respectively, indicating lower performance compared to the other models. Analyzing the results dataset by dataset, NEURALDE records the smallest error in several cases. For instance, in the AIRFOIL dataset, it achieves an error of 0.0014, outperforming all the other models. PRUNE also shows strong performance here, with an error of 0.002, close to NEURALDE. In the CONCRETE dataset, NEURALDE achieves the lowest error of 0.003, while PRUNE achieves the second-lowest error of 0.0077, further confirming its reliability. Similarly, in the LASER dataset, NEURALDE delivers an exceptionally low error of 0.0026, followed closely by PRUNE with an error of 0.007. In the TREASURY dataset, NEURALDE demonstrates superior performance with an error of 1.021, better than all other models, while PRUNE also achieves a competitive error of 13.76, outperforming NEAT and ADAM. PRUNE shows notable performance in several datasets where it surpasses other models. For instance, in the STOCK dataset, PRUNE records an error of 39.08, which is lower than NEAT (215.82) and ADAM (180.89), though slightly higher than NEURALDE (3.40). In the BASEBALL dataset, PRUNE achieves 94.50, outperforming ADAM (77.90) and NEAT (100.39). In the MORTGAGE dataset, PRUNE achieves an error of 12.96, which, while not the lowest, still outperforms several other models including NEAT (14.11) and ADAM (9.24). Overall, while NEURALDE stands out as the most effective model, PRUNE emerges as a highly competitive alternative, consistently delivering strong results across multiple datasets. GENETIC and RBF also perform well in specific cases, but their performance is less consistent compared to NEURALDE and PRUNE. ADAM, NEAT, while BFGS generally underperforms relative to the other methods.
In
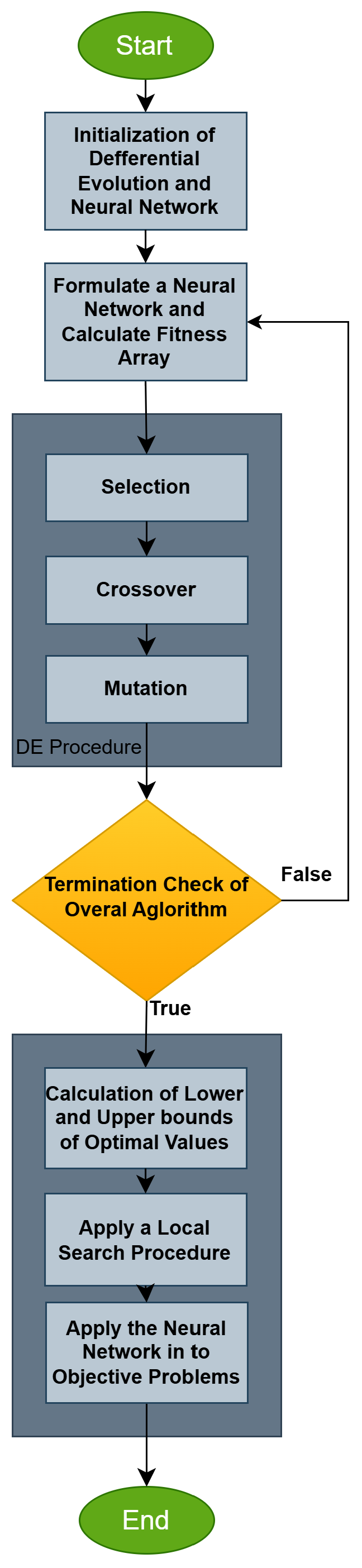
Figure 2, the comparison of the proposed machine learning model NEURALDE with the other models for the classification datasets is illustrated, based on the values of the critical parameter
p, which indicate the levels of statistical significance. The results show exceptionally low
p-values, suggesting that the differences in the performance of NEURALDE compared to other models are not random and are statistically highly significant. Specifically, the comparison of NEURALDE with ADAM yielded
p = 4.7 ×
, with BFGS
p = 1 ×
, with GENETIC
p = 2.9 ×
, with NEAT
p = 2.3 ×
, with RBF
p = 1.3 ×
, and with PRUNE
p = 5.8 ×
. These values are well below the common thresholds of significance (
p < 0.05: significant), indicating that the superiority of NEURALDE is statistically well founded. The results confirm the high efficiency of the proposed model compared to other machine learning methods, demonstrating its reliability and effectiveness in classification problems.
As expected, the proposed method requires significantly more computation time than other method due to its complexity in the calculations and the requirement of the periodical application of a local optimization method.
3.2.1. Experiments with Differential Weight
Another series of experiments was carried out in order to determine the effect that the specific differential weight calculation mechanism has on the accuracy of the proposed technique. For this reason, the following methods were used for the calculation of the differential weight (parameter F):
Denoted as MIGRANT in the experimental tables: In this case, the differential weight mechanism proposed in [
110] was used.
Denoted as ADAPTIVE in the experimental tables: This method used the differential weight calculation proposed in [
111].
Represented as RANDOM in the experimental tables: This method was the default method, suggested by Charilogis [
59].
Table 4 provides the performance of three differential weight computation methods (MIGRANT, ADAPTIVE, and RANDOM) on the series of classification datasets. The analysis reveals that the MIGRANT method achieves the smallest average error rate of 19.34%, indicating the best overall performance among the three methods. The RANDOM method follows closely, with an average error rate of 19.78%, while the ADAPTIVE method has the highest average error rate of 19.98%, suggesting that it is the least effective on average. Examining the individual datasets, MIGRANT is found to outperform the other methods in several cases. For example, in the HEART dataset, MIGRANT records an error rate of 19.01%, better than ADAPTIVE (17.29%) and RANDOM (17.60%). Similarly, in the MAGIC dataset, MIGRANT achieves a competitive error rate of 11.83%, which is close to RANDOM (11.73%) but higher than ADAPTIVE (11.28%). MIGRANT also has the lowest error in datasets such as ALCOHOL (19.62%), REGIONS2 (26.26%), and SPAMBASE (5.87%). ADAPTIVE demonstrates strong performance in specific datasets, achieving the lowest error rates in cases such as MAGIC (11.28%) and HEART (17.29%). However, in many datasets, it performs worse than MIGRANT or RANDOM, such as in SPAMBASE, where it records a lower error rate of 4.78% but is generally outperformed by the other critical datasets. The RANDOM method exhibits competitive performance in several datasets, achieving the lowest error rates in examples like ZOO (6.63%) and ZONF_S (2.41%). However, its performance fluctuates more significantly, with higher error rates in datasets such as HEARTATTACK (19.70%) and REGIONS2 (28.77%), where it is outperformed by MIGRANT and ADAPTIVE. Notable observations include cases where the error rates are very close across the methods, such as in datasets like CIRCULAR, where the differences are minimal (MIGRANT at 4.28%, ADAPTIVE at 4.34%, and RANDOM at 4.23%). In other cases, the differences are more pronounced, as in SEGMENT, where MIGRANT achieves 9.71%, significantly better than ADAPTIVE (15.99%) and RANDOM (15.60%). Overall, MIGRANT demonstrates the most consistent and robust performance across the datasets, with the lowest average error and competitive results in many individual datasets. RANDOM and ADAPTIVE have comparable average errors but show more variability in their performance.
The
Table 5 presents the performance of three differential weight computation methods (MIGRANT, ADAPTIVE, and RANDOM) on the regression datasets. The RANDOM method achieves the lowest average error, 5.56, making it the most effective overall. ADAPTIVE follows with an average error of 6.23, while MIGRANT exhibits the highest average error at 8.03, indicating the least effective performance. For specific datasets, RANDOM often outperforms the others. For instance, in the HOUSING dataset, it records an error of 24.82, lower than MIGRANT (14.58) and ADAPTIVE (32.62). Similarly, in the PLASTIC dataset, RANDOM achieves the smallest error of 3.27 compared to MIGRANT (5.36) and ADAPTIVE (3.57). RANDOM also shows superior performance in datasets like MORTGAGE (0.54) and STOCK (3.4). ADAPTIVE demonstrates strong performance in certain datasets, such as FRIEDMAN, where its error is 1.32, outperforming MIGRANT (1.66) and RANDOM (1.22). In the QUAKE dataset, ADAPTIVE records the lowest error of 0.041. However, its performance is noticeably weaker in other datasets, like HOUSING, where it registers the highest error of the three. MIGRANT outperforms in a few cases, such as the BL dataset, where its error is 0.007, significantly lower than ADAPTIVE (0.019) and RANDOM (0.016). However, it generally records higher errors in many datasets, such as AUTO (45.37) and BASEBALL (79.24), where the other methods perform better. In some cases, such as the LASER dataset, all methods perform similarly, with errors of 0.0026 for MIGRANT and RANDOM, and 0.0027 for ADAPTIVE. In the QUAKE dataset, the differences are also minimal, with RANDOM having the highest error (0.042), but very close to the others. Overall, RANDOM emerges as the most reliable and effective method, with the lowest average error and frequent superiority across the individual datasets. ADAPTIVE performs well in selected datasets but shows greater variability, while MIGRANT demonstrates the least effective performance, with higher errors across numerous datasets.
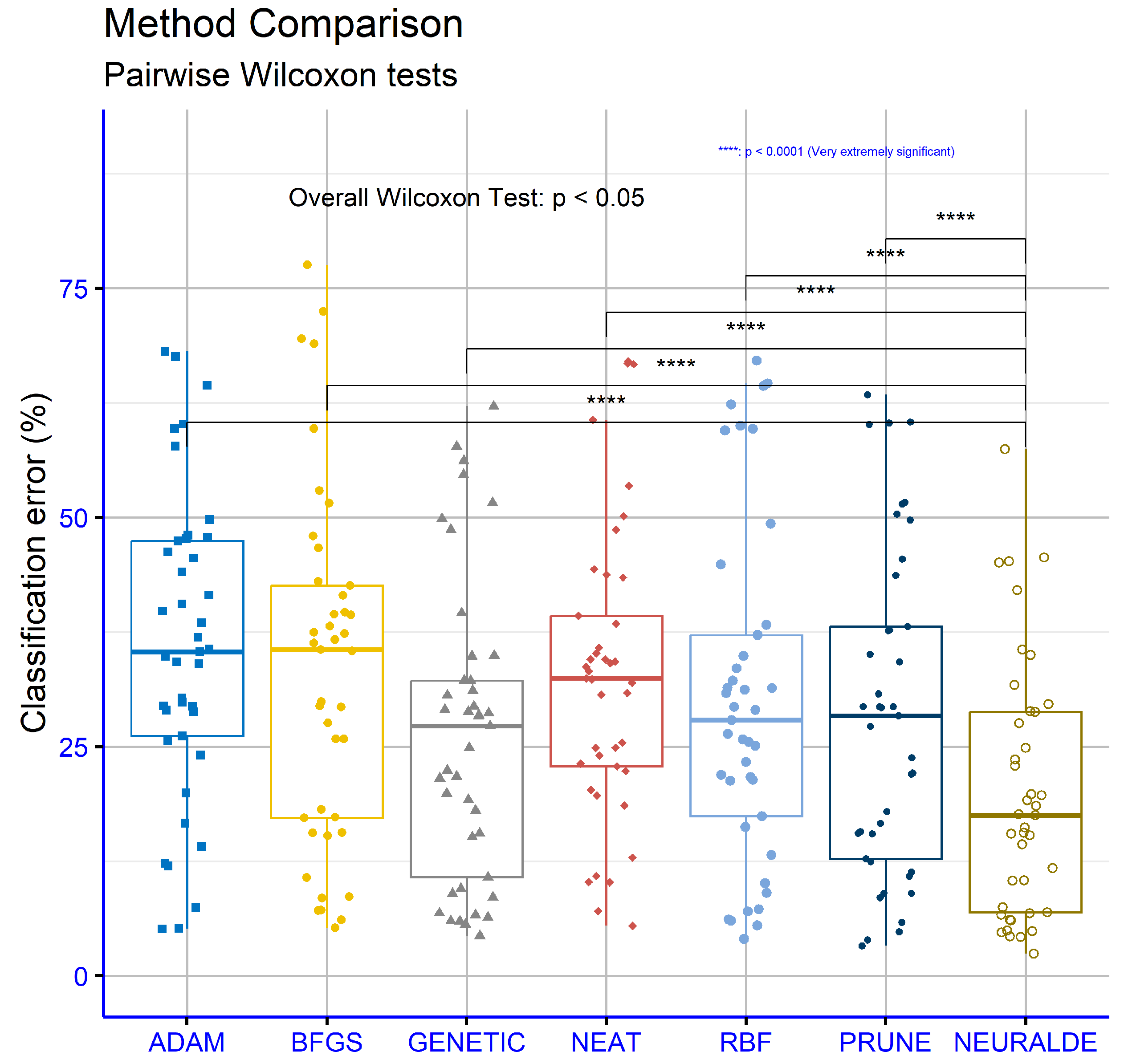
An analysis was conducted to compare different methods for computing differential weights in a proposed machine learning approach, focusing on classification error. The analysis employed the Wilcoxon Test for pairwise comparisons and was applied to the classification datasets. The goal was to examine whether statistically significant differences exist among the three approaches—MIGRANT, ADAPTIVE, and RANDOM (
Figure 7). The results show that the comparison between MIGRANT and ADAPTIVE yielded a
p-value of 0.085. Although this value suggests a trend of differentiation, it does not fall below the conventional significance threshold of 0.05, indicating that the difference is not statistically significant. Similarly, the comparison between MIGRANT and RANDOM produced a
p-value of 0.064, which, while closer to significance, also does not meet the required threshold. Finally, the comparison between ADAPTIVE and RANDOM resulted in a
p-value of 0.23, indicating no statistically significant difference between these two methods. Overall, the findings suggest that while there are indications of differences in behavior among the three approaches, none of these differences are statistically significant based on the data analyzed. This implies that the three methods for computing differential weights may be considered equivalent in terms of their impact on classification error, at least for the datasets used in this study.
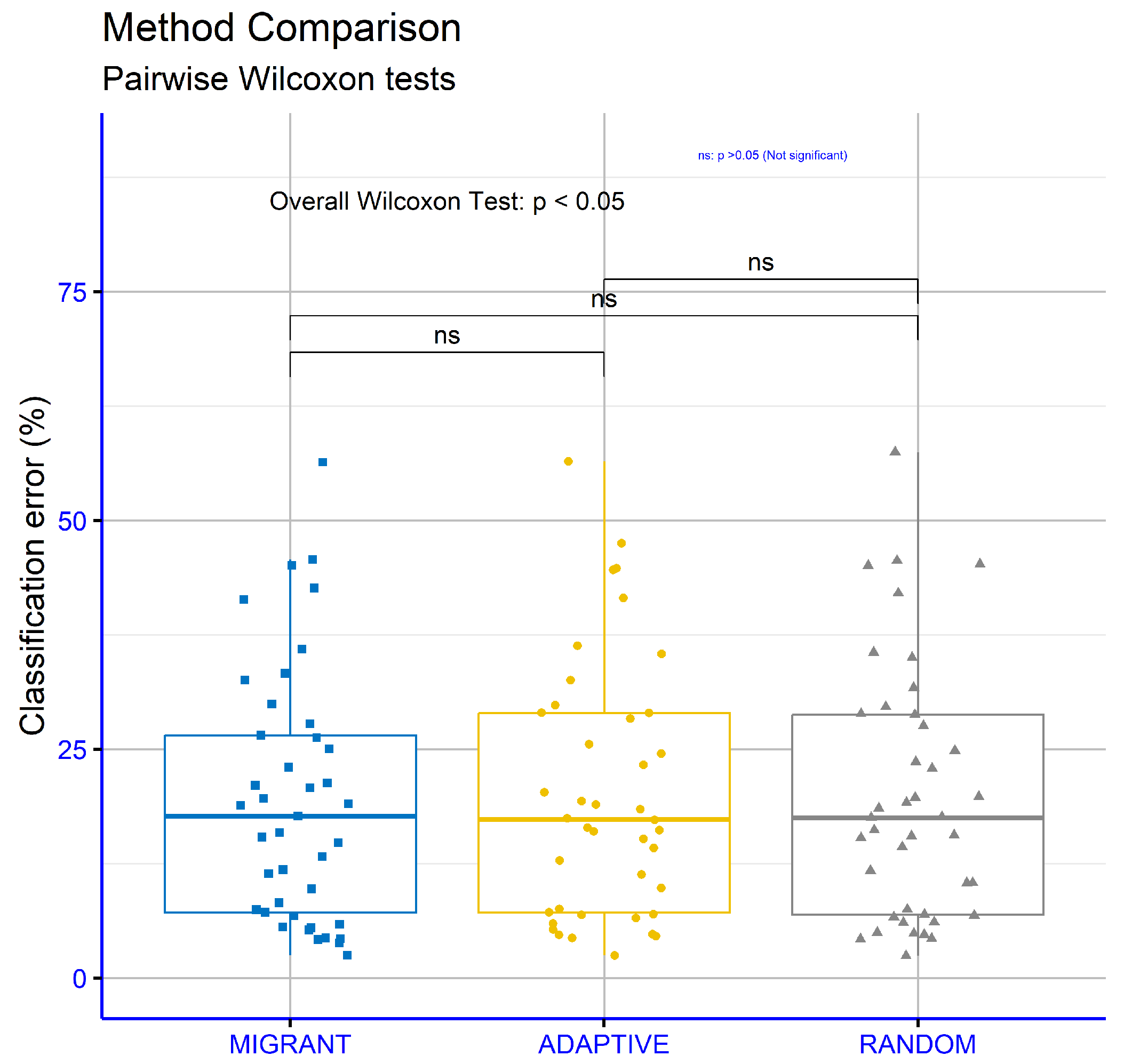
An analysis was conducted to compare the different methods for calculating differential weight in the proposed machine learning approach to the regression datasets. The Wilcoxon Test was used for pairwise comparison and applied across the series of regression datasets, with a focus on regression error. The aim was to determine the statistical significance of the observed differences between the methods (
Figure 8). The results showed that the comparison between MIGRANT and ADAPTIVE yielded a
p-value of 0.68, indicating no statistically significant difference between these two methods. Similarly, the comparison between MIGRANT and RANDOM resulted in a
p-value of 0.9, confirming the absence of a statistically significant difference in this case as well. However, the comparison between ADAPTIVE and RANDOM produced a
p-value of 0.00095, which is below the standard threshold for statistical significance (commonly 0.05). This suggests a statistically significant difference between these two approaches. Overall, the findings indicate that the MIGRANT and ADAPTIVE methods, as well as the MIGRANT and RANDOM methods, exhibit similar performance regarding regression error. In contrast, the ADAPTIVE method shows a clear and statistically significant differentiation from the RANDOM method.
3.2.2. Experiments with the Factor a
In the next series of experiments, the effect of the parameter a on the behavior of the method used on the classification and data fitting data was studied. In this series of experiments, different values of this parameter were studied. This parameter determines the allowable range of values within which the local minimization method can vary the parameters of the artificial neural network.
Table 6 presents the error percentages across the used classification datasets for different values of the parameter
a (1.25, 1.5, 2, 4, and 8). The lowest overall average error is observed for
, with an average of 19.78%. This suggests that
provides the most balanced performance across datasets. The other parameter values—1.25, 1.5, 4, and 8—yield average errors of 19.98%, 19.95%, 20.00%, and 19.95%, respectively. These results indicate that
has a slight advantage in minimizing the overall error compared to the other values. For individual datasets, the performance varies depending on the parameter value. For example, in the ALCOHOL dataset, the smallest error is achieved at
with 19.15%, while the other values result in slightly higher errors. Similarly, in the SEGMENT dataset, the error is minimized at
with 14.27%. However, in datasets like SPIRAL, the errors across all parameter values are close, with no significant advantage for any specific value. In certain datasets, there is a clear trend in error reduction as the parameter value changes. For example, in the SEGMENT dataset, errors decrease consistently from
to
. Conversely, in the ZOO dataset, the error decreases for lower values of a but increases again at higher values, indicating that performance is not always linear with changes in
a. Some datasets exhibit minimal variability in performance across parameter values. For example, the error for the LIVERDISORDER dataset remains relatively stable, ranging from 31.41% to 32.33%. Similarly, the SPIRAL dataset shows little variation, with errors consistently around 42%. In other cases, specific parameter values consistently underperform. For instance, in the Z_F_S dataset,
achieves the lowest error of 6.67%, while
and
result in slightly higher errors. Similarly,
achieves the smallest error for certain datasets like SEGMENT but performs poorly for datasets like WINE. Overall,
emerges as the most effective parameter value, yielding the smallest average error across datasets. While other values of
a perform well in individual cases, their overall performance is less consistent.
Table 7 presents the error rates for the regression datasets across different values of the critical parameter
a (1.25, 1.5, 2, 4, and 8). The lowest average error is observed for
, with an average of 5.39, suggesting that it offers the best overall performance.
ranks second with an average of 5.46, and
slightly higher at 5.56. The values
and
show a marked increase in errors, with averages of 5.94 and 7.44, respectively. This indicates that higher values of
a are less effective compared to smaller ones. For individual datasets, performance varies with the value of
a. For instance, in the AIRFOIL dataset, the error consistently decreases as
a increases, with the smallest error (0.0007) observed at
. Conversely, in the FRIEDMAN dataset, the error rises sharply at
(19.74), signaling a significant performance decline. In some datasets, such as BASEBALL, the error steadily increases with higher values of
a rising from 57.67 at
to 72.37 at
. In contrast, in the HOUSING dataset, the smallest error occurs at
(22.86), although larger values like
show only a slight increase. Certain datasets exhibit minimal sensitivity to changes in
a. For example, in the LASER dataset, the errors remain almost constant regardless of the parameter value, ranging from 0.0025 to 0.0035. Similarly, in the DEE dataset, the errors are nearly identical across all values of
a showing minimal variation. Interestingly, the PLASTIC dataset shows a gradual decrease in error from 3.37 at
to 2.8 at
, indicating improved performance with increasing
a. Conversely, in the STOCK dataset, the error increases significantly from 2.82 at
to 5.42 at
. In conclusion,
emerges as the optimal choice for minimizing overall error in this analysis. Higher values of
a demonstrate reduced effectiveness, especially at
, where the average error rises notably. However, the results reveal that the impact of
a varies by dataset, with some benefiting from smaller values and others exhibiting relative insensitivity to parameter changes.
In
Figure 9, a comparison of the various values of the parameter
a, which defines the bounds of the optimal values in the proposed machine learning method, is presented. The comparisons were conducted using the Wilcoxon Test across the series of used datasets to evaluate the classification error. The results indicate that the
p-values for all pairwise comparisons between the different values of
a are above the significance level of 0.05, suggesting no statistically significant differences. For instance, comparisons between
and the other values (1.5, 2, 4, 8) yielded
p-values ranging from 0.6 to 0.73, while comparisons between
and other values (2, 4, 8) produced
p-values ranging from 0.32 to 0.8. Similarly, comparisons between
and higher values (4 and 8) resulted in
p-values of 0.22 and 0.31, respectively, whereas the comparison between
and
yielded a
p-value of 0.69. In conclusion, the results suggest that variations in the parameter
a within the specified range do not lead to statistically significant differences in classification error, as all the
p-values remain well above the conventional significance threshold. Therefore, the choice of a specific value for
a is likely to have little or no impact on the method’s performance, based on the current data.
In
Figure 10, the comparison of the different values of parameter
a for the regression error is presented. The results show that statistically significant differences were observed in some comparisons, as the
p-values were below the significance level of 0.05. For example, the comparison between
and
yielded
p = 0.024, while between
and
,
p = 0.017 was observed. Similarly, the comparison between
and
resulted in
p = 0.0021, indicating strong statistical significance. Conversely, some comparisons did not show statistically significant differences, as the
p-values were above the significance level. For instance, the comparison between
and
yielded
p = 0.22, while the comparison between
and
resulted in
p = 0.15. In conclusion, the results suggest that the choice of parameter
a affects the regression error in certain cases, with statistically significant differences primarily observed in comparisons between the smaller and larger values of the parameter. However, the differences are not always consistent and depend on the specific combinations of the values being compared.
3.2.3. Experiments with the Local Search Rate
In the last series of experiments, the effects of the execution rate of the local optimization methods were studied, focusing on values ranging from 0.25% to 2%.
Table 8 provides the error rates for the classification datasets used under the different values of the periodic local search
(0.0025, 0.005, 0.01, and 0.02). The analysis reveals that the lowest average error observed is
, with an error rate of 19.43%, suggesting that this value yields the best overall performance. This is closely followed by
, with an average error of 19.66%, and
with 19.78%. The highest average error occurs at
with 20.48%, indicating comparatively poorer performance. Examining the individual datasets, the impact of
appears to vary. For instance, in the ALCOHOL dataset, the smallest error is observed at
with 16.49%, whereas larger values such as
and
result in higher errors (21.81% and 19.15%, respectively). Conversely, in the SEGMENT dataset, the error consistently decreases as
increases, reaching its lowest value at
with 11.70%. This trend is also evident in datasets such as SPAMBASE and WINE, where the errors decrease significantly with higher
values. Some datasets exhibit minimal sensitivity to changes in
. For example, in the HEART and LIVERDISORDER datasets, the error rates remain relatively stable across all
values, showing only marginal fluctuations. In other cases, such as CIRCULAR and HOUSEVOTES, the variations in error rates are similarly minor. However, certain datasets show exceptions to the general trend. For example, in the SONAR dataset, the error rate is lowest at
with 18.53%, but higher
values like
produce increased errors (20.38%). Similarly, the REGIONS2 dataset achieves its best performance at
with an error rate of 28.02%, but other
values yield comparable results, such as 28.77% at both
and
. The data suggest that higher
values, particularly
, generally result in improved performance across most datasets. Nevertheless, the optimal
value may vary depending on specific dataset characteristics, and some datasets show negligible or inconsistent responses to changes in
. Overall,
is recommended for achieving the lowest average error.
Table 9 presents error rates for the regression datasets used under the different values of the periodic local search
(0.0025, 0.005, 0.01, and 0.02). The analysis indicates that the lowest average error is observed at
, with a value of 5.04, suggesting that this setting offers the best overall performance. This is followed by
, with an average error of 5.10,
with 5.56, and
with 6.34, which has the highest average error and the poorest performance. Examining individual datasets reveals variations in the impact of different
values. In the AUTO dataset, the error decreases steadily as
increases, dropping from 17.63 at
to its lowest point of 10.11 at
. Similarly, in the HOUSING dataset, the error significantly reduces from 32.72 at
to 14.02 at
. This decreasing trend is also evident in other datasets, such as MORTGAGE, where the error falls from 1.22 at
to just 0.032 at
, and STOCK, where the error reduces from 6.58 to 1.47. In some datasets, the
parameter has minimal impact. For instance, in the CONCRETE dataset, the error remains constant across all
values at 0.003. Similar stability is observed in the QUAKE and SN datasets, where variations are minimal. However, some datasets exhibit less predictable trends. In the BASEBALL dataset, the error initially decreases from 63.05 at
to 60.56 at
but then increases again to 71.93 at
. Similar inconsistent results are observed in the PY and BL datasets. Overall, the analysis suggests that
delivers the best average performance. Nevertheless, the effect of the
parameter varies across the datasets, with some benefiting more from higher or lower
values.
In
Figure 11, the pairwise comparisons using the Wilcoxon Test are presented to examine the impact of the different values of the local optimization parameter (
) on the proposed machine learning method, based on a series of well-known datasets, with the aim of evaluating the classification error. The Wilcoxon Test results showed that comparisons between
and
,
, and
demonstrated statistically significant differences, as the
p-values were lower than the significance level of 0.05. Specifically, the
p-value for the comparison between
and
was 0.0001, between
and
was 0.00034, and between
and
was 0.00048. Conversely, the comparisons between
and
, as well as between
and
, did not show statistically significant differences, with
p-values of 0.21 and 0.57, respectively. The comparison between
and
yielded a
p-value of 0.052, indicating a marginal lack of significance. Overall, the results suggest that the choice of the pl parameter value affects the classification error primarily in comparisons involving the value
, while the other comparisons do not exhibit clear statistically significant differences.
In
Figure 12, the regression error for the different values of the periodic local search parameter is presented. In the comparison between
and
, the
p-value was 0.011, indicating a statistically significant difference, as it is below the significance level of 0.05. In contrast, the comparisons between
and
, between
and
, between
and
, and between
and
did not show statistically significant differences, with
p-values of 0.12, 0.18, 0.22, and 0.097, respectively, all of which are above the significance level of 0.05. Finally, the comparison between
and
yielded a
p-value of 0.086, which is close to but above the significance level of 0.05, suggesting borderline non-significance. Overall, the results indicate that only the comparison between
and
showed a statistically significant difference, while the remaining comparisons did not present clear statistically significant differences.
3.2.4. Experiments with the Local Search Method
Another test was conducted, where the local optimization method of the proposed technique was altered. In this test, two local optimization methods—Limited Memory BFGS (LBFGS) [
112] and Adam [
17]—were used and the results were compared against those obtained by the application of the BFGS local optimization methods. The experimental results for the classification datasets are depicted in
Table 10, while the experimental results for the regression datasets are shown in
Table 11.
The analysis of
Table 10 regarding the local search methods BFGS, LBFGS, and ADAM for the NEURALDE model presents their performance on the classification datasets, with the values representing error percentages. According to the results, the BFGS method achieves the lowest average error rate at 19.78%, followed by LBFGS with 20.06% and ADAM with 21.48%. This highlights the relative superiority of BFGS in minimizing error compared to the other methods. When analyzing the individual datasets, BFGS records the lowest error rates in several cases. For instance, in the HOUSEVOTES dataset, both BFGS and LBFGS yield similar results, while ADAM shows the lowest error at 5.39%. In the SPAMBASE dataset, ADAM outperforms the others with an error rate of 3.91%, compared to 4.95% and 4.78% for BFGS and LBFGS, respectively. Similarly, in the STUDENT dataset, ADAM performs best with the lowest error at 3.82%. However, in datasets like SEGMENT, BFGS excels with a significantly lower error of 15.60% compared to 19.46% for LBFGS and 27.47% for ADAM. Notable differences are observed in datasets such as DERMATOLOGY, where LBFGS records the lowest error at 9.09%, compared to 10.38% for BFGS and 11.34% for ADAM. In the MAGIC dataset, BFGS and LBFGS deliver comparable performance, with error rates of 11.73% and 11.83%, respectively, while ADAM presents a higher error of 12.72%. Similarly, in the CIRCULAR dataset, BFGS achieves the lowest error at 4.23%, closely followed by LBFGS at 4.29%, and ADAM at 4.52%. Overall, BFGS demonstrates superior performance in most cases, maintaining the lowest variation in error rates and achieving the smallest average error. However, LBFGS delivers competitive results with a slightly higher average error and outperforms in specific datasets like DERMATOLOGY. While ADAM records lower error rates in isolated datasets such as SPAMBASE and STUDENT, it exhibits a higher overall error, indicating greater variability in its performance. In conclusion, the BFGS method emerges as the most efficient overall, while LBFGS offers consistently competitive performance. ADAM shows advantages in certain datasets but has a higher overall average error rate.
The evaluation presented in
Table 11 highlights the performance of the local search methods—BFGS, LBFGS, and ADAM—applied to the regression datasets within the NEURALDE model. The reported values correspond to absolute errors. Among the three methods, BFGS achieves the lowest mean error of 5.56, demonstrating a clear advantage over LBFGS and ADAM, which have mean errors of 6.03 and 8.27, respectively. Examining individual datasets, BFGS achieves superior results in several instances. For example, in the AIRFOIL dataset, it records the smallest error at 0.0014, while LBFGS and ADAM show slightly higher errors of 0.0024 and 0.0031, respectively. On the other hand, LBFGS outperforms in the BASEBALL dataset, achieving the lowest error at 49.78, compared to 60.56 for BFGS and 78.73 for ADAM. Similarly, LBFGS demonstrates the best performance in the BL dataset with an error of 0.01, outperforming BFGS and ADAM, which exhibit errors of 0.016 and 0.024, respectively. In the PLASTIC dataset, LBFGS again achieves the smallest error at 2.53, followed by BFGS with 3.27 and ADAM with 3.4. Conversely, in the MORTGAGE dataset, BFGS performs best, with an error of 0.54, while LBFGS and ADAM display larger errors of 1.54 and 2.01. In the STOCK dataset, BFGS also demonstrates superior performance, recording the lowest error at 3.4, whereas LBFGS and ADAM show significantly higher errors of 8.9 and 11.27, respectively. Certain datasets exhibit negligible differences in performance among the methods. For instance, in the CONCRETE dataset, all three methods produce identical errors of 0.003. Similarly, in the LASER dataset, the errors are nearly identical, with ADAM achieving 0.0025, BFGS achieving 0.0026, and LBFGS achieving 0.0027. Overall, BFGS emerges as the most effective method, delivering the lowest average error and consistently strong results across numerous datasets. LBFGS, while slightly less efficient on average, demonstrates competitive performance in specific datasets such as BASEBALL, BL, and PLASTIC. ADAM, though occasionally effective, as observed in datasets like LASER, shows greater variability and a higher mean error, limiting its overall reliability. In conclusion, BFGS stands out as the most robust and dependable method, with LBFGS showing significant potential in select contexts.
In
Figure 13, the critical parameter
p, which indicates the level of statistical significance, showed no statistically significant difference between the BFGS and LBFGS methods, with
p = 0.78. This high value does not meet the common significance threshold (
p < 0.05). In contrast, the comparison between BFGS and ADAM revealed a statistically significant difference, with
p = 0.0017, indicating clear differentiation between the two methods. Similarly, the comparison between LBFGS and ADAM recorded
p = 0.0031, which is also below the significance level, proving that the two methods exhibit statistically significant differences in classification error performance.
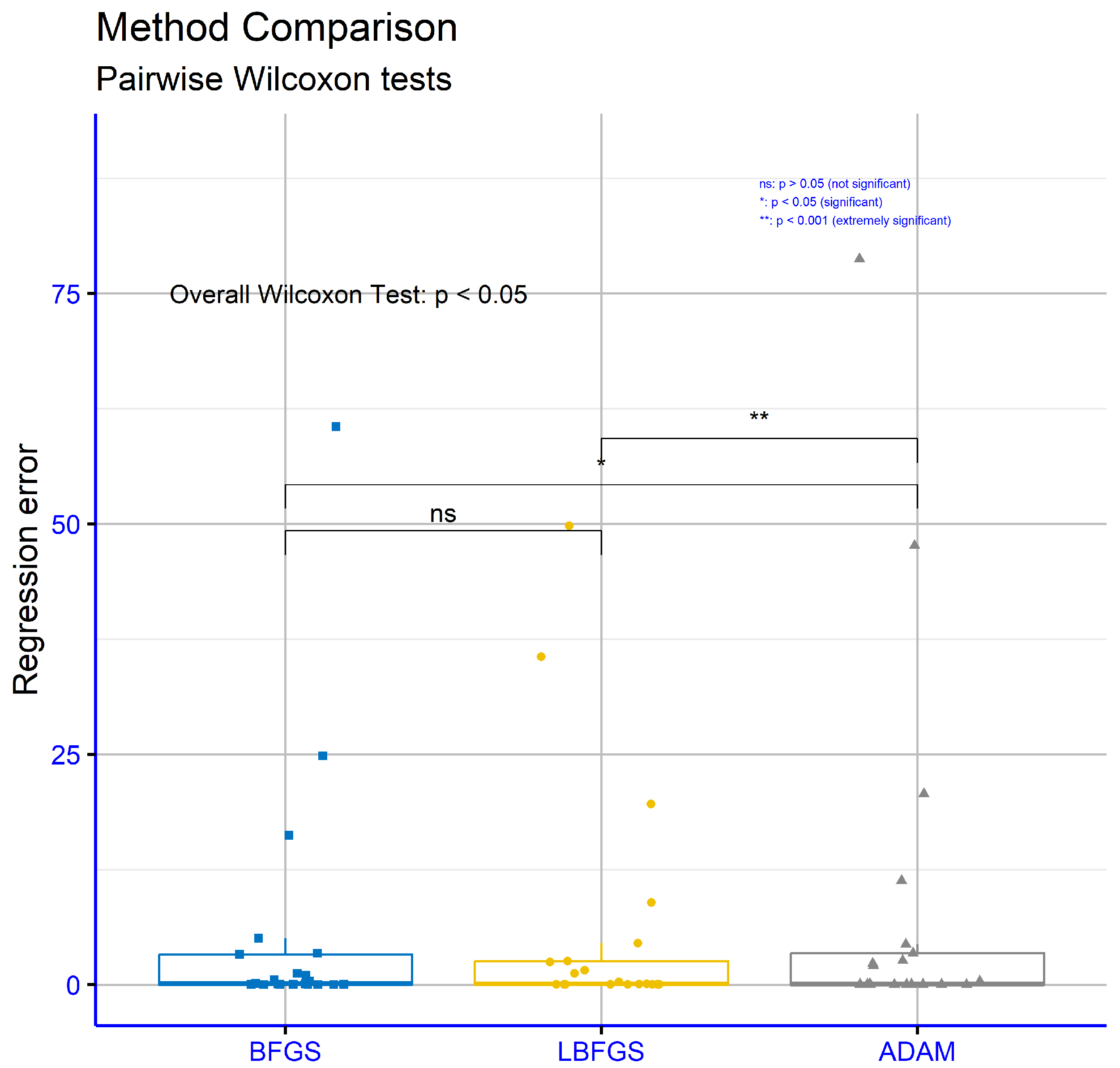
In
Figure 14, the results showed that the BFGS and LBFGS methods have no statistically significant difference, with
p = 1.0, suggesting that the two methods perform essentially the same regarding regression error. However, the comparison between BFGS and ADAM resulted in
p = 0.037, which is below the 0.05 threshold, highlighting a statistically significant difference favoring BFGS. Lastly, the comparison between LBFGS and ADAM recorded
p = 0.0054, confirming a statistically significant differentiation between these two methods as well. In summary, the results of the Wilcoxon Test demonstrate that BFGS and LBFGS have comparable performances in regression error, while they do not exhibit statistically significant differences in classification error. On the other hand, the differences between ADAM and the other two methods (BFGS and LBFGS) are statistically significant in both cases, indicating that ADAM shows distinct performance characteristics in both classification and regression error.