Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of the Compounds 1–7
2.2. Cell Lines
2.3. Viruses
2.4. Viral Replication
2.5. Viral Titration
2.6. Cytotoxicity Assay
2.7. Antiviral Activity Assay
2.8. Target Prediction
2.9. Binding Analysis of Ligands by Molecular Docking
2.10. Molecular Dynamics Simulations
3. Results
3.1. Antiviral Activity of Curcumin Analogues
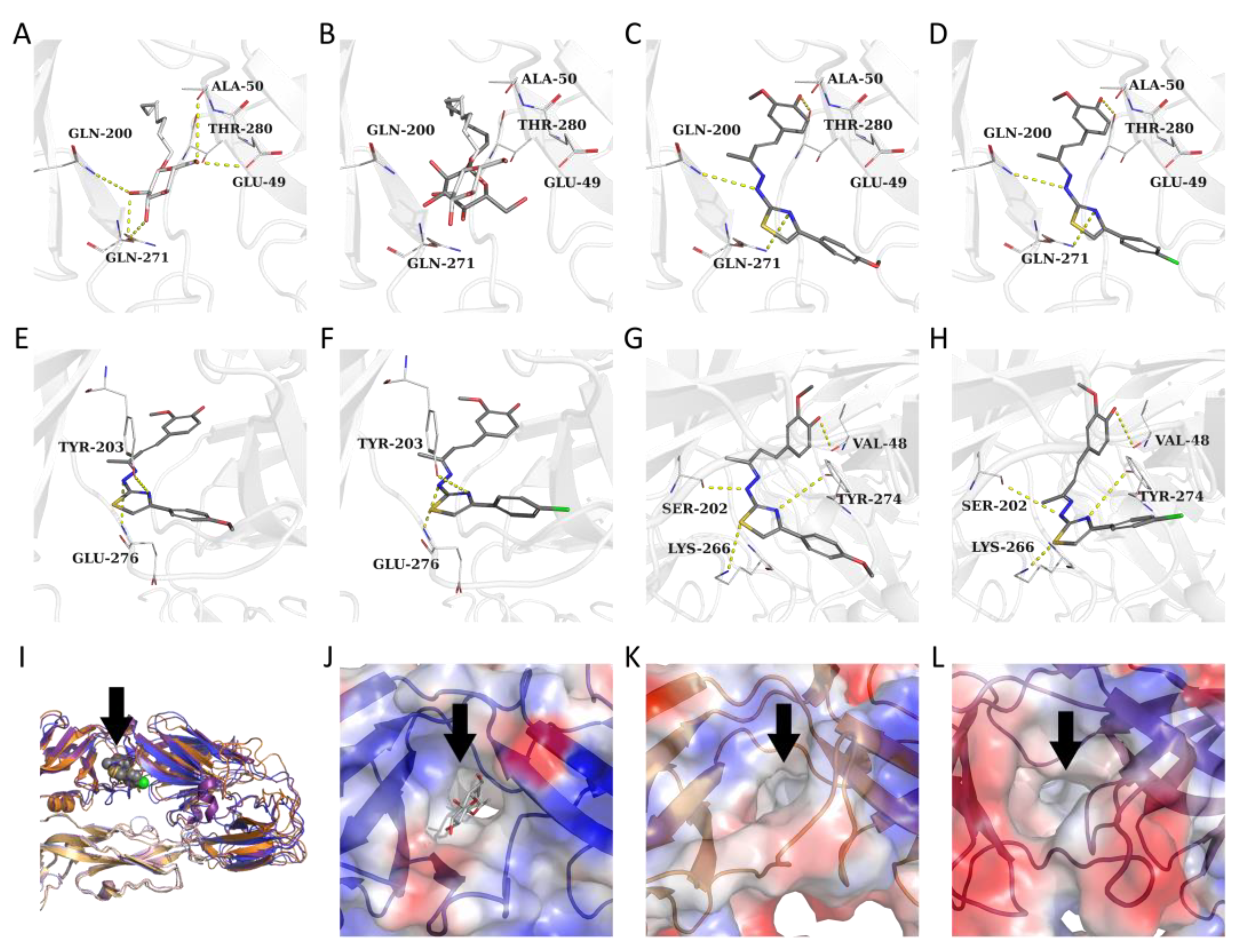
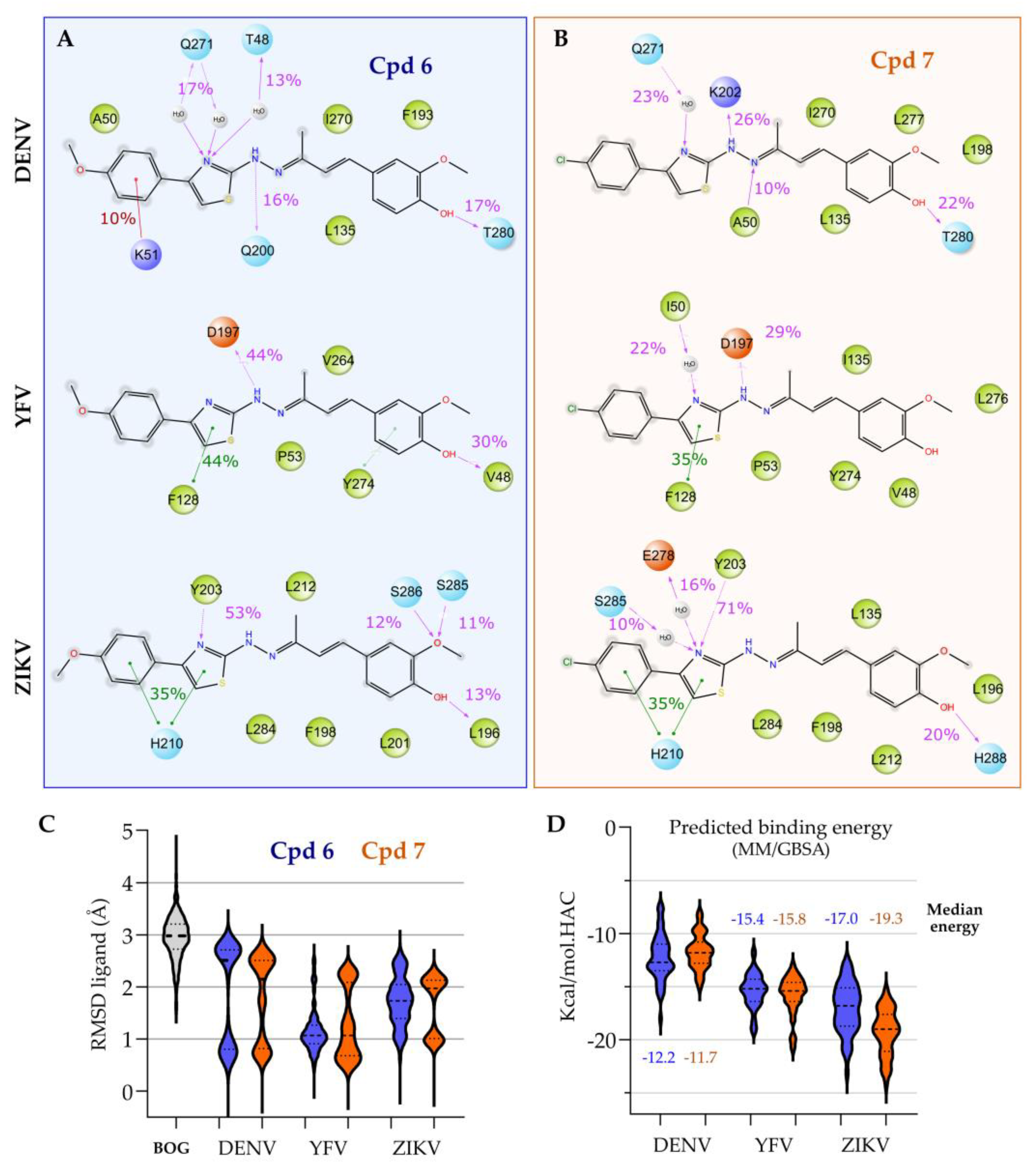
3.2. Target Prediction, Molecular Docking and Dynamics Simulations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef] [PubMed]
- Padilla-S, L.; Rodríguez, A.; Gonzales, M.M.; Gallego-G, J.C.; Castaño-O, J.C. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Arch. Virol. 2014, 159, 573–579. [Google Scholar] [CrossRef]
- Marbawati, D.; Umniyati, S.R. Effects of curcumin and pentagamavunon-0 against dengue-2 virus Infection in vero cells; an in vitro study. Procedia Environ. Sci. 2015, 23, 215–221. [Google Scholar] [CrossRef]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral Res. 2017, 142, 148–157. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Prioritizing Diseases for Research and Development in Emergency Contexts. Available online: https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts (accessed on 7 February 2023).
- World Health Organization (WHO). Surveillance. Available online: https://www.who.int/westernpacific/emergencies/surveillance (accessed on 7 February 2023).
- Nabila, N.; Suada, N.K.; Denis, D.; Yohan, B.; Adi, A.C.; Veterini, A.S.; Anindya, A.L.; Sasmono, R.T.; Rachmawati, H. Antiviral Action of Curcumin Encapsulated in Nanoemulsion against Four Serotypes of Dengue Virus. Pharm. Nanotechnol. 2020, 8, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Liu, W.; Zhai, Y.; Heng, X.; Che, F.Y.; Chen, W.; Sun, D.; Zhai, G. Oral bioavailability of curcumin: Problems and advancements. J. Drug Target. 2016, 24, 694–702. [Google Scholar] [CrossRef]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The Essential Medicinal Chemistry of Curcumin. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar] [CrossRef] [PubMed]
- Shoba, G.; Joy, D.; Joseph, T.; Majeed, M.; Rajendran, R.; Srinivas, P.S. Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med. 1998, 64, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Ratnatilaka Na Bhuket, P.; El-Magboub, A.; Haworth, I.S.; Rojsitthisak, P. Enhancement of Curcumin Bioavailability Via the Prodrug Approach: Challenges and Prospects. Eur. J. Drug Metab. Pharmacokinet. 2017, 42, 341–353. [Google Scholar] [CrossRef]
- Parvathy, K.S.; Negi, P.S.; Srinivas, P. Curcumin–amino acid conjugates: Synthesis, antioxidant and antimutagenic attributes. Food Chem. 2010, 120, 523–530. [Google Scholar] [CrossRef]
- Nimiya, Y.; Wang, W.; Du, Z.; Sukamtoh, E.; Zhu, J.; Decker, E.; Zhang, G. Redox modulation of curcumin stability: Redox active antioxidants increase chemical stability of curcumin. Mol. Nutr. Food Res. 2016, 60, 487–494. [Google Scholar] [CrossRef]
- Reid, J.M.; Buhrow, S.A.; Gilbert, J.A.; Jia, L.; Shoji, M.; Snyder, J.P.; Ames, M.M. Mouse pharmacokinetics and metabolism of the curcumin analog, 4-Piperidione,3,5-bis[(2-fluorophenyl)methylene]-acetate(3E,5E) (EF-24; NSC 716993). Cancer Chemother. Pharmacol. 2014, 73, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Khudhayer Oglah, M.; Fakri Mustafa, Y. Curcumin analogs: Synthesis and biological activities. Med. Chem. Res. 2020, 29, 479–486. [Google Scholar] [CrossRef]
- Braga, S.F.P.; Alves, É.V.P.; Ferreira, R.S.; Fradico, J.R.B.; Lage, P.S.; Duarte, M.C.; Ribeiro, T.G.; Júnior, P.A.S.; Romanha, A.J.; Tonini, M.L.; et al. Synthesis and evaluation of the antiparasitic activity of bis-(arylmethylidene) cycloalkanones. Eur. J. Med. Chem. 2014, 71, 282–289. [Google Scholar] [CrossRef]
- Costa, S.O.A.M.; Rodrigues, I.B.; Braga, A.V.; Barbosa, B.C.M.; Silva, R.R.L.; Rodrigues, F.F.; Melo, I.S.F.; Morais, M.Í.; Castro, B.F.M.; Cunha, A.S., Jr.; et al. RI75, a curcumin analogue, inhibits tumor necrosis factor-α and interleukin-6 production and exhibits antiallodynic and antiedematogenic activities in mice. Inflammopharmacology 2022, 30, 505–515. [Google Scholar] [CrossRef]
- Lino, C.I.; Gonçalves de Souza, I.; Borelli, B.M.; Silvério Matos, T.T.; Santos Teixeira, I.N.; Ramos, J.P.; Maria de Souza Fagundes, E.; de Oliveira Fernandes, P.; Maltarollo, V.G.; Johann, S.; et al. Synthesis, molecular modeling studies and evaluation of antifungal activity of a novel series of thiazole derivatives. Eur. J. Med. Chem. 2018, 151, 248–260. [Google Scholar] [CrossRef]
- Turan-Zitouni, G.; Özdemir, A.; Kaplancıklı, Z.A.; Fehrentz, J.-A.; Martinez, J.; Chevallet, P.; Dusart, G. Preparation of Some Thiazolyl Hydrazone Derivatives and Evaluation of Their Antibacterial Activities. Phosphorus Sulfur Silicon Relat. Elem. 2009, 184, 2613–2623. [Google Scholar] [CrossRef]
- Kaplancıklı, Z.A.; Sever, B.; Altıntop, M.D.; Atlı, O.; Baysal, M.; Ozdemir, A. Synthesis and Evaluation of New Thiazolyl Hydrazone Derivatives as Potential Anticancer Agents. Lett. Drug Des. Discov. 2017, 14, 672–677. [Google Scholar] [CrossRef]
- Cui, M.-Y.; Nie, J.-X.; Yan, Z.-Z.; Xiao, M.-W.; Lin, D.; Ye, J.; Hu, A.-X. Design, synthesis, bioactivity, and DFT calculation of 2-thiazolyl-hydrazone derivatives as influenza neuraminidase inhibitors. Med. Chem. Res. 2019, 28, 938–947. [Google Scholar] [CrossRef]
- Njoroge, F.G.; Chen, K.X.; Shih, N.-Y.; Piwinski, J.J. Challenges in modern drug discovery: A case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection. Acc. Chem. Res. 2008, 41, 50–59. [Google Scholar] [CrossRef]
- Wlodawer, A. Rational approach to AIDS drug design through structural biology. Annu. Rev. Med. 2002, 53, 595–614. [Google Scholar] [CrossRef] [PubMed]
- Serafim, M.S.M.; Santos, V.S.d., Jr.; Gertrudes, J.C.; Maltarollo, V.G.; Honorio, K.M. Machine learning techniques applied to the drug design and discovery of new antivirals: A brief look over the past decade. Expert Opin. Drug Discov. 2021, 16, 961–975. [Google Scholar] [CrossRef]
- Donald, C.L.; Brennan, B.; Cumberworth, S.L.; Rezelj, V.V.; Clark, J.J.; Cordeiro, M.T.; França, R.F.O.; Pena, L.J.; Wilkie, G.S.; Filipe, A.S.; et al. Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. Plos. Negl. Trop. Dis. 2016, 10, e0005048. [Google Scholar] [CrossRef]
- Figueiredo, L.B.; Sakamoto, T.; Coelho, L.F.L.; Rocha, E.S.O.; Gomes Cota, M.M.; Ferreira, G.P.; de Oliveira, J.G.; Kroon, E.G. Dengue Virus 2 American-Asian Genotype Identified during the 2006/2007 Outbreak in Piauí, Brazil Reveals a Caribbean Route of Introduction and Dissemination of Dengue Virus in Brazil. PLoS ONE 2014, 8, e104516. [Google Scholar] [CrossRef]
- Coelho, S.V.A.; Neris, R.L.S.; Papa, M.P.; Schnellrath, L.C.; Meuren, L.M.; Tschoeke, D.A.; Leomil, L.; Verçoza, B.R.F.; Miranda, M.; Thompson, F.L.; et al. Development of standard methods for Zika virus propagation, titration, and purification. J. Virol. Methods 2017, 246, 65–74. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Vallone, A.; D’Alessandro, S.; Brogi, S.; Brindisi, M.; Chemi, G.; Alfano, G.; Lamponi, S.; Lee, S.G.; Jez, J.M.; Koolen, K.J.M.; et al. Antimalarial agents against both sexual and asexual parasites stages: Structure-activity relationships and biological studies of the Malaria Box compound 1-[5-(4-bromo-2-chlorophenyl)furan-2-yl]-N-[(piperidin-4-yl)methyl]methanamine (MMV019918) and analogues. Eur. J. Med. Chem. 2018, 150, 698–718. [Google Scholar] [CrossRef]
- Serafim, M.S.M.; Lavorato, S.N.; Kronenberger, T.; Sousa, Y.V.; Oliveira, G.P.; dos Santos, S.G.; Kroon, E.G.; Maltarollo, V.G.; Alves, R.J.; Mota, B.E.F. Antibacterial activity of synthetic 1,3-bis(aryloxy)propan-2-amines against Gram-positive bacteria. Microbiologyopen 2019, 8, e814. [Google Scholar] [CrossRef] [PubMed]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the protein databank and cambridge structural database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- OpenEye, Cadence Molecular Sciences. OMEGA 3.1.2—Applications. Available online: https://docs.eyesopen.com/applications/omega/releasenotes/version3_1_2.html (accessed on 7 February 2023).
- Hawkins, P.C.D.; Skillman, A.G.; Nicholls, A. Comparison of Shape-Matching and Docking as Virtual Screening Tools. J. Med. Chem. 2007, 50, 74–82. [Google Scholar] [CrossRef] [PubMed]
- OpenEye, Cadence Molecular Sciences. ROCS 3.3.0—Applications. Available online: https://docs.eyesopen.com/applications/rocs/releasenotes/version3_3_0.html (accessed on 7 February 2023).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Azevedo, L.; Serafim, M.S.M.; Maltarollo, V.G.; Grabrucker, A.M.; Granato, D. Atherosclerosis fate in the era of tailored functional foods: Evidence-based guidelines elicited from structure- and ligand-based approaches. Trends Food Sci. Technol. 2022, 128, 75–89. [Google Scholar] [CrossRef]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 2003, 100, 6986–6991. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Song, J.; Lu, X.; Deng, Y.-Q.; Musyoki, A.M.; Cheng, H.; Zhang, Y.; Yuan, Y.; Song, H.; Haywood, J.; et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xiao, H.; Li, S.; Pang, X.; Song, J.; Liu, S.; Cheng, H.; Li, Y.; Wang, X.; Huang, C.; et al. Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope. Cell Rep. 2019, 26, 438–446.e5. [Google Scholar] [CrossRef]
- Dror, R.O.; Jensen, M.Ø.; Borhani, D.W.; Shaw, D.E. Exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. J. Gen. Physiol. 2010, 135, 555–562. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tuckerman, M.E.; Tobias, D.J.; Klein, M.L. Explicit reversible integrators for extended systems dynamics. Mol. Phys. 1996, 87, 1117–1157. [Google Scholar] [CrossRef]
- Tuckerman, M.E.; Marx, D.; Klein, M.L.; Parrinello, M. Efficient and general algorithms for path integral Car–Parrinello molecular dynamics. J. Chem. Phys. 1996, 104, 5579–5588. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef]
- Adasme-Carreño, F.; Muñoz-Gutierrez, C.; Caballero, J.; Alzate-Morales, J.H. Performance of the MM/GBSA scoring using a binding site hydrogen bond network-based frame selection: The protein kinase case. Phys. Chem. Chem. Phys. 2014, 16, 14047–14058. [Google Scholar] [CrossRef]
- Jadav, S.S.; Kaptein, S.; Timiri, A.; De Burghgraeve, T.; Badavath, V.N.; Ganesan, R.; Sinha, B.N.; Neyts, J.; Leyssen, P.; Jayaprakash, V. Design, synthesis, optimization and antiviral activity of a class of hybrid dengue virus E protein inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 1747–1752. [Google Scholar] [CrossRef]
- Tuberculosis Drug Screening Program. Search for New Drugs for Treatment of Tuberculosis. Antimicrob. Agents Chemother. 2001, 45, 1943. [Google Scholar] [CrossRef]
- Nyström, K.; Waldenström, J.; Tang, K.-W.; Lagging, M. Ribavirin: Pharmacology, multiple modes of action and possible future perspectives. Future Virol. 2019, 14, 153–160. [Google Scholar] [CrossRef]
- Talemi, S.R.; Bartenschlager, M.; Schmid, B.; Ruggieri, A.; Bartenschlager, R.; Höfer, T. Dengue virus is sensitive to inhibition prior to productive replication. Cell Rep. 2021, 37, 109801. [Google Scholar] [CrossRef] [PubMed]
- Pires de Mello, C.P.; Tao, X.; Kim, T.H.; Bulitta, J.B.; Rodriquez, J.L.; Pomeroy, J.J.; Brown, A.N. Zika Virus Replication Is Substantially Inhibited by Novel Favipiravir and Interferon Alpha Combination Regimens. Antimicrob. Agents Chemother. 2018, 62, e01983-17. [Google Scholar] [CrossRef]
- Nyström, K.; Wanrooij, P.H.; Waldenström, J.; Adamek, L.; Brunet, S.; Said, J.; Nilsson, S.; Wind-Rotolo, M.; Hellstrand, K.; Norder, H.; et al. Inosine Triphosphate Pyrophosphatase Dephosphorylates Ribavirin Triphosphate and Reduced Enzymatic Activity Potentiates Mutagenesis in Hepatitis C Virus. J. Virol. 2018, 92, e01087-18. [Google Scholar] [CrossRef]
- Wiji Prasetyaningrum, P.; Bahtiar, A.; Hayun, H. Synthesis and Cytotoxicity Evaluation of Novel Asymmetrical Mono-Carbonyl Analogs of Curcumin (AMACs) against Vero, HeLa, and MCF7 Cell Lines. Sci. Pharm. 2018, 86, 25. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, A.; Pilankatta, R.; Teramoto, T.; Sajith, A.M.; Nwulia, E.; Kulkarni, A.; Padmanabhan, R. Inhibition of dengue virus by curcuminoids. Antiviral Res. 2019, 162, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Yusufzai, S.K.; Osman, H.; Khan, M.S.; Razik, B.M.A.; Mohamad, S.; Sulaiman, O.; Gansau, J.A.; Johansah, N.; Ezzat, M.O.; Parumasivam, T.; et al. Synthesis, X-ray crystallographic study, pharmacology and docking of hydrazinyl thiazolyl coumarins as dengue virus NS2B/NS3 serine protease inhibitors. Med. Chem. Res. 2018, 27, 1647–1665. [Google Scholar] [CrossRef]
- Singh, R.K.; Rai, D.; Yadav, D.; Bhargava, A.; Balzarini, J.; De Clercq, E. Synthesis, antibacterial and antiviral properties of curcumin bioconjugates bearing dipeptide, fatty acids and folic acid. Eur. J. Med. Chem. 2010, 45, 1078–1086. [Google Scholar] [CrossRef]
- Rajitha, G.; Ravibabu, V.; Ramesh, G.; Rajitha, B.; Jobina, R.; Siddhardha, B.; Vijaya Laxmi, S. One-pot multicomponent synthesis of novel thiazolylhydrazone derivatives and their antimicrobial activity. Res. Chem. Intermed. 2015, 41, 9703–9713. [Google Scholar] [CrossRef]
- Cruz, L.I.B.; Lopes, L.F.F.; de Camargo Ribeiro, F.; de Sá, N.P.; Lino, C.I.; Tharmalingam, N.; de Oliveira, R.B.; Rosa, C.A.; Mylonakis, E.; Fuchs, B.B.; et al. Anti-Candida albicans Activity of Thiazolylhydrazone Derivatives in Invertebrate and Murine Models. J. Fungi Basel Switz. 2018, 4, 134. [Google Scholar] [CrossRef]
- Nazim, U.; Ahmed, M.; Ali, B.; Khan, K.M.; Ali, M.; Kobarfard, F.; Arshia; Ayatollahi, S.A.-M.; Hassan, A.; Jabeen, A.; et al. Thiosemicarbazone and thiazolylhydrazones of 1-indanone: As a new class of nonacidic anti-inflammatory and antiplatelet aggregation agents. Pak. J. Pharm. Sci. 2019, 32, 15–19. [Google Scholar] [PubMed]
- Zhang, Y.; Fu, X.; Yan, Y.; Liu, J. Microwave-assisted synthesis and biological evaluation of new thiazolylhydrazone derivatives as tyrosinase inhibitors and antioxidants. J. Heterocycl. Chem. 2020, 57, 991–1002. [Google Scholar] [CrossRef]
- de Noronha, M.C.; Cardoso, R.R.; dos Santos D’Almeida, C.T.; Vieira do Carmo, M.A.; Azevedo, L.; Maltarollo, V.G.; Ribeiro, J.I., Jr.; Eller, M.R.; Cameron, L.C.; Ferreira, M.S.L.; et al. Black tea kombucha: Physicochemical, microbiological and comprehensive phenolic profile changes during fermentation, and antimalarial activity. Food Chem. 2022, 384, 132515. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.-Y.; Wouters, R.; Schor, S.; Rozenski, J.; Barouch-Bentov, R.; Prugar, L.I.; O’Brien, C.M.; Brannan, J.M.; Dye, J.M.; Herdewijn, P.; et al. Optimization of Isothiazolo[4,3-b]pyridine-Based Inhibitors of Cyclin G Associated Kinase (GAK) with Broad-Spectrum Antiviral Activity. J. Med. Chem. 2018, 61, 6178–6192. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-Y.; Chen, D.-Y.; Wen, H.-W.; Ou, J.-L.; Chiou, S.-S.; Chen, J.-M.; Wong, M.-L.; Hsu, W.-L. Inhibition of Enveloped Viruses Infectivity by Curcumin. PLoS ONE 2013, 8, e62482. [Google Scholar] [CrossRef]
- Shakour, N.; Cabezas, R.; Santos, J.G.; Barreto, G.E.; Jamialahmadi, T.; Hadizadeh, F.; Sahebkar, A. Curcumin Can Bind and Interact with CRP: An in silico Study. Adv. Exp. Med. Biol. 2021, 1308, 91–100. [Google Scholar] [CrossRef]
- Colpitts, C.C.; Schang, L.M.; Rachmawati, H.; Frentzen, A.; Pfaender, S.; Behrendt, P.; Brown, R.J.P.; Bankwitz, D.; Steinmann, J.; Ott, M.; et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut 2014, 63, 1137–1149. [Google Scholar] [CrossRef]
- Miller, R.H.; Purcell, R.H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 1990, 87, 2057–2061. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cortese, M.; Acosta, E.G.; Bartenschlager, R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018, 16, 125–142. [Google Scholar] [CrossRef]
- Ferreira, L.L.C.; Abreu, M.P.; Costa, C.B.; Leda, P.O.; Behrens, M.D.; dos Santos, E.P. Curcumin and Its Analogs as a Therapeutic Strategy in Infections Caused by RNA Genome Viruses. Food Environ. Virol. 2022, 14, 120–137. [Google Scholar] [CrossRef] [PubMed]
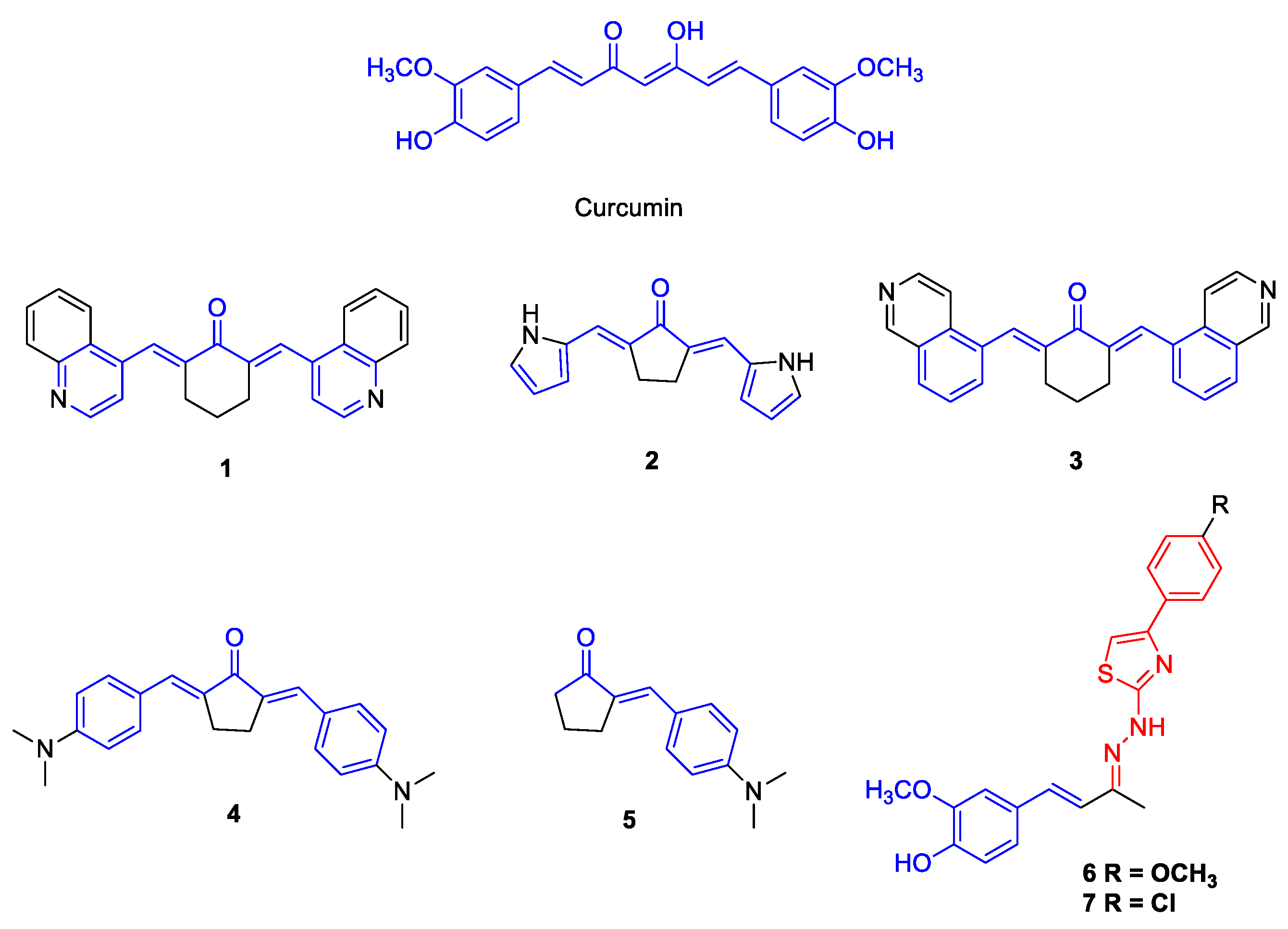
| Compounds | CC50 (µM) | ZIKV (PE243) | YFV (17DD) | DENV-2 (PI59) | |||
|---|---|---|---|---|---|---|---|
| EC50 (µM) | SI a | EC50 (µM) | SI a | EC50 (µM) | SI a | ||
| Curcumin | 22.76 ± 2.84 | 17.12 ± 1.2 | 1.33 | - | - | 13.63 ± 0.95 | 1.67 |
| 1 | <6.25 | - | - | - | - | - | - |
| 2 | >100 | 32.45 ± 1.58 | >3.08 | - | - | - | - |
| 3 | <6.25 | - | - | - | - | - | - |
| 4 | >100 | - | - | - | - | - | - |
| 5 | >100 | - | - | - | - | - | - |
| 6 | 17.7 ± 0.7 | 8.61 ± 0.41 | 2.06 | 10.85 ± 0.13 | 1.63 | 12.5 * | 1.42 |
| 7 | 17.98 ± 2.47 | 4.04 ± 0.38 | 4.45 | 11.94 ± 0.39 | 1.51 | 15 * | 1.2 |
| Ribavirin | >100 | 4.1 ± 0.35 | >24.39 | 40.9 ± 3.49 | >2.44 | - | - |
| TanimotoCombo | |||||
|---|---|---|---|---|---|
| ChEMBL ID | 6 | 7 | Related Target | Reference | PDB ID |
 CHEMBL3401565 | 1.08 | - | E protein | Jadav et al. 2015 [54] | 1OKE |
 CHEMBL3401563 | - | 1.03 | |||
 CHEMBL3401564 | - | 1.03 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serafim, M.S.M.; Kronenberger, T.; de Oliveira, R.B.; Kroon, E.G.; Abrahão, J.S.; Mota, B.E.F.; Maltarollo, V.G. Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety. Future Pharmacol. 2023, 3, 364-378. https://doi.org/10.3390/futurepharmacol3020022
Serafim MSM, Kronenberger T, de Oliveira RB, Kroon EG, Abrahão JS, Mota BEF, Maltarollo VG. Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety. Future Pharmacology. 2023; 3(2):364-378. https://doi.org/10.3390/futurepharmacol3020022
Chicago/Turabian StyleSerafim, Mateus Sá Magalhães, Thales Kronenberger, Renata Barbosa de Oliveira, Erna Geessien Kroon, Jônatas Santos Abrahão, Bruno Eduardo Fernandes Mota, and Vinícius Gonçalves Maltarollo. 2023. "Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety" Future Pharmacology 3, no. 2: 364-378. https://doi.org/10.3390/futurepharmacol3020022
APA StyleSerafim, M. S. M., Kronenberger, T., de Oliveira, R. B., Kroon, E. G., Abrahão, J. S., Mota, B. E. F., & Maltarollo, V. G. (2023). Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety. Future Pharmacology, 3(2), 364-378. https://doi.org/10.3390/futurepharmacol3020022






