Seasonal and Spatial Microbial Community Dynamics Along the Shallow Southwest Florida Continental Shelf
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Water Quality
2.3. Elemental and Stable Isotope Measurements
2.4. 16S rRNA Amplicon Sequencing and Data Processing
2.5. Statistical Analysis
3. Results
3.1. Environmental Parameters
3.1.1. Water Quality and Nutrient Data
3.1.2. Sinking Particles and Surface Sediment
3.2. Diversity Overview
3.2.1. Alpha Diversity
3.2.2. Beta Diversity
3.3. Seasonal Comparison of Taxonomic Composition Among Microbial Habitats
3.3.1. Major Taxonomic Groups
3.3.2. Functional Taxonomic Groups
3.4. Shared ASV Communities
3.4.1. Inshore–Offshore Spatiotemporal Patterns of ASVs Among Three Microbial Habitats
3.4.2. Dominant ASVs Spatiotemporal Comparison of Sediment and Water Column Associated Dominant ASVs Between Three Microbial Habitats
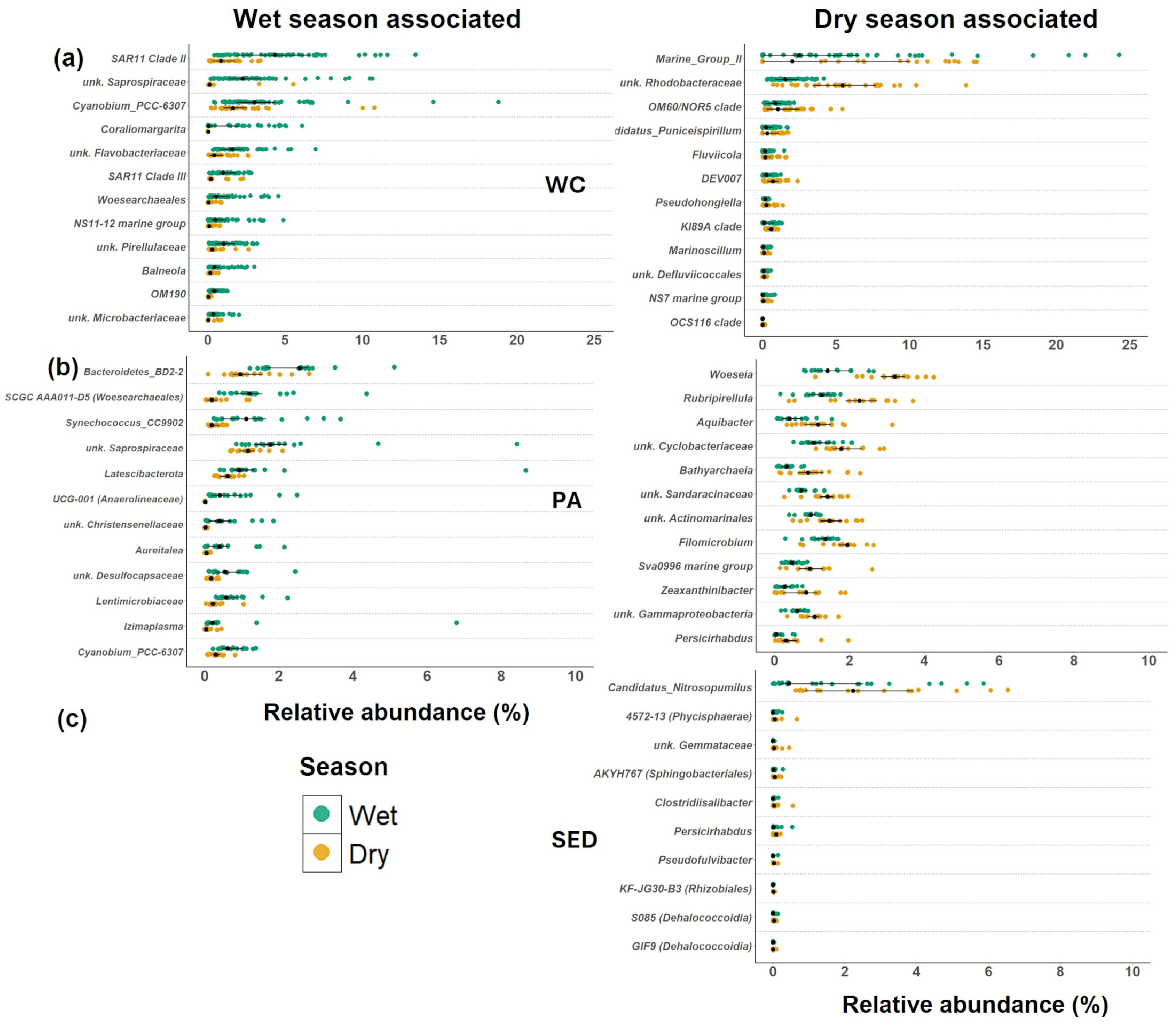
3.4.3. Indicator Bacteria in Inshore–Offshore Gradient
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AOA | Ammonia-Oxidizing Archaea |
| AOB | Ammonia-Oxidizing Bacteria |
| ASV | Amplicon Sequence Variant |
| CDOM | Chromophoric Dissolved Organic Matter |
| Chl-a | Chlorophyll-a |
| CR | Caloosahatchee River |
| CRE | Caloosahatchee River Estuary |
| CSTB | Cross-Shelf Transport Barrier |
| DO | Dissolved Oxygen |
| DOM | Dissolved Organic Matter |
| FGCU | Florida Gulf Coast University |
| FIO | Florida Institute of Oceanography |
| LEfSe | Linear Discriminant Analysis Effect Size |
| PA | Particle-Associated |
| SED | Surface Sediment |
| SPOM | Suspended Particulate Organic Matter |
| SRB | Sulfate-Reducing Bacteria |
| SWFL | Southwest Florida |
| TN | Total Nitrogen |
| TOC | Total Organic Carbon |
| TP | Total Phosphorous |
| WC | Water Column |
References
- Häder, D.-P.; Banaszak, A.T.; Villafañe, V.E.; Narvarte, M.A.; González, R.A.; Helbling, E.W. Anthropogenic pollution of aquatic ecosystems: Emerging problems with global implications. Sci. Total Environ. 2020, 713, 136586. [Google Scholar] [CrossRef]
- Malone, T.C.; Newton, A. The globalization of cultural eutrophication in the coastal ocean: Causes and consequences. Front. Mar. Sci. 2020, 7, 670. [Google Scholar] [CrossRef]
- Doering, P.H.; Chamberlain, R.H. Water quality and source of freshwater discharge to the Caloosahatchee estuary, Florida. J. Am. Water Resour. Assoc. 1999, 35, 793–806. [Google Scholar] [CrossRef]
- Barnes, T. Caloosahatchee estuary conceptual ecological model. Wetlands 2005, 25, 884–897. [Google Scholar] [CrossRef]
- Brewton, R.A.; Kreiger, L.B.; Tyre, K.N.; Baladi, D.; Wilking, L.E.; Herren, L.W.; Lapointe, B.E. Septic system–groundwater–surface water couplings in waterfront communities contribute to harmful algal blooms in southwest Florida. Sci. Total Environ. 2022, 837, 155319. [Google Scholar] [CrossRef]
- Shaddox, T.W.; Unruh, J.B.; Johnson, M.E.; Brown, C.D.; Stacey, G. Nutrient use and management practices on United States golf courses. Horttechnology 2023, 33, 79–97. [Google Scholar] [CrossRef]
- Buzzelli, C.; Wachnicka, A.; Zheng, F.; Chen, Z.; Baldwin, L.; Kahn-Dickens, A. Chapter 8C: St. Lucie and Caloosahatchee River watershed research and water quality monitoring results and activities. In South Florida Environmental Report; South Florida Water Management District (SFWMD): West Palm Beach, FL, USA, 2018; Volume 1, pp. 1–48. [Google Scholar]
- Rumbold, D.G.; Doering, P.H. Water quality and source of freshwater discharge to the Caloosahatchee estuary, Florida: 2009–2018. Fla. Sci. 2020, 83, 1–20. [Google Scholar]
- Olascoaga, M.J.; Rypina, I.I.; Brown, M.G.; Beron-Vera, F.J.; Koçak, H.; Brand, L.E.; Halliwell, G.R.; Shay, L.K. Persistent transport barrier on the West Florida Shelf. Geophys. Res. Lett. 2006, 33, 2006GL027800. [Google Scholar] [CrossRef]
- Yang, H.; Weisberg, R.H.; Niiler, P.P.; Sturges, W.; Johnson, W. Lagrangian circulation and forbidden zone on the West Florida Shelf. Cont. Shelf Res. 1999, 19, 1221–1245. [Google Scholar] [CrossRef]
- Satinsky, B.M.; Smith, C.B.; Sharma, S.; Landa, M.; Medeiros, P.M.; Coles, V.J.; Yager, P.L.; Crump, B.C.; Moran, M.A. Expression patterns of elemental cycling genes in the Amazon River plume. ISME J. 2017, 11, 1852–1864. [Google Scholar] [CrossRef]
- Romero, E.; Garnier, J.; Lassaletta, L.; Billen, G.; Le Gendre, R.; Riou, P.; Cugier, P. Large-scale patterns of river inputs in southwestern Europe: Seasonal and interannual variations and potential eutrophication effects at the coastal zone. Biogeochemistry 2013, 113, 481–505. [Google Scholar] [CrossRef]
- Buzzelli, C.; Doering, P.; Wan, Y.; Sun, D. Modeling ecosystem processes with variable freshwater inflow to the Caloosahatchee River Estuary, southwest Florida. II. Nutrient loading, submarine light, and seagrasses. Estuar. Coast. Shelf Sci. 2014, 151, 272–284. [Google Scholar] [CrossRef]
- Douglass, J.G.; Chamberlain, R.H.; Wan, Y.; Doering, P.H. Submerged vegetation responses to climate variation and altered hydrology in a subtropical estuary: Interpreting 33 years of change. Estuaries Coast 2020, 43, 1406–1424. [Google Scholar] [CrossRef] [PubMed]
- Julian II, P.; Thompson, M. Dark waters: Evaluating seagrass community response to optical water quality and freshwater discharges in a highly managed subtropical estuary. Reg. Stud. Mar. Sci. 2024, 69, 103302. [Google Scholar] [CrossRef]
- Brand, L.E.; Compton, A. Long-term increase in Karenia brevis abundance along the southwest Florida coast. Harmful Algae 2007, 6, 232–252. [Google Scholar] [CrossRef] [PubMed]
- Yentsch, C.S.; Lapointe, B.E.; Poulton, N.; Phinney, D.A. Anatomy of a red tide bloom off the southwest coast of Florida. Harmful Algae 2008, 7, 817–826. [Google Scholar] [CrossRef]
- Milbrandt, E.C.; Martignette, A.J.; Thompson, M.A.; Bartleson, R.D.; Phlips, E.J.; Badylak, S.; Nelson, N.G. Geospatial distribution of hypoxia associated with a Karenia brevis bloom. Estuar. Coast Shelf Sci. 2021, 259, 107446. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; DeLong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Fei, C.; Booker, A.; Klass, S.; Vidyarathna, N.K.; Ahn, S.H.; Mohamed, A.R.; Arshad, M.; Glibert, P.M.; Heil, C.A.; Martínez Martínez, J.; et al. Friends and foes: Symbiotic and algicidal bacterial influence on Karenia brevis blooms. ISME Commun. 2025, 5, ycae164. [Google Scholar] [CrossRef]
- Urakawa, H.; Adhikari, P.L.; Urakawa, H.E.; Bartleson, R.D.; Rumbold, D.G. Microbial assessment of the ecological linkage between a red tide of Karenia brevis and bottom water anoxia off the coast of Fort Myers and Sanibel Island, Florida. Reg. Stud. Mar. Sci. 2024, 78, 103765. [Google Scholar] [CrossRef]
- King, G.M.; Smith, C.; Tolar, B.; Hollibaugh, J.T. Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the Northern Gulf of Mexico. Front. Microbiol. 2013, 3, 438. [Google Scholar] [CrossRef] [PubMed]
- Urakawa, H.; Martens-Habbena, W.; Huguet, C.; de la Torre, J.R.; Ingalls, A.E.; Devol, A.H.; Stahl, D.A. Ammonia availability shapes the seasonal distribution and activity of archaeal and bacterial ammonia oxidizers in the Puget Sound estuary. Limnol. Oceanogr. 2014, 59, 1321–1335. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Urakawa, H.; Bernhard, A.E. Wetland management using microbial indicators. Ecol. Eng. 2017, 108, 456–476. [Google Scholar] [CrossRef]
- Wang, Z.; Juarez, D.L.; Pan, J.; Blinebry, S.K.; Gronniger, J.; Clark, J.S.; Johnson, Z.I.; Hunt, D.E. Microbial communities across nearshore to offshore coastal transects are primarily shaped by distance and temperature. Environ. Microbiol. 2019, 21, 3862–3872. [Google Scholar] [CrossRef]
- Basili, M.; Campanelli, A.; Frapiccini, E.; Luna, G.M.; Quero, G.M. Occurrence and distribution of microbial pollutants in coastal areas of the adriatic sea influenced by river discharge. Environ. Pollut. 2021, 285, 117672. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Eisen, J.A.; Penn, K.; Allison, S.D.; Horner-Devine, M.C. Drivers of bacterial β-diversity depend on spatial scale. Proc. Natl. Acad. Sci. USA 2011, 108, 7850–7854. [Google Scholar] [CrossRef]
- Mönnich, J.; Tebben, J.; Bergemann, J.; Case, R.; Wohlrab, S.; Harder, T. Niche-based assembly of bacterial consortia on the diatom thalassiosira rotula is stable and reproducible. ISME J. 2020, 14, 1614–1625. [Google Scholar] [CrossRef]
- Jackrel, S.L.; Yang, J.W.; Schmidt, K.C.; Denef, V.J. Host specificity of microbiome assembly and its fitness effects in phytoplankton. ISME J. 2021, 15, 774–788. [Google Scholar] [CrossRef]
- Baker, D.; Lauer, J.; Ortega, A.; Jackrel, S.L.; Denef, V.J. Effects of phycosphere bacteria on their algal host are host species-specific and not phylogenetically conserved. Microorganisms 2022, 11, 62. [Google Scholar] [CrossRef]
- Xiao, X.; Pei, M.; Liu, X.; Zhao, Y.; Liang, Y. Planktonic algal bloom significantly alters sediment bacterial community structure. J. Soils Sediments 2017, 17, 2547–2556. [Google Scholar] [CrossRef]
- Novitsky, J. Evidence for sedimenting particles as the origin of the microbial community in a coastal marine sediment. Mar. Ecol. Prog. Ser. 1990, 60, 161–167. [Google Scholar] [CrossRef]
- Urakawa, H.; Yoshida, T.; Nishimura, M.; Ohwada, K. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16s rdna-based approaches and quinone profiling. Environ. Microbiol. 2000, 2, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Y.P.; Gao, S.; Wang, X.H.; Shi, B.W.; Zhou, L.; Wang, D.D.; Dai, C.; Li, G.C. Sediment resuspension in tidally dominated coastal environments: New insights into the threshold for initial movement. Ocean Dyn. 2016, 66, 401–417. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rrna primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The silva ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Wheeler, B.; Torchiano, M.; Torchiano, M.M. Package ‘lmPerm’; Version 2.1.0; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2025. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 24 March 2025).
- Wood, S.N.; Pya, N.; Säfken, B. Smoothing parameter and model selection for general smooth models. J. Am. Stat. Assoc. 2016, 111, 1548–1563. [Google Scholar] [CrossRef]
- Gao, C.-H.; Chen, C.; Dusa, A.; Yu, G.; Cao, B.; Cai, P. ggVennDiagram: Intuitive Venn Diagram Software Extended—Gao—2024—iMeta—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/imt2.177 (accessed on 2 March 2025).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Dong, Q.; Wang, D.; Zhang, P.; Liu, Y.; Niu, C. microbiomeMarker: An R/bioconductor package for microbiome marker identification and visualization. Bioinformatics 2022, 38, 4027–4029. [Google Scholar] [CrossRef] [PubMed]
- Tillman, M.C.; Smith, R.M.; Tubbs, T.R.; Catasus, A.B.; Urakawa, H.; Adhikari, P.L.; Douglass, J.G. Acute impacts of hurricane ian on benthic habitats, water quality, and microbial community composition on the southwest Florida shelf. Coasts 2025, 5, 16. [Google Scholar] [CrossRef]
- Waterson, E.J.; Canuel, E.A. Sources of sedimentary organic matter in the Mississippi River and adjacent Gulf of Mexico as revealed by lipid biomarker and δ13CTOC analyses. Org. Geochem. 2008, 39, 422–439. [Google Scholar] [CrossRef]
- Ye, F.; Guo, W.; Shi, Z.; Jia, G.; Wei, G. Seasonal dynamics of particulate organic matter and its response to flooding in the Pearl River Estuary, China, revealed by stable isotope (δ13 C and δ15 N) analyses. J. Geophys. Res. Oceans 2017, 122, 6835–6856. [Google Scholar] [CrossRef]
- Kieft, B.; Crump, B.; White, A.; Goñi, M.; Mueller, R. Winter river plumes shape community composition and activity of heterotrophic microorganisms on the Oregon coast. Aquat. Microb. Ecol. 2020, 84, 15–29. [Google Scholar] [CrossRef]
- Qiu, C.; Wan, Y. Time series modeling and prediction of salinity in the Caloosahatchee River Estuary: Time series modeling of estuarine salinity. Water Resour. Res. 2013, 49, 5804–5816. [Google Scholar] [CrossRef]
- Fortunato, C.S.; Eiler, A.; Herfort, L.; Needoba, J.A.; Peterson, T.D.; Crump, B.C. Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin. ISME J. 2013, 7, 1899–1911. [Google Scholar] [CrossRef]
- Mason, O.U.; Canter, E.J.; Gillies, L.E.; Paisie, T.K.; Roberts, B.J. Mississippi River plume enriches microbial diversity in the northern Gulf of Mexico. Front. Microbiol. 2016, 7, 01048. [Google Scholar] [CrossRef]
- Probandt, D.; Knittel, K.; Tegetmeyer, H.E.; Ahmerkamp, S.; Holtappels, M.; Amann, R. Permeability shapes bacterial communities in sublittoral surface sediments. Environ. Microbiol. 2017, 19, 1584–1599. [Google Scholar] [CrossRef]
- Guo, X.; Yang, Y.; Niu, Z.; Lu, D.-P.; Zhu, C.; Feng, J.; Wu, J.; Chen, Y.; Tou, F.; Liu, M.; et al. characteristics of microbial community indicate anthropogenic impact on the sediments along the Yangtze Estuary and its coastal area, China. Sci. Total Environ. 2019, 648, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zong, Y.; Lloyd, J.M.; Huang, G.; Leng, M.J.; Kendrick, C.; Lamb, A.L.; Yim, W.W.-S. Bulk organic δ13C and C/N as indicators for sediment sources in the Pearl River Delta and Estuary, Southern China. Estuar. Coast Shelf Sci. 2010, 87, 618–630. [Google Scholar] [CrossRef]
- Sampaio, L.; Freitas, R.; Máguas, C.; Rodrigues, A.; Quintino, V. Coastal sediments under the influence of multiple organic enrichment sources: An evaluation using carbon and nitrogen stable isotopes. Mar. Pollut. Bull. 2010, 60, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Tu, C.; Fu, C.; Xue, Y.; Luo, Y. Sources and fate of organic carbon and nitrogen from land to ocean: Identified by coupling stable isotopes with c/n ratio. Estuar. Coast Shelf Sci. 2016, 181, 114–122. [Google Scholar] [CrossRef]
- Liu, X.; Tang, D.; Ge, C. Distribution and sources of organic carbon, nitrogen and their isotopic composition in surface sediments from the southern Yellow Sea, China. Mar. Pollut. Bull. 2020, 150, 110716. [Google Scholar] [CrossRef]
- Marshall, D.A.; La Peyre, M.K.; Palmer, T.A.; Guillou, G.; Sterba-Boatwright, B.D.; Beseres Pollack, J.; Lebreton, B. Freshwater inflow and responses from estuaries across a climatic gradient: An assessment of northwestern Gulf of Mexico estuaries based on stable isotopes. Limnol. Oceanogr. 2021, 66, 3568–3581. [Google Scholar] [CrossRef]
- He, R.; Weisberg, R.H. A loop current intrusion case study on the west Florida shelf. J. Phys. Oceanogr. 2003, 33, 465–477. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.M.; Bennke, C.M.; Krüger, K.; Chafee, M.; Kappelmann, L.; Reintjes, G.; Waldmann, J.; Quast, C.; Glöckner, F.O.; et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 2016, 5, e11888. [Google Scholar] [CrossRef]
- Mestre, M.; Höfer, J.; Sala, M.M.; Gasol, J.M. Seasonal variation of bacterial diversity along the marine particulate matter continuum. Front. Microbiol. 2020, 11, 1590. [Google Scholar] [CrossRef]
- Wang, F.-Q.; Bartosik, D.; Sidhu, C.; Siebers, R.; Lu, D.-C.; Trautwein-Schult, A.; Becher, D.; Huettel, B.; Rick, J.; Kirstein, I.V.; et al. Particle-attached bacteria act as gatekeepers in the decomposition of complex phytoplankton polysaccharides. Microbiome 2024, 12, 32. [Google Scholar] [CrossRef]
- Korlević, M.; Markovski, M.; Herndl, G.J.; Najdek, M. Temporal variation in the prokaryotic community of a nearshore marine environment. Sci. Rep. 2022, 12, 16859. [Google Scholar] [CrossRef]
- Marín-Vindas, C.; Sebastián, M.; Ruiz-González, C.; Balagué, V.; Vega-Corrales, L.; Gasol, J.M. Shifts in bacterioplankton community structure between dry and wet seasons in a tropical estuary strongly affected by riverine discharge. Sci. Total Environ. 2023, 903, 166104. [Google Scholar] [CrossRef] [PubMed]
- Pollet, T.; Berdjeb, L.; Garnier, C.; Durrieu, G.; Poupon, C.L.; Misson, B.; Briand, J.-F. Prokaryotic community successions and interactions in marine biofilms: The key role of Flavobacteriia. FEMS Microbiol. Ecol. 2018, 94, fiy083. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Tanaka, R.; Hirai, N.; Ochiai, T.; Ohashi, R.; Fujimoto, K.; Akatsuka, Y.; Suzuki, M. investigation of biofilms formed on steelmaking slags in marine environments for water depuration. Int. J. Mol. Sci. 2020, 21, 6945. [Google Scholar] [CrossRef] [PubMed]
- Briand, J.-F.; Pollet, T.; Misson, B.; Garnier, C.; Lejars, M.; Maintenay, M.; Barry-Martinet, R.; Portas, A.; Ghiglione, J.-F.; Bressy, C. Surface characteristics together with environmental conditions shape marine biofilm dynamics in coastal nw mediterranean locations. Front. Mar. Sci. 2022, 8, 746383. [Google Scholar] [CrossRef]
- Ul-Hasan, S.; Bowers, R.M.; Figueroa-Montiel, A.; Licea-Navarro, A.F.; Beman, J.M.; Woyke, T.; Nobile, C.J. Community ecology across bacteria, archaea and microbial eukaryotes in the sediment and seawater of coastal Puerto Nuevo, Baja California. PLoS ONE 2019, 14, e0212355. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef]
- Orellana, L.H.; Francis, T.B.; Ferraro, M.; Hehemann, J.-H.; Fuchs, B.M.; Amann, R.I. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 2022, 16, 630–641. [Google Scholar] [CrossRef]
- Morris, R.M.; Longnecker, K.; Giovannoni, S.J. Pirellula and OM43 are among the dominant lineages identified in an oregon coast diatom bloom. Environ. Microbiol. 2006, 8, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Cruz, B.N.; Neuer, S. Heterotrophic bacteria enhance the aggregation of the marine picocyanobacteria Prochlorococcus and Synechococcus. Front. Microbiol. 2019, 10, 1864. [Google Scholar] [CrossRef]
- Wang, T.; Li, J.; Xu, Y.; Zou, T.; Qin, S. Aggregating Synechococcus contributes to particle organic carbon export in coastal estuarine waters: Its lineage features and assembly processes. Sci. Total Environ. 2024, 917, 170368. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.R.; Doyle, L.J.; Davis, R.A.; DeWitt, N.T.; Suthard, B.C. Patterns and controls of surface sediment distribution: West-central Florida inner shelf. Mar. Geol. 2003, 200, 307–324. [Google Scholar] [CrossRef]
- Tréguer, P.; Bowler, C.; Moriceau, B.; Dutkiewicz, S.; Gehlen, M.; Aumont, O.; Bittner, L.; Dugdale, R.; Finkel, Z.; Iudicone, D.; et al. Influence of diatom diversity on the ocean biological carbon pump. Nature Geosci. 2018, 11, 27–37. [Google Scholar] [CrossRef]
- De Martini, F.; Neuer, S.; Hamill, D.; Robidart, J.; Lomas, M.W. Clade and strain specific contributions of Synechococcus and Prochlorococcus to carbon export in the Sargasso Sea. Limnol. Oceanogr. 2018, 63, S448–S457. [Google Scholar] [CrossRef]
- Schoellhamer, D.H. Sediment resuspension mechanisms in Old Tampa Bay, Florida. Estuar. Coast Shelf Sci. 1995, 40, 603–620. [Google Scholar] [CrossRef]
- Huang, S.; Sherman, A.; Chen, C.; Jaffé, P.R. Tropical cyclone effects on water and sediment chemistry and the microbial community in estuarine ecosystems. Environ. Pollut. 2021, 286, 117228. [Google Scholar] [CrossRef]
- Wang, F.-Y.; Liu, M.-Y. Microbial community diversity of coral reef sediments on Liuqiu Island, southwestern Taiwan. J. Mar. Sci. Eng. 2023, 11, 85. [Google Scholar] [CrossRef]
- Bianucci, L.; Balaguru, K.; Smith, R.W.; Leung, L.R.; Moriarty, J.M. Contribution of hurricane-induced sediment resuspension to coastal oxygen dynamics. Sci. Rep. 2018, 8, 15740. [Google Scholar] [CrossRef]
- Petro, C.; Starnawski, P.; Schramm, A.; Kjeldsen, K. Microbial community assembly in marine sediments. Aquat. Microb. Ecol. 2017, 79, 177–195. [Google Scholar] [CrossRef]
- Govindarajan, A.; Crum, M.; Adolacion, J.; Kiaghadi, A.; Acuña-Gonzalez, E.; Rifai, H.S.; Willson, R.C. Sediment and their bacterial communities in an industrialized estuary after Hurricane Harvey. Mar. Pollut. Bull. 2022, 175, 113359. [Google Scholar] [CrossRef] [PubMed]
- Garrison, C.E.; Roozbehi, S.; Mitra, S.; Corbett, D.R.; Field, E.K. Coastal microbial communities disrupted during the 2018 hurricane season in Outer Banks, North Carolina. Front. Microbiol. 2022, 13, 816573. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, T.S. The role of terrestrially derived organic carbon in the coastal ocean: A changing paradigm and the priming effect. Proc. Natl. Acad. Sci. USA 2011, 108, 19473–19481. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.A.R.; Silveira, W.B.D.; Passos, F.M.L.; Zucchi, T.D. Laccases from Actinobacteria—what we have and what to expect. Adv. Microbiol. 2014, 04, 285–296. [Google Scholar] [CrossRef]
- McKew, B.A.; Dumbrell, A.J.; Taylor, J.D.; McGenity, T.J.; Underwood, G.J.C. Differences between aerobic and anaerobic degradation of microphytobenthic biofilm-derived organic matter within intertidal sediments. FEMS Microbiol. Ecol. 2013, 84, 495–509. [Google Scholar] [CrossRef]
- Feng, B.-W.; Li, X.-R.; Wang, J.-H.; Hu, Z.-Y.; Meng, H.; Xiang, L.-Y.; Quan, Z.-X. Bacterial diversity of water and sediment in the Changjiang Estuary and coastal area of the East China Sea. FEMS. Microbiol. Ecol. 2009, 70, 236–248. [Google Scholar] [CrossRef]
- Li, Y.; Huang, D.; Sun, W.; Sun, X.; Yan, G.; Gao, W.; Lin, H. Characterizing sediment bacterial community and identifying the biological indicators in a seawater-freshwater transition zone during the wet and dry seasons. Environ. Sci. Pollut. Res. 2022, 29, 41219–41230. [Google Scholar] [CrossRef]
- Zinger, L.; Amaral-Zettler, L.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Huse, S.M.; Welch, D.B.M.; Martiny, J.B.H.; Sogin, M.; Boetius, A.; Ramette, A. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE 2011, 6, e24570. [Google Scholar] [CrossRef]
- Giovannoni, S.J. SAR11 bacteria: The most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 2017, 9, 231–255. [Google Scholar] [CrossRef]
- O’Reilly, S.S.; Pentlavalli, P.; Flanagan, P.V.; Allen, C.C.R.; Monteys, X.; Szpak, M.T.; Murphy, B.T.; Jordan, S.F.; Kelleher, B.P. Abundance and diversity of sedimentary bacterial communities in a coastal productive setting in the western Irish Sea. Cont. Shelf Res. 2016, 113, 1–9. [Google Scholar] [CrossRef]
- Durham, B.P.; Grote, J.; Whittaker, K.A.; Bender, S.J.; Luo, H.; Grim, S.L.; Brown, J.M.; Casey, J.R.; Dron, A.; Florez-Leiva, L.; et al. Draft genome sequence of marine Alphaproteobacterial strain HIMB11, the first cultivated representative of a unique lineage within the Roseobacter clade possessing an unusually small genome. Stand. Genomic Sci. 2014, 9, 632–645. [Google Scholar] [CrossRef]
- Hoarfrost, A.; Nayfach, S.; Ladau, J.; Yooseph, S.; Arnosti, C.; Dupont, C.L.; Pollard, K.S. Global ecotypes in the ubiquitous marine clade SAR86. ISME J. 2020, 14, 178–188. [Google Scholar] [CrossRef]
- Halsey, K.H.; Carter, A.E.; Giovannoni, S.J. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ. Microbiol. 2012, 14, 630–640. [Google Scholar] [CrossRef]
- Takeuchi, M.; Katayama, T.; Yamagishi, T.; Hanada, S.; Tamaki, H.; Kamagata, Y.; Oshima, K.; Hattori, M.; Marumo, K.; Nedachi, M.; et al. Methyloceanibacter caenitepidi Gen. Nov., Sp. Nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2014, 64, 462–468. [Google Scholar] [CrossRef]
- Henriques, A.C.; De Marco, P. Complete genome sequences of two strains of “Candidatus Filomicrobium marinum,” a methanesulfonate-degrading species. Genome Announc. 2015, 3, e00160-15. [Google Scholar] [CrossRef] [PubMed]
- Vekeman, B.; Kerckhof, F.; Cremers, G.; De Vos, P.; Vandamme, P.; Boon, N.; Op Den Camp, H.J.M.; Heylen, K. New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 2016, 18, 4523–4536. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, K.-Y.; Hahm, Y.-T.; Kim, B.-S.; Chun, J.; Cha, C.-J. Actibacter sediminis Gen. Nov., Sp. Nov., a marine bacterium of the family Flavobacteriaceae isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2008, 58, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Pinnaka, A.K.; Tanuku, N.R.S. The family Cyclobacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 551–575. ISBN 978-3-642-38953-5. [Google Scholar]
- Lee, Y.; Jeong, H.I.; Jeong, S.E.; Jeon, C.O. Zeaxanthinibacter aestuarii Sp. Nov., isolated from estuary sediment and emended description of the genus Zeaxanthinibacter Asker et al. 2007. Int. J. Syst. Evol. Microbiol. 2016, 66, 3264–3269. [Google Scholar] [CrossRef]
- Xuan, X.-Q.; Mao, R.-Y.; Yu, W.-X.; An, J.; Du, Z.-J.; Mu, D.-S. Robiginitalea marina Sp. Nov., isolated from coastal sediment. Arch. Microbiol. 2022, 204, 644. [Google Scholar] [CrossRef]
- Du, Z.-J.; Wang, Z.-J.; Zhao, J.-X.; Chen, G.-J. Woeseia oceani Gen. Nov., Sp. Nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 107–112. [Google Scholar] [CrossRef]
- Mußmann, M.; Pjevac, P.; Krüger, K.; Dyksma, S. Genomic repertoire of the Woeseiaceae /JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 2017, 11, 1276–1281. [Google Scholar] [CrossRef] [PubMed]
- Buongiorno, J.; Sipes, K.; Wasmund, K.; Loy, A.; Lloyd, K.G. Woeseiales transcriptional response to shallow burial in arctic fjord surface sediment. PLoS ONE 2020, 15, e0234839. [Google Scholar] [CrossRef]
- Jørgensen, B.B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 1982, 296, 643–645. [Google Scholar] [CrossRef]
- Plugge, C.M.; Zhang, W.; Scholten, J.C.M.; Stams, A.J.M. Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2011, 2, 81. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Leung, P.M.; Cook, P.L.M.; Wong, W.W.; Hutchinson, T.; Eate, V.; Kessler, A.J.; Greening, C. Hydrodynamic disturbance controls microbial community assembly and biogeochemical processes in coastal sediments. ISME J. 2022, 16, 750–763. [Google Scholar] [CrossRef]
- Ward, L.M.; Bertran, E.; Johnston, D.T. Expanded genomic sampling refines current understanding of the distribution and evolution of sulfur metabolisms in the Desulfobulbales. Front. Microbiol. 2021, 12, 666052. [Google Scholar] [CrossRef]
- Liesirova, T.; Aarenstrup-Launbjerg, T.; Hallstrøm, S.; Bittner, M.J.; Riemann, L.; Voss, M. Nitrogen-fixing sulfate reducing bacteria in shallow coastal sediments under simulated resuspension. Estuar. Coast Shelf Sci. 2023, 280, 108165. [Google Scholar] [CrossRef]
- Sipler, R.; Bronk, D.; Seitzinger, S.; Lauck, R.; McGuinness, L.; Kirkpatrick, G.; Heil, C.; Kerkhof, L.; Schofield, O. Trichodesmium-derived dissolved organic matter is a source of nitrogen capable of supporting the growth of toxic red tide Karenia brevis. Mar. Ecol. Prog. Ser. 2013, 483, 31–45. [Google Scholar] [CrossRef]
- Jabir, T.; Vipindas, P.V.; Jesmi, Y.; Divya, P.S.; Adarsh, B.M.; Nafeesathul Miziriya, H.S.; Mohamed Hatha, A.A. Influence of environmental factors on benthic nitrogen fixation and role of sulfur reducing diazotrophs in a eutrophic tropical estuary. Mar. Pollut. Bull. 2021, 165, 112126. [Google Scholar] [CrossRef]
- Begmatov, S.; Savvichev, A.S.; Kadnikov, V.V.; Beletsky, A.V.; Rusanov, I.I.; Klyuvitkin, A.A.; Novichkova, E.A.; Mardanov, A.V.; Pimenov, N.V.; Ravin, N.V. Microbial Communities involved in methane, sulfur, and nitrogen cycling in the sediments of the barents sea. Microorganisms 2021, 9, 2362. [Google Scholar] [CrossRef]
- Wei, H.; Lin, X. Shifts in the relative abundance and potential rates of sediment ammonia-oxidizing archaea and bacteria along environmental gradients of an urban river–estuary–adjacent sea continuum. Sci. Total Environ. 2021, 771, 144824. [Google Scholar] [CrossRef] [PubMed]
- Martens-Habbena, W.; Berube, P.M.; Urakawa, H.; De La Torre, J.R.; Stahl, D.A. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 2009, 461, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Hink, L.; Gubry-Rangin, C.; Nicol, G.W. Nitrous oxide production by ammonia oxidizers: Physiological diversity, niche differentiation and potential mitigation strategies. Glob. Change Biol. 2020, 26, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Hink, L.; Gubry-Rangin, C.; Nicol, G.W.; Prosser, J.I. The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J. 2018, 12, 1084–1093. [Google Scholar] [CrossRef]
- Ivey, J.E.; Wolny, J.L.; Heil, C.A.; Murasko, S.M.; Brame, J.A.; Parks, A.A. Urea inputs drive picoplankton blooms in Sarasota Bay, Florida, U.S.A. Water 2020, 12, 2755. [Google Scholar] [CrossRef]
- Li, F.; Burger, A.; Eppley, J.M.; Poff, K.E.; Karl, D.M.; DeLong, E.F. Planktonic microbial signatures of sinking particle export in the open ocean’s interior. Nat. Commun. 2023, 14, 7177. [Google Scholar] [CrossRef]
- Che, Y.; Lin, C.; Li, S.; Liu, J.; Zhu, L.; Yu, S.; Wang, N.; Li, H.; Bao, M.; Zhou, Y.; et al. Influences of hydrodynamics on microbial community assembly and organic carbon composition of resuspended sediments in shallow marginal seas. Water Res. 2024, 248, 120882. [Google Scholar] [CrossRef]
- Chistoserdova, L. Methylotrophs in natural habitats: Current insights through metagenomics. Appl. Microbiol. Biotechnol. 2015, 99, 5763–5779. [Google Scholar] [CrossRef]












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tubbs, T.R.; Smith, R.M.; Catasus, A.B.; Adhikari, P.L.; Douglass, J.G.; Urakawa, H. Seasonal and Spatial Microbial Community Dynamics Along the Shallow Southwest Florida Continental Shelf. Coasts 2025, 5, 47. https://doi.org/10.3390/coasts5040047
Tubbs TR, Smith RM, Catasus AB, Adhikari PL, Douglass JG, Urakawa H. Seasonal and Spatial Microbial Community Dynamics Along the Shallow Southwest Florida Continental Shelf. Coasts. 2025; 5(4):47. https://doi.org/10.3390/coasts5040047
Chicago/Turabian StyleTubbs, Trevor R., Robert Marlin Smith, Adam B. Catasus, Puspa L. Adhikari, James G. Douglass, and Hidetoshi Urakawa. 2025. "Seasonal and Spatial Microbial Community Dynamics Along the Shallow Southwest Florida Continental Shelf" Coasts 5, no. 4: 47. https://doi.org/10.3390/coasts5040047
APA StyleTubbs, T. R., Smith, R. M., Catasus, A. B., Adhikari, P. L., Douglass, J. G., & Urakawa, H. (2025). Seasonal and Spatial Microbial Community Dynamics Along the Shallow Southwest Florida Continental Shelf. Coasts, 5(4), 47. https://doi.org/10.3390/coasts5040047






