Preanalytical Strategies for Native Mass Spectrometry Analysis of Protein Modifications, Complexes, and Higher-Order Structures
Abstract
1. Introduction
2. General Considerations for Native Mass Spectrometry Analysis of Protein Species
2.1. Sample Preparation
2.2. Instrument Parameters for Data Acquisition
2.3. Data Analysis
3. Offline Strategies for Isolation, Separation, and Enrichment of Intact Protein Species for nMS Analysis
3.1. Immunoprecipitation (IP)
3.2. Gel Electrophoresis (GE)
3.3. Free-Flow Electrophoresis (FFE)
4. Online Strategies for Isolation, Separation, and Enrichment of Intact Protein Species for nMS Analysis
4.1. Liquid Chromatography (LC)
4.1.1. Size Exclusion Chromatography (SEC)
4.1.2. Ion Exchange Chromatography (IEC)
4.1.3. Hydrophobic Interaction Chromatography (HIC)
4.1.4. Affinity Liquid Chromatography (ALC)
4.2. Capillary Electrophoresis (CE)
4.2.1. Capillary Zone Electrophoresis (CZE)
4.2.2. Mobility Capillary Electrophoresis (MoCE)
4.2.3. Affinity Capillary Electrophoresis (ACE)
4.2.4. Capillary Isoelectric Focusing (cIEF)
5. New Frontiers of Native Mass Spectrometry and Proteomics
5.1. Automated Purification, Buffer Exchange, and Individual Ion Mass Spectrometry
5.2. Ion Mobility Spectrometry (IMS)
5.3. Ambient Surface Mass Spectrometry (ASMS)
6. Assessment of nMS Sample Resemblance to the Native State
7. Importance and Challenges of Native Mass Spectrometry Analysis of Protein Species
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Tak, I.-u.-R.; Ali, F.; Dar, J.S.; Magray, A.R.; Ganai, B.A.; Chishti, M.Z. Chapter 1-Posttranslational Modifications of Proteins and Their Role in Biological Processes and Associated Diseases. In Protein Modificomics; Dar, T.A., Singh, L.R., Eds.; Academic Press: New York, NY, USA, 2019; pp. 1–35. [Google Scholar]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ji, B. Protein conformational transitions coupling with ligand interactions: Simulations from molecules to medicine. Med. Nov. Technol. Devices 2019, 3, 100026. [Google Scholar] [CrossRef]
- Ramazi, S.; Dadzadi, M.; Darvazi, M.; Seddigh, N.; Allahverdi, A. Protein modification in neurodegenerative diseases. MedComm 2024, 5, e674. [Google Scholar] [CrossRef]
- Xiong, D.; Lee, D.; Li, L.; Zhao, Q.; Yu, H. Implications of disease-related mutations at protein-protein interfaces. Curr. Opin. Struct. Biol. 2022, 72, 219–225. [Google Scholar] [CrossRef]
- Lagassé, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef]
- Jenkins, N. Modifications of therapeutic proteins: Challenges and prospects. Cytotechnology 2007, 53, 121–125. [Google Scholar] [CrossRef]
- Schmidt, C.; Robinson, C.V. Dynamic protein ligand interactions—Insights from MS. FEBS J. 2014, 281, 1950–1964. [Google Scholar] [CrossRef]
- Visser, N.F.C.; Lingeman, H.; Irth, H. Sample preparation for peptides and proteins in biological matrices prior to liquid chromatography and capillary zone electrophoresis. Anal. Bioanal. Chem. 2005, 382, 535–558. [Google Scholar] [CrossRef]
- Filipe, V.; Hawe, A.; Carpenter, J.F.; Jiskoot, W. Analytical approaches to assess the degradation of therapeutic proteins. TrAC Trends Anal. Chem. 2013, 49, 118–125. [Google Scholar] [CrossRef]
- Kaulich, P.T.; Jeong, K.; Kohlbacher, O.; Tholey, A. Influence of different sample preparation approaches on proteoform identification by top-down proteomics. Nat. Methods 2024, 21, 2397–2407. [Google Scholar] [CrossRef] [PubMed]
- Lorber, B.; Fischer, F.; Bailly, M.; Roy, H.; Kern, D. Protein analysis by dynamic light scattering: Methods and techniques for students. Biochem. Mol. Biol. Educ. 2012, 40, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.M.; Raman, C.S.; Nall, B.T. Isothermal Titration Calorimetry of Protein–Protein Interactions. Methods 1999, 19, 213–221. [Google Scholar] [CrossRef]
- Papageorgiou, A.C.; Poudel, N.; Mattsson, J. Protein Structure Analysis and Validation with X-Ray Crystallography. Methods Mol. Biol. 2021, 2178, 377–404. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, K.; He, L.; Zhang, X.; Jiang, B.; Jiang, L.; Li, C.; Wang, G.; Yang, Y.; Liu, M. NMR-Based Methods for Protein Analysis. Anal. Chem. 2021, 93, 1866–1879. [Google Scholar] [CrossRef]
- Parois, P.; Cooper, R.I.; Thompson, A.L. Crystal structures of increasingly large molecules: Meeting the challenges with CRYSTALS software. Chem. Cent. J. 2015, 9, 30. [Google Scholar] [CrossRef]
- Huang, Y.; Eliezer, D. Sharpening the lens of NMR spectroscopy to study large proteins. Nat. Chem. 2025, 17, 786–788. [Google Scholar] [CrossRef]
- Artigues, A.; Nadeau, O.W.; Rimmer, M.A.; Villar, M.T.; Du, X.; Fenton, A.W.; Carlson, G.M. Protein Structural Analysis via Mass Spectrometry-Based Proteomics. In Modern Proteomics–Sample Preparation, Analysis and Practical Applications; Mirzaei, H., Carrasco, M., Eds.; Springer International Publishing: Cham, Germany, 2016; pp. 397–431. [Google Scholar]
- Edwards, A.N.; Hsu, K.-L. Emerging opportunities for intact and native protein analysis using chemical proteomics. Anal. Chim. Acta 2025, 1338, 343551. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Yin, S.; Loo, J.A. Top-Down ESI-ECD-FT-ICR Mass Spectrometry Localizes Noncovalent Protein-Ligand Binding Sites. J. Am. Chem. Soc. 2006, 128, 14432–14433. [Google Scholar] [CrossRef]
- Yin, S.; Loo, J.A. Top-down mass spectrometry of supercharged native protein–ligand complexes. Int. J. Mass Spectrom. 2011, 300, 118–122. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, W.; Wen, J.; Blankenship, R.E.; Gross, M.L. Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J. Am. Soc. Mass Spectrom. 2010, 21, 1966–1968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, W.; Wen, J.; Blankenship, R.E.; Gross, M.L. Native Electrospray and Electron-Capture Dissociation FTICR Mass Spectrometry for Top-Down Studies of Protein Assemblies. Anal. Chem. 2011, 83, 5598–5606. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.J.; Damoc, E.; Denisov, E.; Makarov, A.; Heck, A.J.R. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods 2012, 9, 1084–1086. [Google Scholar] [CrossRef] [PubMed]
- Michalski, W.P.; Shiell, B.J. Strategies for analysis of electrophoretically separated proteins and peptides. Anal. Chim. Acta 1999, 383, 27–46. [Google Scholar] [CrossRef]
- Hernández, H.; Robinson, C.V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007, 2, 715–726. [Google Scholar] [CrossRef]
- Sobott, F.; Hernández, H.; McCammon, M.G.; Tito, M.A.; Robinson, C.V. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 2002, 74, 1402–1407. [Google Scholar] [CrossRef]
- Xu, T.; Wang, Q.; Wang, Q.; Sun, L. Mass spectrometry-intensive top-down proteomics: An update on technology advancements and biomedical applications. Anal. Methods 2024, 16, 4664–4682. [Google Scholar] [CrossRef]
- Robinson, C.V. Protein complexes take flight. Nat. Struct. Biol. 2002, 9, 505–506. [Google Scholar] [CrossRef]
- Pukala, T.L.; Ruotolo, B.T.; Zhou, M.; Politis, A.; Stefanescu, R.; Leary, J.A.; Robinson, C.V. Subunit architecture of multiprotein assemblies determined using restraints from gas-phase measurements. Structure 2009, 17, 1235–1243. [Google Scholar] [CrossRef]
- Zhou, M.; Lantz, C.; Brown, K.A.; Ge, Y.; Paša-Tolić, L.; Loo, J.A.; Lermyte, F. Higher-order structural characterisation of native proteins and complexes by top-down mass spectrometry. Chem. Sci. 2020, 11, 12918–12936. [Google Scholar] [CrossRef]
- Blackwell, A.E.; Dodds, E.D.; Bandarian, V.; Wysocki, V.H. Revealing the Quaternary Structure of a Heterogeneous Noncovalent Protein Complex through Surface-Induced Dissociation. Anal. Chem. 2011, 83, 2862–2865. [Google Scholar] [CrossRef][Green Version]
- Romano, C.A.; Zhou, M.; Song, Y.; Wysocki, V.H.; Dohnalkova, A.C.; Kovarik, L.; Paša-Tolić, L.; Tebo, B.M. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nat. Commun. 2017, 8, 746. [Google Scholar] [CrossRef]
- Ayon, N.J. Features, roles and chiral analyses of proteinogenic amino acids. Aims Mol. Sci. 2020, 7, 229–268. [Google Scholar] [CrossRef]
- Vemula, H.; Ayon, N.J.; Gutheil, W.G. Cytoplasmic peptidoglycan intermediate levels in Staphylococcus aureus. Biochimie 2016, 121, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Vemula, H.; Ayon, N.J.; Burton, A.; Gutheil, W.G. Antibiotic Effects on Methicillin-Resistant Staphylococcus aureus Cytoplasmic Peptidoglycan Intermediate Levels and Evidence for Potential Metabolite Level Regulatory Loops. Antimicrob. Agents Chemother. 2017, 61, 10. [Google Scholar] [CrossRef] [PubMed]
- Ayon, N.J.; Sharma, A.D.; Gutheil, W.G. LC-MS/MS-Based Separation and Quantification of Marfey’s Reagent Derivatized Proteinogenic Amino Acid dl-Stereoisomers. J. Am. Soc. Mass Spectrom. 2019, 30, 448–458. [Google Scholar] [CrossRef]
- Chen, A.; Aquino, R.M.; Vidal, H.A.; Wong, C.V.; Luo, R.Y. A liquid chromatography-high-resolution mass spectrometry method for separation and identification of hemoglobin variant subunits with mass shifts less than 1 Da. J. Mass Spectrom. Adv. Clin. Lab 2025, 35, 1–7. [Google Scholar] [CrossRef]
- El Kennani, S.; Crespo, M.; Govin, J.; Pflieger, D. Proteomic Analysis of Histone Variants and Their PTMs: Strategies and Pitfalls. Proteomes 2018, 6, 29. [Google Scholar] [CrossRef]
- Bults, P.; Spanov, B.; Olaleye, O.; van de Merbel, N.C.; Bischoff, R. Intact protein bioanalysis by liquid chromatography–High-resolution mass spectrometry. J. Chromatogr. B 2019, 1110–1111, 155–167. [Google Scholar] [CrossRef]
- Hajba, L.; Jeong, S.; Chung, D.S.; Guttman, A. Capillary Gel Electrophoresis of Proteins: Historical overview and recent advances. TrAC Trends Anal. Chem. 2023, 162, 117024. [Google Scholar] [CrossRef]
- Barth, M.; Schmidt, C. Native mass spectrometry—A valuable tool in structural biology. J. Mass Spectrom. 2020, 55, e4578. [Google Scholar] [CrossRef] [PubMed]
- Masson, G.R.; Burke, J.E.; Ahn, N.G.; Anand, G.S.; Borchers, C.; Brier, S.; Bou-Assaf, G.M.; Engen, J.R.; Englander, S.W.; Faber, J.; et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 2019, 16, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Walzthoeni, T.; Kahraman, A.; Herzog, F.; Rinner, O.; Beck, M.; Aebersold, R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteom. MCP 2010, 9, 1634–1649. [Google Scholar] [CrossRef]
- Ozohanics, O.; Ambrus, A. Hydrogen-Deuterium Exchange Mass Spectrometry: A Novel Structural Biology Approach to Structure, Dynamics and Interactions of Proteins and Their Complexes. Life 2020, 10, 286. [Google Scholar] [CrossRef]
- Leitner, A.; Faini, M.; Stengel, F.; Aebersold, R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem. Sci. 2016, 41, 20–32. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Yu, B.; Wang, S.; Shen, Y.; Cong, H. Recent advances on protein separation and purification methods. Adv. Colloid Interface Sci. 2020, 284, 102254. [Google Scholar] [CrossRef]
- Thomas, S.L.; Thacker, J.B.; Schug, K.A.; Maráková, K. Sample preparation and fractionation techniques for intact proteins for mass spectrometric analysis. J. Sep. Sci. 2021, 44, 211–246. [Google Scholar] [CrossRef]
- Masse, F.; Parat, M.; Matte, A.; Thauvette, L.; Hélie, G.; Durocher, Y.; Vercauteren, F. Parallelized protein purification: Opportunities and challenges in earlystage biotherapeutics research & development. Am. Pharm. Rev. 2017, 20. Available online: https://www.americanpharmaceuticalreview.com/Featured-Articles/345445-Parallelized-Protein-Purification-Opportunities-and-Challenges-in-EarlyStage-Biotherapeutics-Research-Development/ (accessed on 31 August 2025).
- Matte, A. Chapter 9-High-throughput, parallelized and automated protein purification for therapeutic antibody development. In Approaches to the Purification, Analysis and Characterization of Antibody-Based Therapeutics; Matte, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 181–198. [Google Scholar]
- Donnelly, D.P.; Rawlins, C.M.; DeHart, C.J.; Fornelli, L.; Schachner, L.F.; Lin, Z.; Lippens, J.L.; Aluri, K.C.; Sarin, R.; Chen, B.; et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat. Methods 2019, 16, 587–594. [Google Scholar] [CrossRef]
- Heck, A.J.R. Native mass spectrometry: A bridge between interactomics and structural biology. Nat. Methods 2008, 5, 927–933. [Google Scholar] [CrossRef]
- Kafader, J.O.; Melani, R.D.; Schachner, L.F.; Ives, A.N.; Patrie, S.M.; Kelleher, N.L.; Compton, P.D. Native vs Denatured: An in Depth Investigation of Charge State and Isotope Distributions. J. Am. Soc. Mass Spectrom. 2020, 31, 574–581. [Google Scholar] [CrossRef]
- Hyung, S.-J.; Ruotolo, B.T. Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics 2012, 12, 1547–1564. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, I.D.G.; Loo, J.A. Evolution of Mass Spectrometers for High m/z Biological Ion Formation, Transmission, Analysis and Detection: A Personal Perspective. J. Am. Soc. Mass Spectrom. 2025, 36, 632–652. [Google Scholar] [CrossRef] [PubMed]
- Kirshenbaum, N.; Michaelevski, I.; Sharon, M. Analyzing large protein complexes by structural mass spectrometry. J. Vis. Exp. JoVE 2010, 1954. [Google Scholar] [CrossRef]
- Tamara, S.; den Boer, M.A.; Heck, A.J.R. High-Resolution Native Mass Spectrometry. Chem. Rev. 2022, 122, 7269–7326. [Google Scholar] [CrossRef]
- Susa, A.C.; Xia, Z.; Williams, E.R. Native Mass Spectrometry from Common Buffers with Salts That Mimic the Extracellular Environment. Angew. Chem. Int. Ed. 2017, 56, 7912–7915. [Google Scholar] [CrossRef]
- Konermann, L.; Liu, Z.; Haidar, Y.; Willans, M.J.; Bainbridge, N.A. On the Chemistry of Aqueous Ammonium Acetate Droplets during Native Electrospray Ionization Mass Spectrometry. Anal. Chem. 2023, 95, 13957–13966. [Google Scholar] [CrossRef]
- Ventouri, I.K.; Malheiro, D.B.A.; Voeten, R.L.C.; Kok, S.; Honing, M.; Somsen, G.W.; Haselberg, R. Probing Protein Denaturation during Size-Exclusion Chromatography Using Native Mass Spectrometry. Anal. Chem. 2020, 92, 4292–4300. [Google Scholar] [CrossRef]
- Konermann, L. Addressing a Common Misconception: Ammonium Acetate as Neutral pH “Buffer” for Native Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 1827–1835. [Google Scholar] [CrossRef]
- Pacholarz, K.J.; Barran, P.E. Use of a charge reducing agent to enable intact mass analysis of cysteine-linked antibody-drug-conjugates by native mass spectrometry. EuPA Open Proteom. 2016, 11, 23–27. [Google Scholar] [CrossRef]
- Zhuang, X.; Gavriilidou, A.F.M.; Zenobi, R. Influence of Alkylammonium Acetate Buffers on Protein–Ligand Noncovalent Interactions Using Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Wagner, N.D.; Yan, J.; Li, J.; Huang, R.Y.C.; Balog, A.J.; Newitt, J.A.; Chen, G.; Gross, M.L. Native mass spectrometry and gas-phase fragmentation provide rapid and in-depth topological characterization of a PROTAC ternary complex. Cell Chem. Biol. 2021, 28, 1528–1538.e1524. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Gabelica, V. Native Electrospray Mass Spectrometry of DNA G-Quadruplexes in Potassium Solution. J. Am. Soc. Mass Spectrom. 2014, 25, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, D.; Marie, G.; Serani, L.; Laprévote, O. Stabilization of Gas-Phase Noncovalent Macromolecular Complexes in Electrospray Mass Spectrometry Using Aqueous Triethylammonium Bicarbonate Buffer. Anal. Chem. 2001, 73, 1699–1706. [Google Scholar] [CrossRef]
- Hadavi, D.; Ng, C.Y.; Zhao, Y.; Mathew, A.; Anthony, I.G.M.; Cillero-Pastor, B.; Cuypers, E.; Siegel, T.P.; Honing, M. Buffer 4-Ethylmorpholinium/Acetate: Exploring a New Alternative Buffer for Native Mass Spectrometry. Rapid Commun. Mass Spectrom. 2025, 39, e10048. [Google Scholar] [CrossRef]
- Brown, K.A.; Melby, J.A.; Roberts, D.S.; Ge, Y. Top-down proteomics: Challenges, innovations, and applications in basic and clinical research. Expert Rev. Proteom. 2020, 17, 719–733. [Google Scholar] [CrossRef]
- Vimer, S.; Ben-Nissan, G.; Sharon, M. Mass Spectrometry Analysis of Intact Proteins from Crude Samples. Anal. Chem. 2020, 92, 12741–12749. [Google Scholar] [CrossRef]
- Benesch, J.L.P.; Ruotolo, B.T.; Simmons, D.A.; Robinson, C.V. Protein Complexes in the Gas Phase: Technology for Structural Genomics and Proteomics. Chem. Rev. 2007, 107, 3544–3567. [Google Scholar] [CrossRef]
- Breuker, K.; McLafferty, F.W. Stepwise evolution of protein native structure with electrospray into the gas phase, 10−12 to 102. Proc. Natl. Acad. Sci. USA 2008, 105, 18145–18152. [Google Scholar] [CrossRef]
- Stiving, A.Q.; VanAernum, Z.L.; Busch, F.; Harvey, S.R.; Sarni, S.H.; Wysocki, V.H. Surface-Induced Dissociation: An Effective Method for Characterization of Protein Quaternary Structure. Anal. Chem. 2019, 91, 190–209. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Reading, E.; Hopper, J.T.S.; Robinson, C.V. Mass spectrometry of intact membrane protein complexes. Nat. Protoc. 2013, 8, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Aquilina, J.A.; Benesch, J.L.P.; Bateman, O.A.; Slingsby, C.; Robinson, C.V. Polydispersity of a mammalian chaperone: Mass spectrometry reveals the population of oligomers in αB-crystallin. Proc. Natl. Acad. Sci. USA 2003, 100, 10611–10616. [Google Scholar] [CrossRef] [PubMed]
- Rostom, A.A.; Sunde, M.; Richardson, S.J.; Schreiber, G.; Jarvis, S.; Bateman, R.; Dobson, C.M.; Robinson, C.V. Dissection of multi-protein complexes using mass spectrometry: Subunit interactions in transthyretin and retinol-binding protein complexes. Proteins Struct. Funct. Bioinform. 1998, 33, 3–11. [Google Scholar] [CrossRef]
- Schwartz, B.L.; Bruce, J.E.; Anderson, G.A.; Hofstadler, S.A.; Rockwood, A.L.; Smith, R.D.; Chilkoti, A.; Stayton, P.S. Dissociation of tetrameric ions of noncovalent streptavidin complexes formed by electrospray ionization. J. Am. Soc. Mass Spectrom. 1995, 6, 459–465. [Google Scholar] [CrossRef]
- Fändrich, M.; Tito, M.A.; Leroux, M.R.; Rostom, A.A.; Hartl, F.U.; Dobson, C.M.; Robinson, C.V. Observation of the noncovalent assembly and disassembly pathways of the chaperone complex MtGimC by mass spectrometry. Proc. Natl. Acad. Sci. USA 2000, 97, 14151–14155. [Google Scholar] [CrossRef]
- Ilag, L.L.; Videler, H.; McKay, A.R.; Sobott, F.; Fucini, P.; Nierhaus, K.H.; Robinson, C.V. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. USA 2005, 102, 8192–8197. [Google Scholar] [CrossRef]
- Lantz, C.; Wei, B.; Zhao, B.; Jung, W.; Goring, A.K.; Le, J.; Miller, J.; Loo, R.R.O.; Loo, J.A. Native Top-Down Mass Spectrometry with Collisionally Activated Dissociation Yields Higher-Order Structure Information for Protein Complexes. J. Am. Chem. Soc. 2022, 144, 21826–21830. [Google Scholar] [CrossRef]
- Busch, F.; VanAernum, Z.L.; Ju, Y.; Yan, J.; Gilbert, J.D.; Quintyn, R.S.; Bern, M.; Wysocki, V.H. Localization of Protein Complex Bound Ligands by Surface-Induced Dissociation High-Resolution Mass Spectrometry. Anal. Chem. 2018, 90, 12796–12801. [Google Scholar] [CrossRef]
- Quintyn, R.S.; Zhou, M.; Yan, J.; Wysocki, V.H. Surface-Induced Dissociation Mass Spectra as a Tool for Distinguishing Different Structural Forms of Gas-Phase Multimeric Protein Complexes. Anal. Chem. 2015, 87, 11879–11886. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Kelleher, N.L.; McLafferty, F.W. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J. Am. Chem. Soc. 1998, 120, 3265–3266. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef]
- Searfoss, R.M.; Zahn, E.; Lin, Z.; Garcia, B.A. Establishing a Top-Down Proteomics Platform on a Time-of-Flight Instrument with Electron-Activated Dissociation. J. Proteome Res. 2025, 24, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.B.; Li, W.; Holden, D.D.; Zhang, Y.; Griep-Raming, J.; Fellers, R.T.; Early, B.P.; Thomas, P.M.; Kelleher, N.L.; Brodbelt, J.S. Complete Protein Characterization Using Top-Down Mass Spectrometry and Ultraviolet Photodissociation. J. Am. Chem. Soc. 2013, 135, 12646–12651. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.M.; Lössl, P.; Liu, F.; Huguet, R.; Mullen, C.; Yamashita, M.; Zabrouskov, V.; Makarov, A.; Altelaar, A.F.M.; Heck, A.J.R. Benchmarking Multiple Fragmentation Methods on an Orbitrap Fusion for Top-down Phospho-Proteoform Characterization. Anal. Chem. 2015, 87, 4152–4158. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.P.; Li, W.; Zhang, Y.; Brodbelt, J.S. Characterization of Native Protein Complexes Using Ultraviolet Photodissociation Mass Spectrometry. J. Am. Chem. Soc. 2014, 136, 12920–12928. [Google Scholar] [CrossRef]
- Reid, G.E.; McLuckey, S.A. ‘Top down’ protein characterization via tandem mass spectrometry. J. Mass Spectrom. 2002, 37, 663–675. [Google Scholar] [CrossRef]
- Qi, Y.; Volmer, D.A. Electron-based fragmentation methods in mass spectrometry: An overview. Mass Spectrom. Rev. 2017, 36, 4–15. [Google Scholar] [CrossRef]
- Fornelli, L.; Srzentić, K.; Toby, T.K.; Doubleday, P.F.; Huguet, R.; Mullen, C.; Melani, R.D.; dos Santos Seckler, H.; DeHart, C.J.; Weisbrod, C.R.; et al. Thorough Performance Evaluation of 213 nm Ultraviolet Photodissociation for Top-down Proteomics. Mol. Cell. Proteom. 2020, 19, 405–420. [Google Scholar] [CrossRef]
- Lanzillotti, M.; Brodbelt, J.S. Comparison of Top-Down Protein Fragmentation Induced by 213 and 193 nm UVPD. J. Am. Soc. Mass Spectrom. 2023, 34, 279–285. [Google Scholar] [CrossRef]
- Wong, D.L. Sensitive Native Mass Spectrometry of Macromolecules Using Standard Flow LC/MS.; Agilent Technologies, Inc.: Santa Clara, CA, USA, 2020. [Google Scholar]
- Zhou, M.; Dagan, S.; Wysocki, V.H. Impact of charge state on gas-phase behaviors of noncovalent protein complexes in collision induced dissociation and surface induced dissociation. Analyst 2013, 138, 1353–1362. [Google Scholar] [CrossRef]
- Lippens, J.L.; Nshanian, M.; Spahr, C.; Egea, P.F.; Loo, J.A.; Campuzano, I.D.G. Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry as a Platform for Characterizing Multimeric Membrane Protein Complexes. J. Am. Soc. Mass Spectrom. 2018, 29, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Liu, H.; Zhang, Z. Native Mass Spectrometry Analysis of Biotherapeutics and Aggregates with Enhanced Sensitivity; SCIEX: Framingham, MA, USA, 2025. [Google Scholar]
- Compton, P.D.; Zamdborg, L.; Thomas, P.M.; Kelleher, N.L. On the Scalability and Requirements of Whole Protein Mass Spectrometry. Anal. Chem. 2011, 83, 6868–6874. [Google Scholar] [CrossRef]
- Roberts, D.S.; Loo, J.A.; Tsybin, Y.O.; Liu, X.; Wu, S.; Chamot-Rooke, J.; Agar, J.N.; Paša-Tolić, L.; Smith, L.M.; Ge, Y. Top-down proteomics. Nat. Rev. Methods Primers 2024, 4, 38. [Google Scholar] [CrossRef]
- Kaltashov, I.A.; Bobst, C.E.; Pawlowski, J.; Wang, G. Mass spectrometry-based methods in characterization of the higher order structure of protein therapeutics. J. Pharm. Biomed. Anal. 2020, 184, 113169. [Google Scholar] [CrossRef]
- Kaltashov, I.A.; Bobst, C.E.; Abzalimov, R.R. Mass spectrometry-based methods to study protein architecture and dynamics. Protein Sci. A Publ. Protein Soc. 2013, 22, 530–544. [Google Scholar] [CrossRef]
- Durbin, K.R.; Robey, M.T.; Voong, L.N.; Fellers, R.T.; Lutomski, C.A.; El-Baba, T.J.; Robinson, C.V.; Kelleher, N.L. ProSight Native: Defining Protein Complex Composition from Native Top-Down Mass Spectrometry Data. J. Proteome Res. 2023, 22, 2660–2668. [Google Scholar] [CrossRef]
- LeDuc, R.D.; Taylor, G.K.; Kim, Y.-B.; Januszyk, T.E.; Bynum, L.H.; Sola, J.V.; Garavelli, J.S.; Kelleher, N.L. ProSight PTM: An integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 2004, 32, W340–W345. [Google Scholar] [CrossRef]
- Fellers, R.T.; Greer, J.B.; Early, B.P.; Yu, X.; LeDuc, R.D.; Kelleher, N.L.; Thomas, P.M. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15, 1235–1238. [Google Scholar] [CrossRef]
- Shankar, V.; Tibshirani, R.; Zare, R.N. MassExplorer: A computational tool for analyzing desorption electrospray ionization mass spectrometry data. Bioinformatics 2021, 37, 3688–3690. [Google Scholar] [CrossRef]
- Park, J.; Piehowski, P.D.; Wilkins, C.; Zhou, M.; Mendoza, J.; Fujimoto, G.M.; Gibbons, B.C.; Shaw, J.B.; Shen, Y.; Shukla, A.K.; et al. Informed-Proteomics: Open-source software package for top-down proteomics. Nat. Methods 2017, 14, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Kou, Q.; Xun, L.; Liu, X. TopPIC: A software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 2016, 32, 3495–3497. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sirotkin, Y.; Shen, Y.; Anderson, G.; Tsai, Y.S.; Ting, Y.S.; Goodlett, D.R.; Smith, R.D.; Bafna, V.; Pevzner, P.A. Protein Identification Using Top-Down Spectra. Mol. Cell. Proteom. 2012, 11, M111-008524. [Google Scholar] [CrossRef]
- Urh, M.; Simpson, D.; Zhao, K. Affinity chromatography: General methods. Methods Enzymol. 2009, 463, 417–438. [Google Scholar] [CrossRef]
- Tchikov, V.; Fritsch, J.; Kabelitz, D.; Schütze, S. 2-Immunomagnetic Isolation of Subcellular Compartments. In Methods in Microbiology; Kabelitz, D., Kaufmann, S.H.E., Eds.; Academic Press: New York, NY, USA, 2010; Volume 37, pp. 21–33. [Google Scholar]
- Husain, A.; Begum, N.A.; Kobayashi, M.; Honjo, T. Native Co-immunoprecipitation Assay to Identify Interacting Partners of Chromatin-associated Proteins in Mammalian Cells. Bio-Protoc. 2020, 10, e3837. [Google Scholar] [CrossRef]
- Cosseau, C.; Grunau, C. Native Chromatin Immunoprecipitation. In Epigenetics Protocols; Tollefsbol, T.O., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 195–212. [Google Scholar]
- Thorne, A.W.; Myers, F.A.; Hebbes, T.R. Native Chromatin Immunoprecipitation. In Epigenetics Protocols; Tollefsbol, T.O., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 21–44. [Google Scholar]
- Solier, C.; Langen, H. Antibody-based proteomics and biomarker research—Current status and limitations. Proteomics 2014, 14, 774–783. [Google Scholar] [CrossRef]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. MCP 2014, 13, 372–387. [Google Scholar] [CrossRef]
- Sebollela, A.; Campos, R.; Carraro, M.; Pinheiro, N.; Bitencour, A.; Spagnol, V.; Cline, E.; Ayon, N.; Mcgee, J.; Viola, K. Antibody assisted biochemical isolation and conformational analysis of native Alzheimer’s relevant Abeta oligomers. J. Neurochem. 2023, 166, 104. [Google Scholar]
- Chang, I.-F. Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 2006, 6, 6158–6166. [Google Scholar] [CrossRef]
- Tomomori-Sato, C.; Sato, S.; Conaway, R.C.; Conaway, J.W. Immunoaffinity Purification of Protein Complexes from Mammalian Cells. In Gene Regulation: Methods and Protocols; Bina, M., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 273–287. [Google Scholar]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Olinares, P.D.B.; Dunn, A.D.; Padovan, J.C.; Fernandez-Martinez, J.; Rout, M.P.; Chait, B.T. A Robust Workflow for Native Mass Spectrometric Analysis of Affinity-Isolated Endogenous Protein Assemblies. Anal. Chem. 2016, 88, 2799–2807. [Google Scholar] [CrossRef]
- Rosenberg, O.S.; Deindl, S.; Comolli, L.R.; Hoelz, A.; Downing, K.H.; Nairn, A.C.; Kuriyan, J. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006, 273, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Antrobus, R.; Borner, G.H.H. Improved Elution Conditions for Native Co-Immunoprecipitation. PLoS ONE 2011, 6, e18218. [Google Scholar] [CrossRef]
- Liu, X.; Abad, L.; Chatterjee, L.; Cristea, I.M.; Varjosalo, M. Mapping protein–protein interactions by mass spectrometry. Mass Spectrom. Rev. 2024; epub ahead of printing. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Tian, M.; Yin, J.; Duan, H.; Tian, Y.; Wang, H.; Xia, C.; Wang, Z.; Zhu, Y.; Wang, Y.; et al. Biofunctionalized dissolvable hydrogel microbeads enable efficient characterization of native protein complexes. Nat. Commun. 2024, 15, 8633. [Google Scholar] [CrossRef] [PubMed]
- Smisek, D.L.; Hoagland, D.A. Agarose gel electrophoresis of high molecular weight, synthetic polyelectrolytes. Macromolecules 1989, 22, 2270–2277. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef]
- Shoji, M.; Kato, M.; Hashizume, S. Electrophoretic recovery of proteins from polyacrylamide gel. J. Chromatogr. A 1995, 698, 145–162. [Google Scholar] [CrossRef]
- Kurien, B.T.; Scofield, R.H. Extraction of Proteins from Gels: A Brief Review. In Protein Electrophoresis: Methods and Protocols; Kurien, B.T., Scofield, R.H., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 403–405. [Google Scholar]
- Ruggiero-Lopez, D.; Louisot, P.; Martin, A. A nondenaturing preparative gel electrophoresis system for the recovery of functional proteins. Application to the identification of an endogenous protein inhibitor of fucosyl-transferase activities. Anal. Biochem. 1993, 212, 247–252. [Google Scholar] [CrossRef]
- Seelert, H.; Krause, F. Preparative isolation of protein complexes and other bioparticles by elution from polyacrylamide gels. Electrophoresis 2008, 29, 2617–2636. [Google Scholar] [CrossRef]
- Vallejos, R.H.; Ceccarelli, E.; Chan, R. Evidence for the existence of a thylakoid intrinsic protein that binds ferredoxin-NADP+ oxidoreductase. J. Biol. Chem. 1984, 259, 8048–8051. [Google Scholar] [CrossRef]
- Rivoal, J.; Smith, C.R.; Moraes, T.F.; Turpin, D.H.; Plaxton, W.C. A Method for Activity Staining after Native Polyacrylamide Gel Electrophoresis Using a Coupled Enzyme Assay and Fluorescence Detection: Application to the Analysis of Several Glycolytic Enzymes. Anal. Biochem. 2002, 300, 94–99. [Google Scholar] [CrossRef]
- Nowakowski, A.B.; Wobig, W.J.; Petering, D.H. Native SDS-PAGE: High resolution electrophoretic separation of proteins with retention of native properties including bound metal ions. Metallomics 2014, 6, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Wittig, I.; Braun, H.-P.; Schägger, H. Blue native PAGE. Nat. Protoc. 2006, 1, 418–428. [Google Scholar] [CrossRef]
- Wittig, I.; Schägger, H. Features and applications of blue-native and clear-native electrophoresis. Proteomics 2008, 8, 3974–3990. [Google Scholar] [CrossRef] [PubMed]
- SchÄGger, H. 5-Blue Native Electrophoresis. In Membrane Protein Purification and Crystallization, 2nd ed.; Hunte, C., Von Jagow, G., SchÄGger, H., Eds.; Academic Press: San Diego, CA, USA, 2003; pp. 105–130. [Google Scholar]
- Pupo, E.; López, C.M.; Alonso, M.; Hardy, E. High-efficiency passive elution of bacterial lipopolysaccharides from polyacrylamide gels. Electrophoresis 2000, 21, 526–530. [Google Scholar] [CrossRef]
- Wittig, I.; Karas, M.; Schägger, H. High Resolution Clear Native Electrophoresis for In-gel Functional Assays and Fluorescence Studies of Membrane Protein Complexes. Mol. Cell. Proteom. 2007, 6, 1215–1225. [Google Scholar] [CrossRef]
- Wittig, I.; Schägger, H. Advantages and limitations of clear-native PAGE. Proteomics 2005, 5, 4338–4346. [Google Scholar] [CrossRef]
- Sarkozy, D.; Guttman, A. Analysis of Peptides and Proteins by Native and SDS Capillary Gel Electrophoresis Coupled to Electrospray Ionization Mass Spectrometry via a Closed-Circuit Coaxial Sheath Flow Reactor Interface. Anal. Chem. 2023, 95, 7082–7086. [Google Scholar] [CrossRef]
- Timón-Gómez, A.; Pérez-Pérez, R.; Nyvltova, E.; Ugalde, C.; Fontanesi, F.; Barrientos, A. Protocol for the Analysis of Yeast and Human Mitochondrial Respiratory Chain Complexes and Supercomplexes by Blue Native Electrophoresis. STAR Protoc. 2020, 1, 100089. [Google Scholar] [CrossRef]
- Morales, V.; Orenday-Tapia, L.; Ieva, R. Analysis of Transmembrane β-Barrel Proteins by Native and Semi-native Polyacrylamide Gel Electrophoresis. In Transmembrane β-Barrel Proteins: Methods and Protocols; Ieva, R., Ed.; Springer: New York, NY, USA, 2024; pp. 133–145. [Google Scholar]
- Wright, E.P.; Partridge, M.A.; Padula, M.P.; Gauci, V.J.; Malladi, C.S.; Coorssen, J.R. Top-down proteomics: Enhancing 2D gel electrophoresis from tissue processing to high-sensitivity protein detection. Proteomics 2014, 14, 872–889. [Google Scholar] [CrossRef]
- Drews, O.; Zong, C.; Ping, P. Exploring proteasome complexes by proteomic approaches. Proteomics 2007, 7, 1047–1058. [Google Scholar] [CrossRef]
- Pieper, R.; Gatlin, C.L.; Makusky, A.J.; Russo, P.S.; Schatz, C.R.; Miller, S.S.; Su, Q.; McGrath, A.M.; Estock, M.A.; Parmar, P.P.; et al. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 2003, 3, 1345–1364. [Google Scholar] [CrossRef]
- Munawar, N.; Olivero, G.; Jerman, E.; Doyle, B.; Streubel, G.; Wynne, K.; Bracken, A.; Cagney, G. Native gel analysis of macromolecular protein complexes in cultured mammalian cells. Proteomics 2015, 15, 3603–3612. [Google Scholar] [CrossRef]
- Ezsias, B.; Goessweiner-Mohr, N.; Siligan, C.; Horner, A.; Vargas, C.; Keller, S.; Pohl, P. Clear Native Gel Electrophoresis for the Purification of Fluorescently Labeled Membrane Proteins in Native Nanodiscs. Anal. Chem. 2025, 97, 16796–16804. [Google Scholar] [CrossRef]
- Bonaventura, C.; Bonaventura, J.; Stevens, R.; Millington, D. Acrylamide in Polyacrylamide Gels Can Modify Proteins during Electrophoresis. Anal. Biochem. 1994, 222, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Besse, S.; Bulteau, A.L.; Boucher, F.; Riou, B.; Swynghedauw, B.; de Leiris, J. Antioxidant treatment prevents cardiac protein oxidation after ischemia-reperfusion and improves myocardial function and coronary perfusion in senescent hearts. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2006, 57, 541–552. [Google Scholar]
- Ernst, A.; Stolzing, A.; Sandig, G.; Grune, T. Antioxidants effectively prevent oxidation-induced protein damage in OLN 93 cells. Arch. Biochem. Biophys. 2004, 421, 54–60. [Google Scholar] [CrossRef]
- Jacobs, E.; Clad, A. Electroelution of fixed and stained membrane proteins from preparative sodium dodecyl sulfate-polyacrylamide gels into a membrane trap. Anal. Biochem. 1986, 154, 583–589. [Google Scholar] [CrossRef]
- Öfverstedt, L.-G.; Sundelin, J.; Johansson, G. Recovery of proteins on a milligram scale from polyacrylamide electrophoresis gels, exemplified by purification of a retinol-binding protein. Anal. Biochem. 1983, 134, 361–367. [Google Scholar] [CrossRef]
- Bernabeu, C.; Conde, F.P.; Vazouez, D. Extraction of pure ribosomal protein and removal of Coomassie blue from acrylamide gels. Anal. Biochem. 1978, 84, 97–102. [Google Scholar] [CrossRef]
- Gulle, H.; Schoel, B.; Kaufmann, S.H. Direct blotting with viable cells of protein mixtures separated by two-dimensional gel electrophoresis. J. Immunol. Methods 1990, 133, 253–261. [Google Scholar] [CrossRef]
- Persidis, A.; Harcombe, A.A. Simultaneous electroelution of proteins from denaturing or native gels into a well matrix. Anal. Biochem. 1992, 201, 185–189. [Google Scholar] [CrossRef]
- Andersen, P.; Heron, I. Simultaneous electroelution of whole SDS-polyacrylamide gels for the direct cellular analysis of complex protein mixtures. J. Immunol. Methods 1993, 161, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.C.; Doucette, A.A. Gel-Eluted Liquid Fraction Entrapment Electrophoresis: An Electrophoretic Method for Broad Molecular Weight Range Proteome Separation. Anal. Chem. 2008, 80, 1568–1573. [Google Scholar] [CrossRef] [PubMed]
- Skinner, O.S.; Do Vale, L.H.; Catherman, A.D.; Havugimana, P.C.; de Sousa, M.V.; Compton, P.D.; Kelleher, N.L. Native GELFrEE: A new separation technique for biomolecular assemblies. Anal. Chem. 2015, 87, 3032–3038. [Google Scholar] [CrossRef] [PubMed]
- Skinner, O.S.; Haverland, N.A.; Fornelli, L.; Melani, R.D.; Do Vale, L.H.F.; Seckler, H.S.; Doubleday, P.F.; Schachner, L.F.; Srzentić, K.; Kelleher, N.L.; et al. Top-down characterization of endogenous protein complexes with native proteomics. Nat. Chem. Biol. 2018, 14, 36–41. [Google Scholar] [CrossRef]
- Durbin, K.R.; Fornelli, L.; Fellers, R.T.; Doubleday, P.F.; Narita, M.; Kelleher, N.L. Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa. J. Proteome Res. 2016, 15, 976–982. [Google Scholar] [CrossRef]
- Catherman, A.D.; Durbin, K.R.; Ahlf, D.R.; Early, B.P.; Fellers, R.T.; Tran, J.C.; Thomas, P.M.; Kelleher, N.L. Large-scale top-down proteomics of the human proteome: Membrane proteins, mitochondria, and senescence. Mol. Cell. Proteom. 2013, 12, 3465–3473. [Google Scholar] [CrossRef]
- Liu, F.; Rijkers, D.T.S.; Post, H.; Heck, A.J.R. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 2015, 12, 1179–1184. [Google Scholar] [CrossRef]
- Bier, M. New Principle of Preparative Electrophoresis. Science 1957, 125, 1084–1085. [Google Scholar] [CrossRef]
- Turgeon, R.T.; Bowser, M.T. Micro free-flow electrophoresis: Theory and applications. Anal. Bioanal. Chem. 2009, 394, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Islinger, M.; Wildgruber, R.; Völkl, A. Preparative free-flow electrophoresis, a versatile technology complementing gradient centrifugation in the isolation of highly purified cell organelles. Electrophoresis 2018, 39, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Raymond, D.E.; Manz, A.; Widmer, H.M. Continuous Separation of High Molecular Weight Compounds Using a Microliter Volume Free-Flow Electrophoresis Microstructure. Anal. Chem. 1996, 68, 2515–2522. [Google Scholar] [CrossRef]
- Islinger, M.; Eckerskorn, C.; Völkl, A. Free-flow electrophoresis in the proteomic era: A technique in flux. Electrophoresis 2010, 31, 1754–1763. [Google Scholar] [CrossRef]
- Lee, Y.; Kwon, J.-S. Microfluidic free-flow electrophoresis: A promising tool for protein purification and analysis in proteomics. J. Ind. Eng. Chem. 2022, 109, 79–99. [Google Scholar] [CrossRef]
- Kohlheyer, D.; Eijkel, J.C.T.; van den Berg, A.; Schasfoort, R.B.M. Miniaturizing free-flow electrophoresis–a critical review. Electrophoresis 2008, 29, 977–993. [Google Scholar] [CrossRef]
- Kohlheyer, D.; Eijkel, J.C.T.; Schlautmann, S.; van den Berg, A.; Schasfoort, R.B.M. Bubble-Free Operation of a Microfluidic Free-Flow Electrophoresis Chip with Integrated Pt Electrodes. Anal. Chem. 2008, 80, 4111–4118. [Google Scholar] [CrossRef]
- Jeon, H.; Kim, Y.; Lim, G. Continuous particle separation using pressure-driven flow-induced miniaturizing free-flow electrophoresis (PDF-induced μ-FFE). Sci. Rep. 2016, 6, 19911. [Google Scholar] [CrossRef]
- Ayan, K.; Ganar, K.; Deshpande, S.; Boom, R.M.; Nikiforidis, C.V. Continuous counter-current electrophoretic separation of oleosomes and proteins from oilseeds. Food Hydrocoll. 2023, 144, 109053. [Google Scholar] [CrossRef]
- Wen, J.; Wilker, E.W.; Yaffe, M.B.; Jensen, K.F. Microfluidic Preparative Free-Flow Isoelectric Focusing: System Optimization for Protein Complex Separation. Anal. Chem. 2010, 82, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Islinger, M.; Weber, P.; Eckerskorn, C.; Völkl, A. Efficient separation and analysis of peroxisomal membrane proteins using free-flow isoelectric focusing. Electrophoresis 2004, 25, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Ouvry-Patat, S.A.; Torres, M.P.; Quek, H.-H.; Gelfand, C.A.; O’Mullan, P.; Nissum, M.; Schroeder, G.K.; Han, J.; Elliott, M.; Dryhurst, D.; et al. Free-flow electrophoresis for top-down proteomics by Fourier transform ion cyclotron resonance mass spectrometry. Proteomics 2008, 8, 2798–2808. [Google Scholar] [CrossRef]
- D’Atri, V.; Murisier, A.; Fekete, S.; Veuthey, J.-L.; Guillarme, D. Current and future trends in reversed-phase liquid chromatography-mass spectrometry of therapeutic proteins. TrAC Trends Anal. Chem. 2020, 130, 115962. [Google Scholar] [CrossRef]
- Kaltashov, I.A.; Pawlowski, J.W.; Yang, W.; Muneeruddin, K.; Yao, H.; Bobst, C.E.; Lipatnikov, A.N. LC/MS at the whole protein level: Studies of biomolecular structure and interactions using native LC/MS and cross-path reactive chromatography (XP-RC) MS. Methods 2018, 144, 14–26. [Google Scholar] [CrossRef]
- Muneeruddin, K.; Thomas, J.J.; Salinas, P.A.; Kaltashov, I.A. Characterization of small protein aggregates and oligomers using size exclusion chromatography with online detection by native electrospray ionization mass spectrometry. Anal. Chem. 2014, 86, 10692–10699. [Google Scholar] [CrossRef]
- Mayer, C.L.; Snyder, W.K.; Swietlicka, M.A.; VanSchoiack, A.D.; Austin, C.R.; McFarland, B.J. Size-exclusion chromatography can identify faster-associating protein complexes and evaluate design strategies. BMC Res. Notes 2009, 2, 135. [Google Scholar] [CrossRef]
- Arakawa, T.; Ejima, D.; Li, T.; Philo, J.S. The critical role of mobile phase composition in size exclusion chromatography of protein pharmaceuticals. J. Pharm. Sci. 2010, 99, 1674–1692. [Google Scholar] [CrossRef]
- van Schaick, G.; Haselberg, R.; Somsen, G.W.; Wuhrer, M.; Domínguez-Vega, E. Studying protein structure and function by native separation–mass spectrometry. Nat. Rev. Chem. 2022, 6, 215–231. [Google Scholar] [CrossRef]
- Ehkirch, A.; Hernandez-Alba, O.; Colas, O.; Beck, A.; Guillarme, D.; Cianférani, S. Hyphenation of size exclusion chromatography to native ion mobility mass spectrometry for the analytical characterization of therapeutic antibodies and related products. J. Chromatogr. 2018, 1086, 176–183. [Google Scholar] [CrossRef]
- Yan, Y.; Xing, T.; Wang, S.; Daly, T.J.; Li, N. Coupling mixed-mode size exclusion chromatography with native mass spectrometry for sensitive detection and quantitation of homodimer impurities in bispecific IgG. Anal. Chem. 2019, 91, 11417–11424. [Google Scholar] [CrossRef] [PubMed]
- Goyon, A.; Beck, A.; Colas, O.; Sandra, K.; Guillarme, D.; Fekete, S. Evaluation of size exclusion chromatography columns packed with sub-3 μm particles for the analysis of biopharmaceutical proteins. J. Chromatogr. A 2017, 1498, 80–89. [Google Scholar] [CrossRef]
- Fekete, S.; Ganzler, K.; Guillarme, D. Critical evaluation of fast size exclusion chromatographic separations of protein aggregates, applying sub-2 μm particles. J. Pharm. Biomed. Anal. 2013, 78, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Ventouri, I.K.; Veelders, S.; Passamonti, M.; Endres, P.; Roemling, R.; Schoenmakers, P.J.; Somsen, G.W.; Haselberg, R.; Gargano, A.F.G. Micro-flow size-exclusion chromatography for enhanced native mass spectrometry of proteins and protein complexes. Anal. Chim. Acta 2023, 1266, 341324. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kou, Q.; Guo, R.; Yang, Z.; Chen, D.; Liu, X.; Hong, H.; Sun, L. Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography–Capillary Zone Electrophoresis–Tandem Mass Spectrometry. Anal. Chem. 2018, 90, 10095–10099. [Google Scholar] [CrossRef]
- Deslignière, E.; Ehkirch, A.; Botzanowski, T.; Beck, A.; Hernandez-Alba, O.; Cianférani, S. Toward Automation of Collision-Induced Unfolding Experiments through Online Size Exclusion Chromatography Coupled to Native Mass Spectrometry. Anal. Chem. 2020, 92, 12900–12908. [Google Scholar] [CrossRef]
- Zhai, Z.; Holmark, T.; van der Zon, A.A.M.; Tseliou, V.; Mutti, F.G.; Astefanei, A.; Gargano, A.F.G. Nanoflow Size Exclusion Chromatography–Native Mass Spectrometry of Intact Proteoforms and Protein Complexes. Anal. Chem. 2025, 97, 12241–12250. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.-L.; Guillarme, D. Ion-exchange chromatography for the characterization of biopharmaceuticals. J. Pharm. Biomed. Anal. 2015, 113, 43–55. [Google Scholar] [CrossRef]
- Füssl, F.; Cook, K.; Scheffler, K.; Farrell, A.; Mittermayr, S.; Bones, J. Charge Variant Analysis of Monoclonal Antibodies Using Direct Coupled pH Gradient Cation Exchange Chromatography to High-Resolution Native Mass Spectrometry. Anal. Chem. 2018, 90, 4669–4676. [Google Scholar] [CrossRef]
- Füssl, F.; Trappe, A.; Cook, K.; Scheffler, K.; Fitzgerald, O.; Bones, J. Comprehensive characterisation of the heterogeneity of adalimumab via charge variant analysis hyphenated on-line to native high resolution Orbitrap mass spectrometry. In MAbs; Taylor & Francis: Abingdon, UK, 2019; Volume 11, pp. 116–128. [Google Scholar] [CrossRef]
- Füssl, F.; Criscuolo, A.; Cook, K.; Scheffler, K.; Bones, J. Cracking Proteoform Complexity of Ovalbumin with Anion-Exchange Chromatography–High-Resolution Mass Spectrometry under Native Conditions. J. Proteome Res. 2019, 18, 3689–3702. [Google Scholar] [CrossRef]
- Ma, F.; Raoufi, F.; Bailly, M.A.; Fayadat-Dilman, L.; Tomazela, D. Hyphenation of strong cation exchange chromatography to native mass spectrometry for high throughput online characterization of charge heterogeneity of therapeutic monoclonal antibodies. In MAbs; Taylor & Francis: Abingdon, UK, 2020; Volume 12, p. 1763762. [Google Scholar] [CrossRef]
- Fischer, M.S.; Rogers, H.T.; Chapman, E.A.; Chan, H.-J.; Krichel, B.; Gao, Z.; Larson, E.J.; Ge, Y. Online Mixed-Bed Ion Exchange Chromatography for Native Top-Down Proteomics of Complex Mixtures. J. Proteome Res. 2024, 23, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Shire, S.J. 2-Analytical tools used in the formulation and assessment of stability of monoclonal antibodies (mAbs). In Monoclonal Antibodies; Shire, S.J., Ed.; Woodhead Publishing: Cambridge, UK, 2015; pp. 17–44. [Google Scholar]
- Queiroz, J.A.; Tomaz, C.T.; Cabral, J.M.S. Hydrophobic interaction chromatography of proteins. J. Biotechnol. 2001, 87, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Fekete, S.; Veuthey, J.-L.; Beck, A.; Guillarme, D. Hydrophobic interaction chromatography for the characterization of monoclonal antibodies and related products. J. Pharm. Biomed. Anal. 2016, 130, 3–18. [Google Scholar] [CrossRef]
- Xiu, L.; Valeja, S.G.; Alpert, A.J.; Jin, S.; Ge, Y. Effective protein separation by coupling hydrophobic interaction and reverse phase chromatography for top-down proteomics. Anal. Chem. 2014, 86, 7899–7906. [Google Scholar] [CrossRef]
- Chen, B.; Peng, Y.; Valeja, S.G.; Xiu, L.; Alpert, A.J.; Ge, Y. Online hydrophobic interaction chromatography–mass spectrometry for top-down proteomics. Anal. Chem. 2016, 88, 1885–1891. [Google Scholar] [CrossRef]
- Chen, B.; Lin, Z.; Alpert, A.J.; Fu, C.; Zhang, Q.; Pritts, W.A.; Ge, Y. Online hydrophobic interaction chromatography–mass spectrometry for the analysis of intact monoclonal antibodies. Anal. Chem. 2018, 90, 7135–7138. [Google Scholar] [CrossRef]
- Gilroy, J.J.; Eakin, C.M. Characterization of drug load variants in a thiol linked antibody-drug conjugate using multidimensional chromatography. J. Chromatogr. B 2017, 1060, 182–189. [Google Scholar] [CrossRef]
- Kempen, T.; Cadang, L.; Fan, Y.; Zhang, K.; Chen, T.; Wei, B. Online native hydrophobic interaction chromatography-mass spectrometry of antibody-drug conjugates. In MAbs; Taylor & Francis: Abingdon, UK, 2025; Volume 17, p. 2446304. [Google Scholar] [CrossRef]
- Wei, B.; Han, G.; Tang, J.; Sandoval, W.; Zhang, Y.T. Native Hydrophobic Interaction Chromatography Hyphenated to Mass Spectrometry for Characterization of Monoclonal Antibody Minor Variants. Anal. Chem. 2019, 91, 15360–15364. [Google Scholar] [CrossRef]
- Rodriguez, E.L.; Poddar, S.; Iftekhar, S.; Suh, K.; Woolfork, A.G.; Ovbude, S.; Pekarek, A.; Walters, M.; Lott, S.; Hage, D.S. Affinity chromatography: A review of trends and developments over the past 50 years. J. Chromatogr. B 2020, 1157, 122332. [Google Scholar] [CrossRef]
- Oliver, C.; Jamur, M.C. Immunocytochemical Methods and Protocols; Methods in Molecular Biology; Humana Press: Hatfield, UK, 2010; Volume 588, pp. 63–66. [Google Scholar] [CrossRef]
- Banks, C.A.S.; Kong, S.E.; Washburn, M.P. Affinity purification of protein complexes for analysis by multidimensional protein identification technology. Protein Expr. Purif. 2012, 86, 105–119. [Google Scholar] [CrossRef]
- LaCava, J.; Molloy, K.R.; Taylor, M.S.; Domanski, M.; Chait, B.T.; Rout, M.P. Affinity Proteomics to Study Endogenous Protein Complexes: Pointers, Pitfalls, Preferences and Perspectives. BioTechniques 2015, 58, 103–119. [Google Scholar] [CrossRef]
- Schlothauer, T.; Rueger, P.; Stracke, J.O.; Hertenberger, H.; Fingas, F.; Kling, L.; Emrich, T.; Drabner, G.; Seeber, S.; Auer, J. Analytical FcRn affinity chromatography for functional characterization of monoclonal antibodies. In MAbs; Taylor & Francis: Abingdon, UK, 2013; Volume 11, pp. 576–586. [Google Scholar]
- Lippold, S.; Nicolardi, S.; Domínguez-Vega, E.; Heidenreich, A.-K.; Vidarsson, G.; Reusch, D.; Haberger, M.; Wuhrer, M.; Falck, D. Glycoform-resolved FcɣRIIIa affinity chromatography–mass spectrometry. In MAbs; Taylor & Francis: Abingdon, UK, 2019; pp. 1191–1196. [Google Scholar]
- Prinston, J.E.; Peng, W.; Provoncha, K.; Moon, Y.; Koufos, E.; Sandu, C.; Fu, Y.; Yan, Y.; Wang, S.; Li, N.; et al. A target affinity enrichment workflow to characterize critical post-translational modifications within therapeutic antibodies. J. Pharm. Sci. 2025, 114, 103710. [Google Scholar] [CrossRef]
- Cotham, V.C.; Liu, A.P.; Wang, S.; Li, N. A generic platform to couple affinity chromatography with native mass spectrometry for the analysis of therapeutic monoclonal antibodies. J. Pharm. Biomed. Anal. 2023, 228, 115337. [Google Scholar] [CrossRef] [PubMed]
- Schwenzer, A.-K.; Kruse, L.; Jooß, K.; Neusüß, C. Capillary electrophoresis-mass spectrometry for protein analyses under native conditions: Current progress and perspectives. Proteomics 2024, 24, 2300135. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Yang, Z.; McCool, E.N.; Lubeckyj, R.A.; Chen, D.; Sun, L. Capillary zone electrophoresis-mass spectrometry for top-down proteomics. TrAC Trends Anal. Chem. 2019, 120, 115644. [Google Scholar] [CrossRef] [PubMed]
- Haselberg, R.; de Jong, G.J.; Somsen, G.W. Capillary electrophoresis–mass spectrometry for the analysis of intact proteins. J. Chromatogr. A 2007, 1159, 81–109. [Google Scholar] [CrossRef]
- Haselberg, R.; Ratnayake, C.K.; de Jong, G.J.; Somsen, G.W. Performance of a sheathless porous tip sprayer for capillary electrophoresis–electrospray ionization-mass spectrometry of intact proteins. J. Chromatogr. A 2010, 1217, 7605–7611. [Google Scholar] [CrossRef]
- Hajba, L.; Guttman, A. Recent advances in column coatings for capillary electrophoresis of proteins. TrAC Trends Anal. Chem. 2017, 90, 38–44. [Google Scholar] [CrossRef]
- Zhang, W.; Xiang, Y.; Xu, W. Probing protein higher-order structures by native capillary electrophoresis-mass spectrometry. TrAC Trends Anal. Chem. 2022, 157, 116739. [Google Scholar] [CrossRef]
- Shen, X.; Liang, Z.; Xu, T.; Yang, Z.; Wang, Q.; Chen, D.; Pham, L.; Du, W.; Sun, L. Investigating native capillary zone electrophoresis-mass spectrometry on a high-end quadrupole-time-of-flight mass spectrometer for the characterization of monoclonal antibodies. Int. J. Mass Spectrom. 2021, 462, 116541. [Google Scholar] [CrossRef] [PubMed]
- Belov, A.M.; Viner, R.; Santos, M.R.; Horn, D.M.; Bern, M.; Karger, B.L.; Ivanov, A.R. Analysis of Proteins, Protein Complexes, and Organellar Proteomes Using Sheathless Capillary Zone Electrophoresis-Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 2614–2634. [Google Scholar] [CrossRef] [PubMed]
- Jooß, K.; McGee, J.P.; Melani, R.D.; Kelleher, N.L. Standard procedures for native CZE-MS of proteins and protein complexes up to 800 kDa. Electrophoresis 2021, 42, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Q.; Qi, Z.; Moeller, W.; Wysocki, V.H.; Sun, L. Native Proteomics by Capillary Zone Electrophoresis-Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2024, 63, e202408370. [Google Scholar] [CrossRef]
- Jooß, K.; Schachner, L.F.; Watson, R.; Gillespie, Z.B.; Howard, S.A.; Cheek, M.A.; Meiners, M.J.; Sobh, A.; Licht, J.D.; Keogh, M.-C.; et al. Separation and Characterization of Endogenous Nucleosomes by Native Capillary Zone Electrophoresis–Top-Down Mass Spectrometry. Anal. Chem. 2021, 93, 5151–5160. [Google Scholar] [CrossRef]
- Drouin, N.; Boccard, J.; Rudaz, S.; González-Ruiz, V. Data Analysis Strategies in CE–MS for Metabolomics. In Capillary Electrophoresis-Mass Spectrometry for Proteomics and Metabolomics; Wiley-VCH GmbH: Weinheim, Germany, 2022; pp. 35–63. [Google Scholar]
- Sadeghi, S.A.; Chen, W.; Wang, Q.; Wang, Q.; Fang, F.; Liu, X.; Sun, L. Pilot Evaluation of the Long-Term Reproducibility of Capillary Zone Electrophoresis–Tandem Mass Spectrometry for Top-Down Proteomics of a Complex Proteome Sample. J. Proteome Res. 2024, 23, 1399–1407. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, R.; Zhang, W.; Hong, J.; Xiang, Y.; Xu, W.J.C.s. Rapid 3-dimensional shape determination of globular proteins by mobility capillary electrophoresis and native mass spectrometry. Chem. Sci. 2020, 11, 4758–4765. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, H.; Zhang, R.; Fang, X.; Xu, W. Structure and effective charge characterization of proteins by a mobility capillary electrophoresis based method. Chem. Sci. 2019, 10, 7779–7787. [Google Scholar] [CrossRef]
- Dubský, P.; Dvořák, M.; Ansorge, M. Affinity capillary electrophoresis: The theory of electromigration. Anal. Bioanal. Chem. 2016, 408, 8623–8641. [Google Scholar] [CrossRef]
- Hong, J.; Wu, H.; Zhang, R.; He, M.; Xu, W. The Coupling of Taylor Dispersion Analysis and Mass Spectrometry to Differentiate Protein Conformations. Anal. Chem. 2020, 92, 5200–5206. [Google Scholar] [CrossRef]
- Domínguez-Vega, E.; Haselberg, R.; Somsen, G.W.; de Jong, G.J. Simultaneous assessment of protein heterogeneity and affinity by capillary electrophoresis-mass spectrometry. Anal. Chem. 2015, 87, 8781–8788. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Weber, S.G. Determination of binding constants by affinity capillary electrophoresis, electrospray ionization mass spectrometry and phase-distribution methods. TrAC Trends Anal. Chem. 2008, 27, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Vuignier, K.; Veuthey, J.L.; Carrupt, P.A.; Schappler, J. Characterization of drug-protein interactions by capillary electrophoresis hyphenated to mass spectrometry. Electrophoresis 2012, 33, 3306–3315. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dou, Z.; Yuan, Y.; Man, S.; Wolfs, K.; Adams, E.; Van Schepdael, A. On-line screening of matrix metalloproteinase inhibitors by capillary electrophoresis coupled to ESI mass spectrometry. J. Chromatogr. B 2013, 930, 48–53. [Google Scholar] [CrossRef]
- Mironov, G.G.; Clouthier, C.M.; Akbar, A.; Keillor, J.W.; Berezovski, M.V. Simultaneous analysis of enzyme structure and activity by kinetic capillary electrophoresis-MS. Nat. Chem. Biol. 2016, 12, 918–922. [Google Scholar] [CrossRef]
- Fermas, S.; Gonnet, F.; Sutton, A.; Charnaux, N.; Mulloy, B.; Du, Y.; Baleux, F.; Daniel, R. Sulfated oligosaccharides (heparin and fucoidan) binding and dimerization of stromal cell-derived factor-1 (SDF-1/CXCL 12) are coupled as evidenced by affinity CE-MS analysis. Glycobiology 2008, 18, 1054–1064. [Google Scholar] [CrossRef]
- Langmajerová, M.; Řemínek, R.; Pelcová, M.; Foret, F.; Glatz, Z. Combination of on-line CE assay with MS detection for the study of drug metabolism by cytochromes P450. Electrophoresis 2015, 36, 1365–1373. [Google Scholar] [CrossRef]
- Davoine, C.; Fillet, M. Hyphenation of Affinity Capillary Electrophoresis with Mass Spectrometry for the Study of Ligand–Protein Interactions: n-Methylmorpholine Acetate Buffer and Polydopamine-Based Coating as Key Assets. Anal. Chem. 2025, 97, 3988–3995. [Google Scholar] [CrossRef]
- Fanigliulo, A.; Bortolotti, F.; Pascali, J.; Tagliaro, F. Chapter 15 Forensic toxicological screening with capillary electrophoresis and related techniques. In Handbook of Analytical Separations; Bogusz, M.J., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2008; Volume 6, pp. 513–534. [Google Scholar]
- Xu, T.; Sun, L. A Mini Review on Capillary Isoelectric Focusing-Mass Spectrometry for Top-Down Proteomics. Front. Chem. 2021, 9, 651757. [Google Scholar] [CrossRef]
- Kristl, T.; Stutz, H.; Wenz, C.; Rozing, G. Principles and Applications of Capillary Isoelectric Focusing; Agilent Technologies, Inc.: Santa Clara, CA, USA, 2014; pp. 1–50. [Google Scholar]
- Lopez-Soto-Yarritu, P.; Díez-Masa, J.C.; Cifuentes, A.; de Frutos, M. Improved capillary isoelectric focusing method for recombinant erythropoietin analysis. J. Chromatogr. A 2002, 968, 221–228. [Google Scholar] [CrossRef]
- Fonslow, B.R.; Kang, S.A.; Gestaut, D.R.; Graczyk, B.; Davis, T.N.; Sabatini, D.M.; Yates, J.R., III. Native capillary isoelectric focusing for the separation of protein complex isoforms and subcomplexes. Anal. Chem. 2010, 82, 6643–6651. [Google Scholar] [CrossRef][Green Version]
- Przybylski, C.; Mokaddem, M.; Prull-Janssen, M.; Saesen, E.; Lortat-Jacob, H.; Gonnet, F.; Varenne, A.; Daniel, R. On-line capillary isoelectric focusing hyphenated to native electrospray ionization mass spectrometry for the characterization of interferon-γ and variants. Analyst 2015, 140, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Han, L.; Sun, L. Automated Capillary Isoelectric Focusing-Mass Spectrometry with Ultrahigh Resolution for Characterizing Microheterogeneity and Isoelectric Points of Intact Protein Complexes. Anal. Chem. 2022, 94, 9674–9682. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Lamp, J.; Xia, Q.; Zhang, Y. Capillary Isoelectric Focusing-Mass Spectrometry Method for the Separation and Online Characterization of Intact Monoclonal Antibody Charge Variants. Anal. Chem. 2018, 90, 2246–2254. [Google Scholar] [CrossRef]
- Naghdi, E.; Reinau, M.E.; Krogh, T.N.; Neusüß, C. Chemical Mobilization-Based Capillary Isoelectric Focusing–Mass Spectrometry Using the nanoCEasy Interface for Pharmaceutical Protein Analysis. Anal. Chem. 2024, 96, 12827–12837. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Diaz, R.; Wehr, T.; Zhu, M. Capillary isoelectric focusing. Electrophoresis 1997, 18, 2134–2144. [Google Scholar] [CrossRef]
- Rodriguez-Diaz, R.; Zhu, M.; Wehr, T. Strategies to improve performance of capillary isoelectric focusing. J. Chromatogr. A 1997, 772, 145–160. [Google Scholar] [CrossRef]
- Wu, J.; Pawliszyn, J. Universal detection for capillary isoelectric focusing without mobilization using concentration gradient imaging system. Anal. Chem. 1992, 64, 224–227. [Google Scholar] [CrossRef]
- Wu, J.; Pawliszyn, J. Capillary isoelectric focusing with a universal concentration gradient imaging system using a charge-coupled photodiode array. Anal. Chem. 1992, 64, 2934–2941. [Google Scholar] [CrossRef]
- Wu, J.; Pawliszyn, J. Dual Detection for Capillary Isoelectric Focusing with Refractive Index Gradient and Absorption Imaging Detectors. Anal. Chem. 1994, 66, 867–873. [Google Scholar] [CrossRef]
- Salas-Solano, O.; Babu, K.; Park, S.S.; Zhang, X.; Zhang, L.; Sosic, Z.; Boumajny, B.; Zeng, M.; Cheng, K.-C.; Reed-Bogan, A.; et al. Intercompany Study to Evaluate the Robustness of Capillary Isoelectric Focusing Technology for the Analysis of Monoclonal Antibodies. Chromatographia 2011, 73, 1137–1144. [Google Scholar] [CrossRef]
- He, X.Z.; Que, A.H.; Mo, J.J. Analysis of charge heterogeneities in mAbs using imaged CE. Electrophoresis 2009, 30, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Kwok, T.; Chan, S.L.; Courtney, M.; Zhou, M.; Huang, T.; Bo, T.; Li, V.; Chen, T. Imaged capillary isoelectric focusing tandem high-resolution mass spectrometry using nano electrospray ionization (ESI) for protein heterogeneity characterization. Anal. Biochem. 2023, 680, 115312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, G.; Du, M.; Bo, T.; Chen, T.; Huang, T. Imaged Capillary Isoelectric Focusing Coupled to High-Resolution Mass Spectrometry (icIEF-MS) for Cysteine-Linked Antibody-Drug Conjugate (ADC) Heterogeneity Characterization Under Native Condition. Electrophoresis 2024, 45, 1915–1926. [Google Scholar] [CrossRef]
- Rustandi, R.R.; Peklansky, B.; Anderson, C.L. Use of imaged capillary isoelectric focusing technique in development of diphtheria toxin mutant CRM197. Electrophoresis 2014, 35, 1065–1071. [Google Scholar] [CrossRef]
- Loughney, J.W.; Minsker, K.; Ha, S.; Rustandi, R.R. Development of an imaged capillary isoelectric focusing method for characterizing the surface charge of mRNA lipid nanoparticle vaccines. Electrophoresis 2019, 40, 2602–2609. [Google Scholar] [CrossRef]
- Bo, T.; Yang, F.; Yan, B.; Michels, D.A.; Huang, T.; Pawliszyn, J. Recent developments of imaged capillary isoelectric focusing technology for in-depth biopharmaceutical characterization. TrAC Trends Anal. Chem. 2025, 187, 118212. [Google Scholar] [CrossRef]
- Ghizzani, V.; Ascione, A.; Gonnella, F.; Massolini, G.; Luciani, F. Exploring imaged capillary isoelectric focusing parameters for enhanced charge variants quality control. Front. Chem. 2025, 13, 1536222. [Google Scholar] [CrossRef]
- Wu, J.; McElroy, W.; Pawliszyn, J.; Heger, C.D. Imaged capillary isoelectric focusing: Applications in the pharmaceutical industry and recent innovations of the technology. TrAC Trends Anal. Chem. 2022, 150, 116567. [Google Scholar] [CrossRef]
- Xian, F.; Hendrickson, C.L.; Marshall, A.G. High Resolution Mass Spectrometry. Anal. Chem. 2012, 84, 708–719. [Google Scholar] [CrossRef]
- Makarov, A.; Denisov, E.; Lange, O.; Horning, S. Dynamic range of mass accuracy in LTQ orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 2006, 17, 977–982. [Google Scholar] [CrossRef]
- Kafader, J.O.; Melani, R.D.; Senko, M.W.; Makarov, A.A.; Kelleher, N.L.; Compton, P.D. Measurement of Individual Ions Sharply Increases the Resolution of Orbitrap Mass Spectra of Proteins. Anal. Chem. 2019, 91, 2776–2783. [Google Scholar] [CrossRef] [PubMed]
- Kafader, J.O.; Melani, R.D.; Durbin, K.R.; Ikwuagwu, B.; Early, B.P.; Fellers, R.T.; Beu, S.C.; Zabrouskov, V.; Makarov, A.A.; Maze, J.T.; et al. Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat. Methods 2020, 17, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.V.; Zydney, A.L. Protein Ultrafiltration. In Encyclopedia of Industrial Biotechnology; Wiley: Hoboken, NJ, USA, 2010; pp. 1–25. [Google Scholar]
- Majors, R.E. Sample Preparation for Large-Scale Protein Purification; LCGC Europe: Chester, UK, 2005; pp. 82–92. [Google Scholar]
- Park, H.-M.; Winton, V.J.; Drader, J.J.; Manalili Wheeler, S.; Lazar, G.A.; Kelleher, N.L.; Liu, Y.; Tran, J.C.; Compton, P.D. Novel Interface for High-Throughput Analysis of Biotherapeutics by Electrospray Mass Spectrometry. Anal. Chem. 2020, 92, 2186–2193. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.P.; Senko, M.W.; Jooß, K.; Des Soye, B.J.; Compton, P.D.; Kelleher, N.L.; Kafader, J.O. Automated Control of Injection Times for Unattended Acquisition of Multiplexed Individual Ion Mass Spectra. Anal. Chem. 2022, 94, 16543–16548. [Google Scholar] [CrossRef]
- Des Soye, B.J.; McGee, J.P.; Hollas, M.A.R.; Forte, E.; Fellers, R.T.; Melani, R.D.; Wilkins, J.T.; Compton, P.D.; Kafader, J.O.; Kelleher, N.L. Automated Immunoprecipitation, Sample Preparation, and Individual Ion Mass Spectrometry Platform for Proteoforms. Anal. Chem. 2024, 96, 13879–13887. [Google Scholar] [CrossRef]
- Horlebein, J.; Moon, E.; Szekeres, G.P.; von Helden, G.; Österlund, N.; Pagel, K. Gas-phase purification enables structural studies of amyloid intermediates. Trends Chem. 2025, 7, 317–332. [Google Scholar] [CrossRef]
- Bernstein, S.L.; Wyttenbach, T.; Baumketner, A.; Shea, J.-E.; Bitan, G.; Teplow, D.B.; Bowers, M.T. Amyloid β-Protein: Monomer Structure and Early Aggregation States of Aβ42 and Its Pro19 Alloform. J. Am. Chem. Soc. 2005, 127, 2075–2084. [Google Scholar] [CrossRef]
- Bernstein, S.L.; Dupuis, N.F.; Lazo, N.D.; Wyttenbach, T.; Condron, M.M.; Bitan, G.; Teplow, D.B.; Shea, J.-E.; Ruotolo, B.T.; Robinson, C.V.; et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009, 1, 326–331. [Google Scholar] [CrossRef]
- Dupuis, N.F.; Wu, C.; Shea, J.-E.; Bowers, M.T. The Amyloid Formation Mechanism in Human IAPP: Dimers Have β-Strand Monomer−Monomer Interfaces. J. Am. Chem. Soc. 2011, 133, 7240–7243. [Google Scholar] [CrossRef]
- Smith, D.P.; Radford, S.E.; Ashcroft, A.E. Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl. Acad. Sci. USA 2010, 107, 6794–6798. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.A.; Berhane, B.; Kaddis, C.S.; Wooding, K.M.; Xie, Y.; Kaufman, S.L.; Chernushevich, I.V. Electrospray ionization mass spectrometry and ion mobility analysis of the 20S proteasome complex. J. Am. Soc. Mass Spectrom. 2005, 16, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, B.T.; Giles, K.; Campuzano, I.; Sandercock, A.M.; Bateman, R.H.; Robinson, C.V. Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science 2005, 310, 1658–1661. [Google Scholar] [CrossRef] [PubMed]
- Politis, A.; Park, A.Y.; Hyung, S.-J.; Barsky, D.; Ruotolo, B.T.; Robinson, C.V. Integrating Ion Mobility Mass Spectrometry with Molecular Modelling to Determine the Architecture of Multiprotein Complexes. PLoS ONE 2010, 5, e12080. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, M.; Sandercock, A.M.; Fraser, C.S.; Ridlova, G.; Stephens, E.; Schenauer, M.R.; Yokoi-Fong, T.; Barsky, D.; Leary, J.A.; Hershey, J.W.; et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. USA 2008, 105, 18139–18144. [Google Scholar] [CrossRef]
- Lane, L.A.; Fernández-Tornero, C.; Zhou, M.; Morgner, N.; Ptchelkine, D.; Steuerwald, U.; Politis, A.; Lindner, D.; Gvozdenovic, J.; Gavin, A.-C.; et al. Mass Spectrometry Reveals Stable Modules in holo and apo RNA Polymerases I and III. Structure 2011, 19, 90–100. [Google Scholar] [CrossRef]
- Uetrecht, C.; Barbu, I.M.; Shoemaker, G.K.; van Duijn, E.; Heck, A.J.R. Interrogating viral capsid assembly with ion mobility–mass spectrometry. Nat. Chem. 2011, 3, 126–132. [Google Scholar] [CrossRef]
- Christofi, E.; Barran, P. Ion Mobility Mass Spectrometry (IM-MS) for Structural Biology: Insights Gained by Measuring Mass, Charge, and Collision Cross Section. Chem. Rev. 2023, 123, 2902–2949. [Google Scholar] [CrossRef]
- Liénard, R.; Duez, Q.; Grayson, S.M.; Gerbaux, P.; Coulembier, O.; De Winter, J. Limitations of ion mobility spectrometry-mass spectrometry for the relative quantification of architectural isomeric polymers: A case study. Mass Spectrom. 2020, 34, e8660. [Google Scholar] [CrossRef]
- Vickers, S.; Aldobiyan, I.; Lowen, S.M.; Irving, J.A.; Lomas, D.A.; Thalassinos, K. Top-Down Ion Mobility Mass Spectrometry Reveals a Disease Associated Conformational Ensemble of Alpha-1-antitrypsin. J. Am. Chem. Soc. 2025, 147, 16909–16921. [Google Scholar] [CrossRef]
- Kocurek, K.I.; Griffiths, R.L.; Cooper, H.J. Ambient ionisation mass spectrometry for in situ analysis of intact proteins. J. Mass Spectrom. 2018, 53, 565–578. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Monge, M.E.; Fernández, F.M. Ambient mass spectrometry in metabolomics. Analyst 2017, 142, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.J.; Griffiths, R.L.; Edwards, R.L.; Cooper, H.J. Native Liquid Extraction Surface Analysis Mass Spectrometry: Analysis of Noncovalent Protein Complexes Directly from Dried Substrates. J. Am. Soc. Mass Spectrom. 2015, 26, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.L.; Cooper, H.J. Direct Tissue Profiling of Protein Complexes: Toward Native Mass Spectrometry Imaging. Anal. Chem. 2016, 88, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.L.; Konijnenberg, A.; Viner, R.; Cooper, H.J. Direct Mass Spectrometry Analysis of Protein Complexes and Intact Proteins up to >70 kDa from Tissue. Anal. Chem. 2019, 91, 6962–6966. [Google Scholar] [CrossRef]
- Hale, O.J.; Cooper, H.J. Native Mass Spectrometry Imaging and In Situ Top-Down Identification of Intact Proteins Directly from Tissue. J. Am. Soc. Mass Spectrom. 2020, 31, 2531–2537. [Google Scholar] [CrossRef]
- Douglass, K.A.; Venter, A.R. Protein analysis by desorption electrospray ionization mass spectrometry and related methods. J. Mass Spectrom. 2013, 48, 553–560. [Google Scholar] [CrossRef]
- Ambrose, S.; Housden, N.G.; Gupta, K.; Fan, J.; White, P.; Yen, H.-Y.; Marcoux, J.; Kleanthous, C.; Hopper, J.T.S.; Robinson, C.V. Native Desorption Electrospray Ionization Liberates Soluble and Membrane Protein Complexes from Surfaces. Angew. Chem. Int. Ed. Engl. 2017, 56, 14463–14468. [Google Scholar] [CrossRef]
- Hale, O.J.; Cooper, H.J. Native Mass Spectrometry Imaging of Proteins and Protein Complexes by Nano-DESI. Anal. Chem. 2021, 93, 4619–4627. [Google Scholar] [CrossRef]
- Saibil, H.R. Cryo-EM in molecular and cellular biology. Mol. Cell 2022, 82, 274–284. [Google Scholar] [CrossRef]
- Sterling, H.J.; Batchelor, J.D.; Wemmer, D.E.; Williams, E.R. Effects of Buffer Loading for Electrospray Ionization Mass Spectrometry of a Noncovalent Protein Complex that Requires High Concentrations of Essential Salts. J. Am. Soc. Mass Spectrom. 2010, 21, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Jordan, J.S.; Williams, E.R. Is Native Mass Spectrometry in Ammonium Acetate Really Native? Protein Stability Differences in Biochemically Relevant Salt Solutions. Anal. Chem. 2024, 96, 17586–17593. [Google Scholar] [CrossRef] [PubMed]
- Behnke, J.-S.; Urner, L.H. Emergence of mass spectrometry detergents for membrane proteomics. Anal. Bioanal. Chem. 2023, 415, 3897–3909. [Google Scholar] [CrossRef]
- Tomioka, Y.; Arakawa, T.; Akuta, T.; Nakagawa, M.; Ishibashi, M. Analysis of proteins by agarose native gel electrophoresis in the presence of solvent additives. Int. J. Biol. Macromol. 2022, 198, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Arakawa, T. Application of native polyacrylamide gel electrophoresis for protein analysis: Bovine serum albumin as a model protein. Int. J. Biol. Macromol. 2019, 125, 566–571. [Google Scholar] [CrossRef]
- Breuker, K.; Oh, H.; Horn, D.M.; Cerda, B.A.; McLafferty, F.W. Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J. Am. Chem. Soc. 2002, 124, 6407–6420. [Google Scholar] [CrossRef]
- Schennach, M.; Breuker, K. Proteins with highly similar native folds can show vastly dissimilar folding behavior when desolvated. Angew. Chem. Int. Ed. Engl. 2014, 53, 164–168. [Google Scholar] [CrossRef]
- Wyttenbach, T.; Bowers, M.T. Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility–Mass Spectrometry Environment. J. Phys. Chem. B 2011, 115, 12266–12275. [Google Scholar] [CrossRef]
- Webb, I.K. Revealing the fates of proteins in the gas phase. Int. J. Mass Spectrom. 2024, 504, 117312. [Google Scholar] [CrossRef]
- Lanucara, F.; Holman, S.W.; Gray, C.J.; Eyers, C.E. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014, 6, 281–294. [Google Scholar] [CrossRef]
- Mikhailov, V.A.; Mize, T.H.; Benesch, J.L.P.; Robinson, C.V. Mass-Selective Soft-Landing of Protein Assemblies with Controlled Landing Energies. Anal. Chem. 2014, 86, 8321–8328. [Google Scholar] [CrossRef] [PubMed]
- Longchamp, J.-N.; Rauschenbach, S.; Abb, S.; Escher, C.; Latychevskaia, T.; Kern, K.; Fink, H.-W. Imaging proteins at the single-molecule level. Proc. Natl. Acad. Sci. USA 2017, 114, 1474–1479. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Hoffmann, W.; Warnke, S.; Bowers, M.T.; Pagel, K.; von Helden, G. Retention of Native Protein Structures in the Absence of Solvent: A Coupled Ion Mobility and Spectroscopic Study. Angew. Chem. Int. Ed. 2016, 55, 14173–14176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Browne, S.J.; Vachet, R.W. Exploring Salt Bridge Structures of Gas-Phase Protein Ions using Multiple Stages of Electron Transfer and Collision Induced Dissociation. J. Am. Soc. Mass Spectrom. 2014, 25, 604–613. [Google Scholar] [CrossRef]
- Happonen, L.J.; Varjosalo, M. State-of-the-Art and Future Directions in Structural Proteomics. Mol. Cell. Proteom. 2025, 24, 101065. [Google Scholar] [CrossRef]
- Edwards, G.B.; Muthurajan, U.M.; Bowerman, S.; Luger, K. Analytical Ultracentrifugation (AUC): An Overview of the Application of Fluorescence and Absorbance AUC to the Study of Biological Macromolecules. Curr. Protoc. Mol. Biol. 2020, 133, e131. [Google Scholar] [CrossRef]
- Rabilloud, T.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: A tutorial. J. Proteom. 2011, 74, 1829–1841. [Google Scholar] [CrossRef]
- Some, D.; Amartely, H.; Tsadok, A.; Lebendiker, M. Characterization of Proteins by Size-Exclusion Chromatography Coupled to Multi-Angle Light Scattering (SEC-MALS). J. Vis. Exp. JoVE 2019, 148, e59615. [Google Scholar] [CrossRef]
- Housmans, J.A.J.; Wu, G.; Schymkowitz, J.; Rousseau, F. A guide to studying protein aggregation. FEBS J. 2023, 290, 554–583. [Google Scholar] [CrossRef]
- Woldeyes, M.A.; Calero-Rubio, C.; Furst, E.M.; Roberts, C.J. Light Scattering to Quantify Protein-Protein Interactions at High Protein Concentrations. Methods Mol. Biol. 2019, 2039, 23–37. [Google Scholar] [CrossRef]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef]
- Goure, W.F.; Krafft, G.A.; Jerecic, J.; Hefti, F. Targeting the proper amyloid-beta neuronal toxins: A path forward for Alzheimer’s disease immunotherapeutics. Alzheimer’s Res. Ther. 2014, 6, 42. [Google Scholar] [CrossRef]
- Cline, E.N.; Bicca, M.A.; Viola, K.L.; Klein, W.L. The Amyloid-β Oligomer Hypothesis: Beginning of the Third Decade. J. Alzheimer’s Dis. 2018, 64, S567–S610. [Google Scholar] [CrossRef]
- O’Neale, C.V.T.; Harvey, S.R.; Chetyrkin, S.; Wysocki, V.H.; Schey, K.L. Endogenous Aquaporin-0 Lipid Binding in Ocular Lens Tissue via Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2025, 36, 1588–1597. [Google Scholar] [CrossRef]
- Jordan, J.S.; Harper, C.C.; Zhang, F.; Kofman, E.; Sam, M.; Zaragoza, J.P.; Bhagwat, B.; Fayadat-Dilman, L.; Williams, E.R. Characterizing Monoclonal Antibody Aggregation Using Charge Detection Mass Spectrometry and Industry Standard Methods. J. Am. Soc. Mass Spectrom. 2025, 36, 1241–1253. [Google Scholar] [CrossRef]
- Devi, S.; Chaturvedi, M.; Fatima, S.; Priya, S. Environmental factors modulating protein conformations and their role in protein aggregation diseases. Toxicology 2022, 465, 153049. [Google Scholar] [CrossRef]
- Singh, A. Understanding protein subcellular compartmentalization signals. Nat. Methods 2025, 22, 651. [Google Scholar] [CrossRef] [PubMed]
- Benfey, P.N.; Mitchell-Olds, T. From Genotype to Phenotype: Systems Biology Meets Natural Variation. Science 2008, 320, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Black, K.A.; Stollar, E.J. Uncovering protein function: From classification to complexes. Essays Biochem. 2022, 66, 255–285. [Google Scholar] [CrossRef]
- Li, L.; Zinovyeva, A.Y. Protein Extract Preparation and Co-immunoprecipitation from Caenorhabditis elegans. J. Vis. Exp. JoVE 2020, e61243. [Google Scholar] [CrossRef]
- Tokmakov, A.A.; Kurotani, A.; Sato, K.I. Protein pI and Intracellular Localization. Front. Mol. Biosci. 2021, 8, 775736. [Google Scholar] [CrossRef] [PubMed]
- Locke, D.K.; Irina, V.; Harris, A.L. Isoelectric points and post-translational modifications of connexin26 and connexin32. FASEB J. 2006, 20, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Mavridou, D.; Damian, M.; Mutti, F.G.; Schoenmakers, P.J.; Gargano, A.F.G. Characterization of Complex Proteoform Mixtures by Online Nanoflow Ion-Exchange Chromatography-Native Mass Spectrometry. Anal. Chem. 2024, 96, 8880–8885. [Google Scholar] [CrossRef] [PubMed]
- Kerr, C.M.; Schneider, O.L.; Tichy, S.; Huge, B.J.; Champion, M.M. Capillary Isoelectric Focusing of Proteins and Peptides Using an In-Line cIEF-ESI Interface with Improved MS Characteristics. Anal. Chem. 2025, 97, 649–657. [Google Scholar] [CrossRef]
- Landreh, M.; Sahin, C.; Gault, J.; Sadeghi, S.; Drum, C.L.; Uzdavinys, P.; Drew, D.; Allison, T.M.; Degiacomi, M.T.; Marklund, E.G. Predicting the Shapes of Protein Complexes through Collision Cross Section Measurements and Database Searches. Anal. Chem. 2020, 92, 12297–12303. [Google Scholar] [CrossRef]
- Ruotolo, B.T. Collision Cross Sections for Native Proteomics: Challenges and Opportunities. J. Proteome Res. 2022, 21, 2–8. [Google Scholar] [CrossRef]
- Batra, J.; Hultquist, J.F.; Liu, D.; Shtanko, O.; Von Dollen, J.; Satkamp, L.; Jang, G.M.; Luthra, P.; Schwarz, T.M.; Small, G.I.; et al. Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 2018, 175, 1917–1930.e1913. [Google Scholar] [CrossRef]
- Henry, N.; Krammer, E.-M.; Stengel, F.; Adams, Q.; Van Liefferinge, F.; Hubin, E.; Chaves, R.; Efremov, R.; Aebersold, R.; Vandenbussche, G.; et al. Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach. PLoS Comput. Biol. 2018, 14, e1006165. [Google Scholar] [CrossRef]
- Zhou, M.; Robinson, C.V. When proteomics meets structural biology. Trends Biochem. Sci. 2010, 35, 522–529. [Google Scholar] [CrossRef]
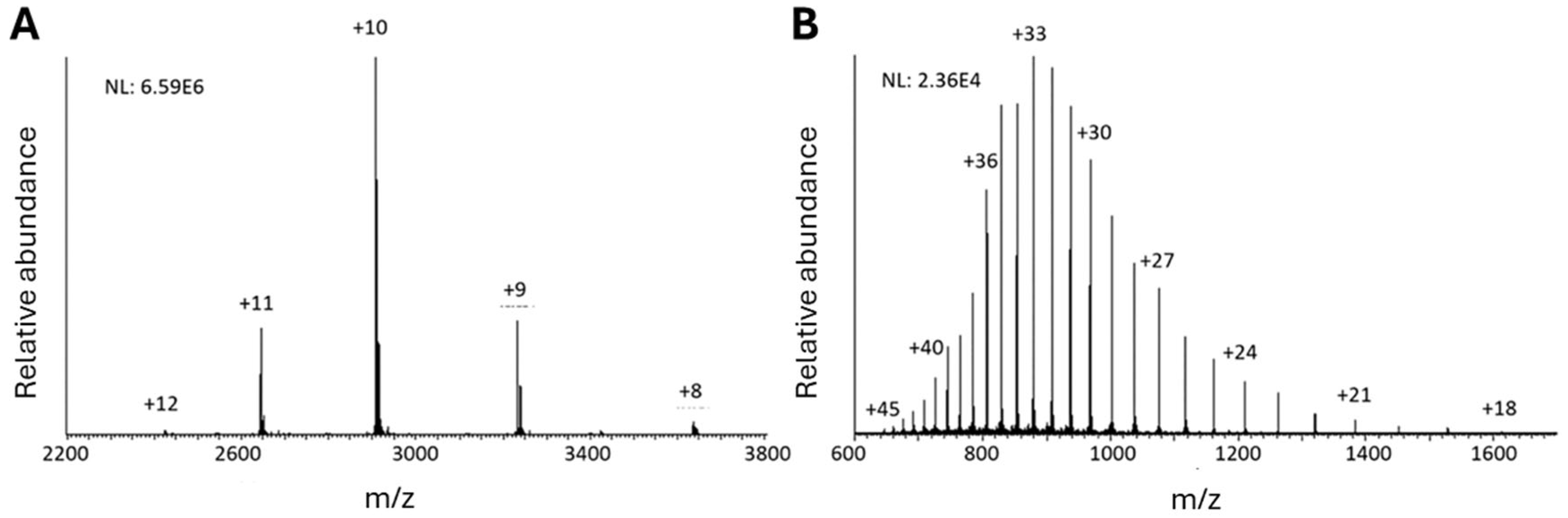
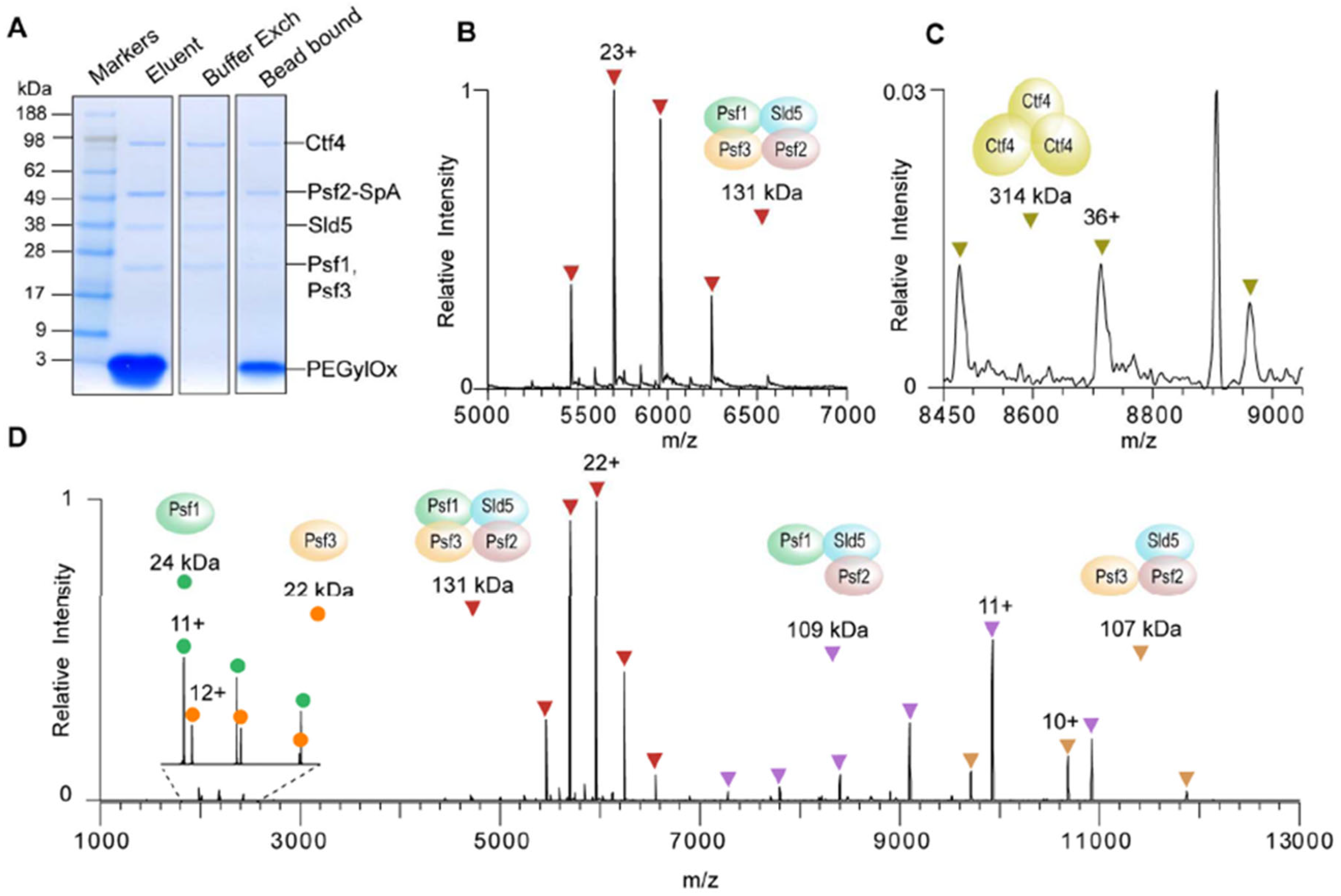
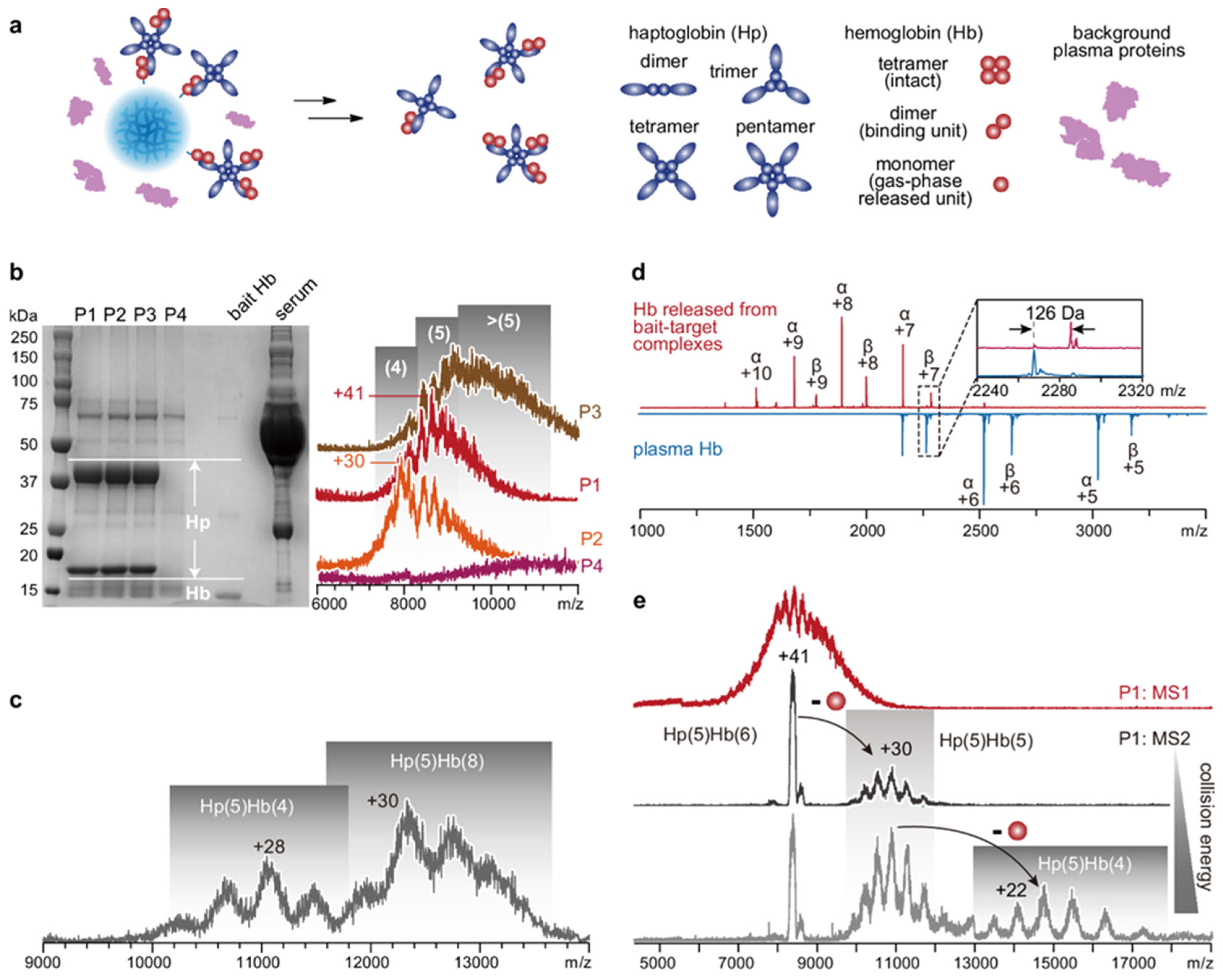
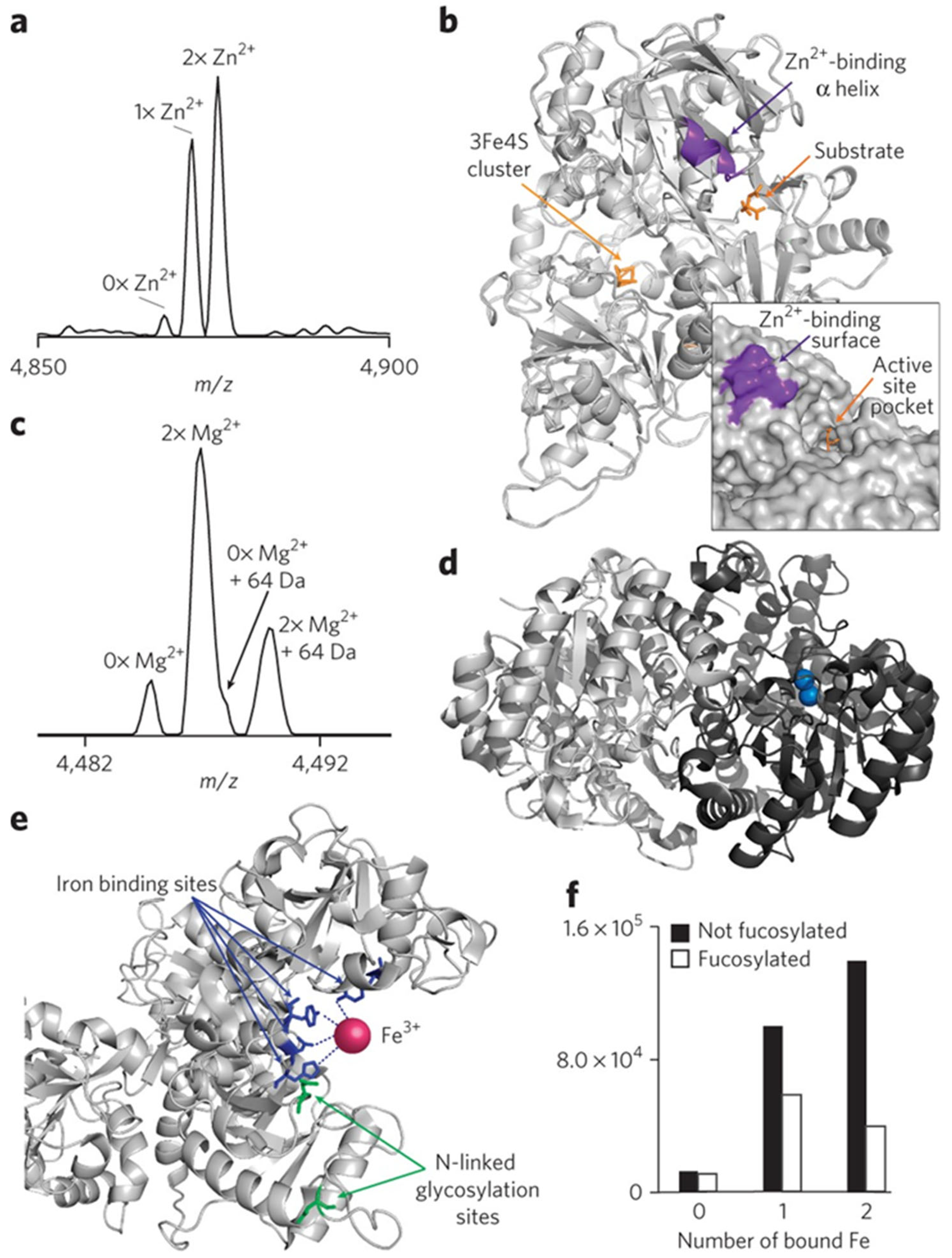
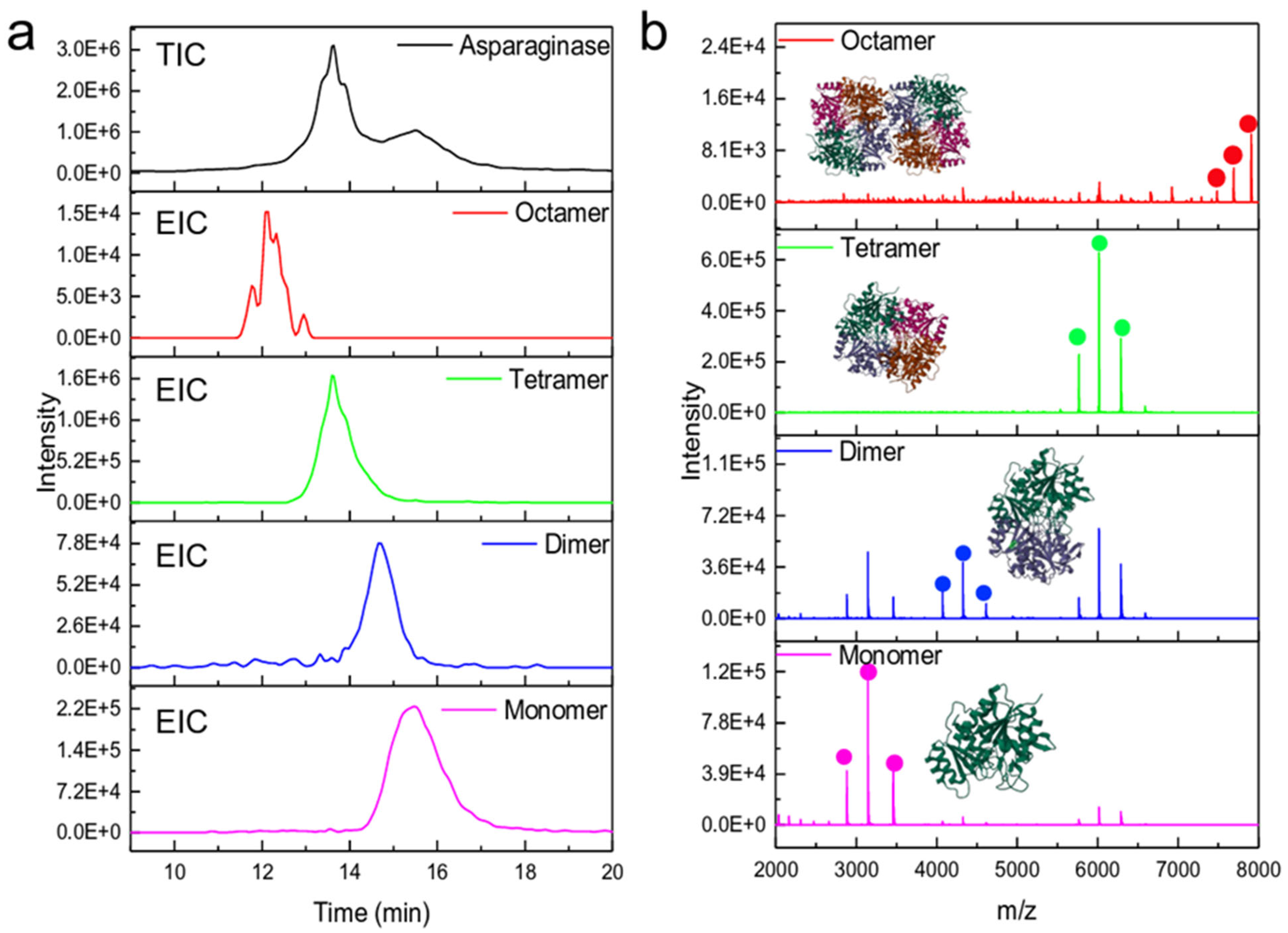
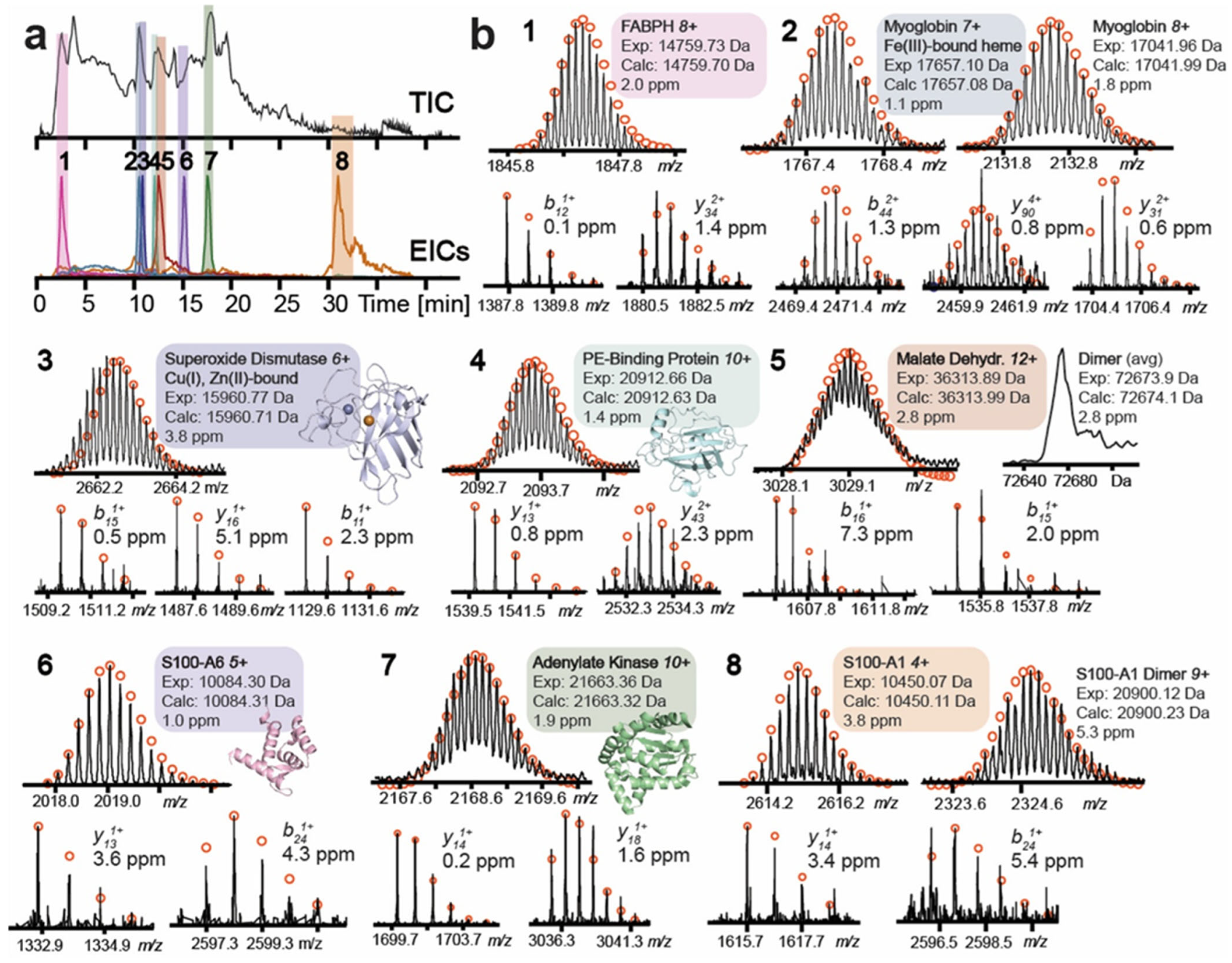
 : fucose;
: fucose;  : N-acetyl-glucosamine (GlcNAc);
: N-acetyl-glucosamine (GlcNAc);  : mannose. (c). FcRn-MS analysis of a mixture of a wild-type mAb and its YTE-format. The insets show the deconvoluted mass spectra for the two TIC peaks. © Elsevier, reprinted with permission [213].
: mannose. (c). FcRn-MS analysis of a mixture of a wild-type mAb and its YTE-format. The insets show the deconvoluted mass spectra for the two TIC peaks. © Elsevier, reprinted with permission [213].
 : fucose;
: fucose;  : N-acetyl-glucosamine (GlcNAc);
: N-acetyl-glucosamine (GlcNAc);  : mannose. (c). FcRn-MS analysis of a mixture of a wild-type mAb and its YTE-format. The insets show the deconvoluted mass spectra for the two TIC peaks. © Elsevier, reprinted with permission [213].
: mannose. (c). FcRn-MS analysis of a mixture of a wild-type mAb and its YTE-format. The insets show the deconvoluted mass spectra for the two TIC peaks. © Elsevier, reprinted with permission [213].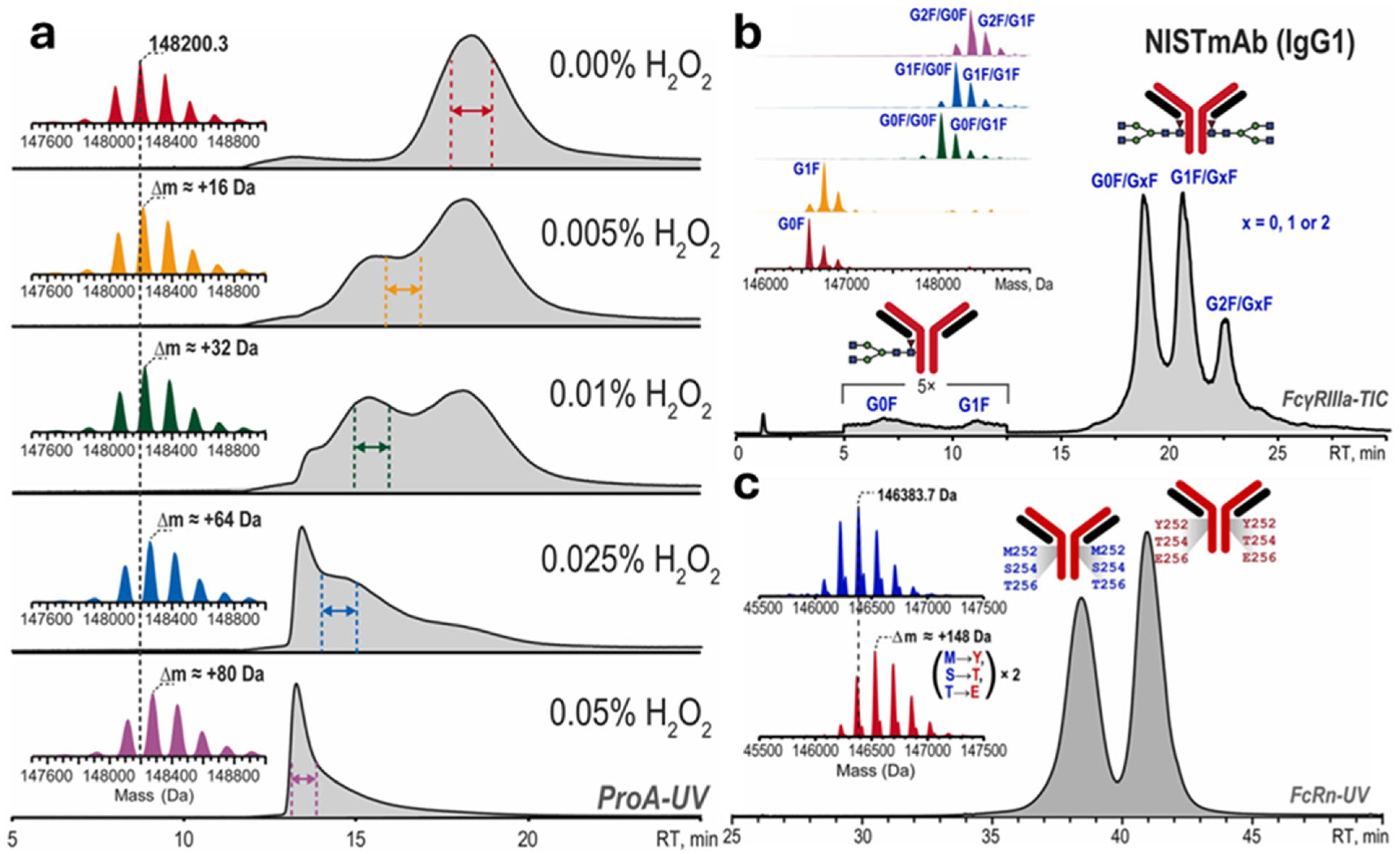
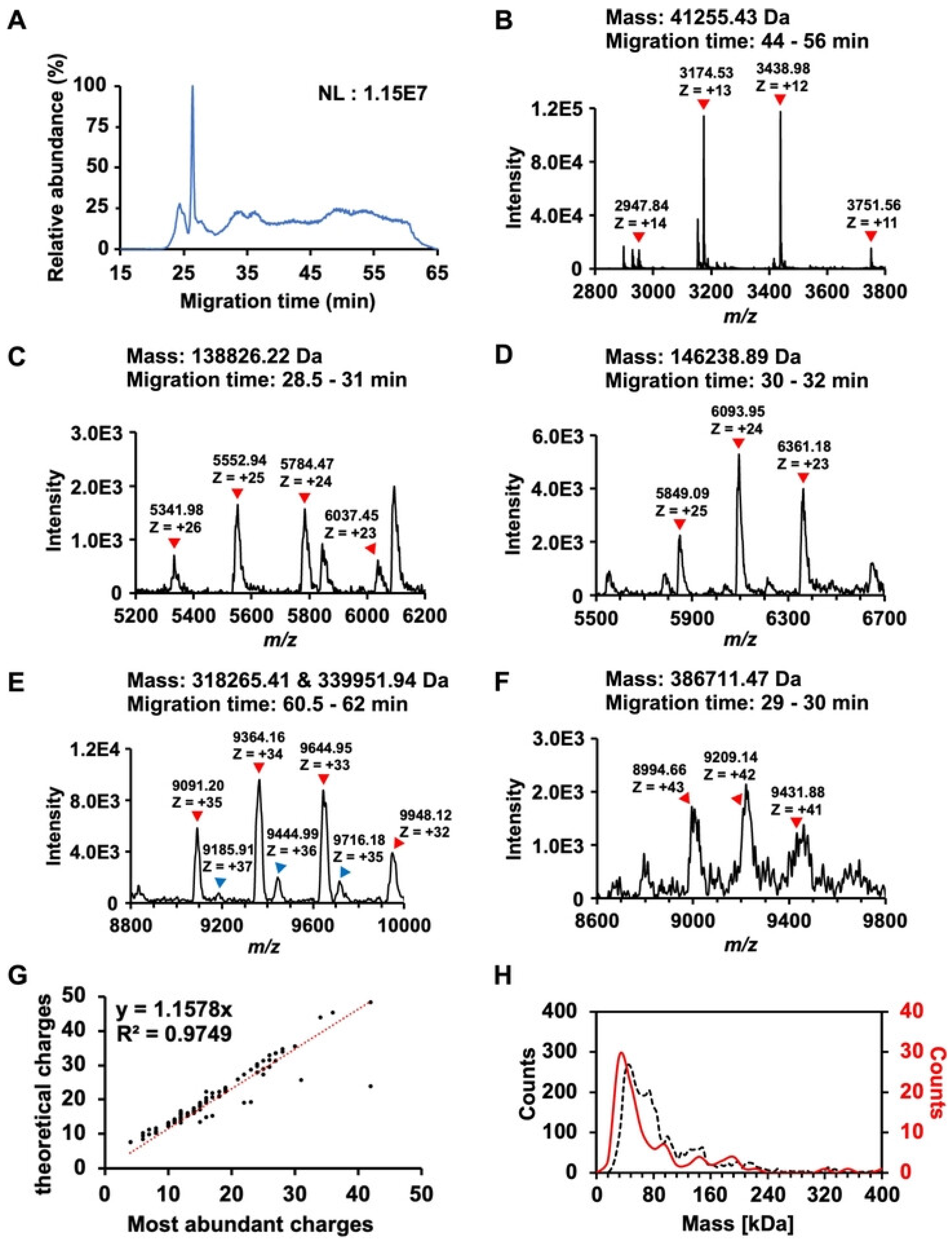
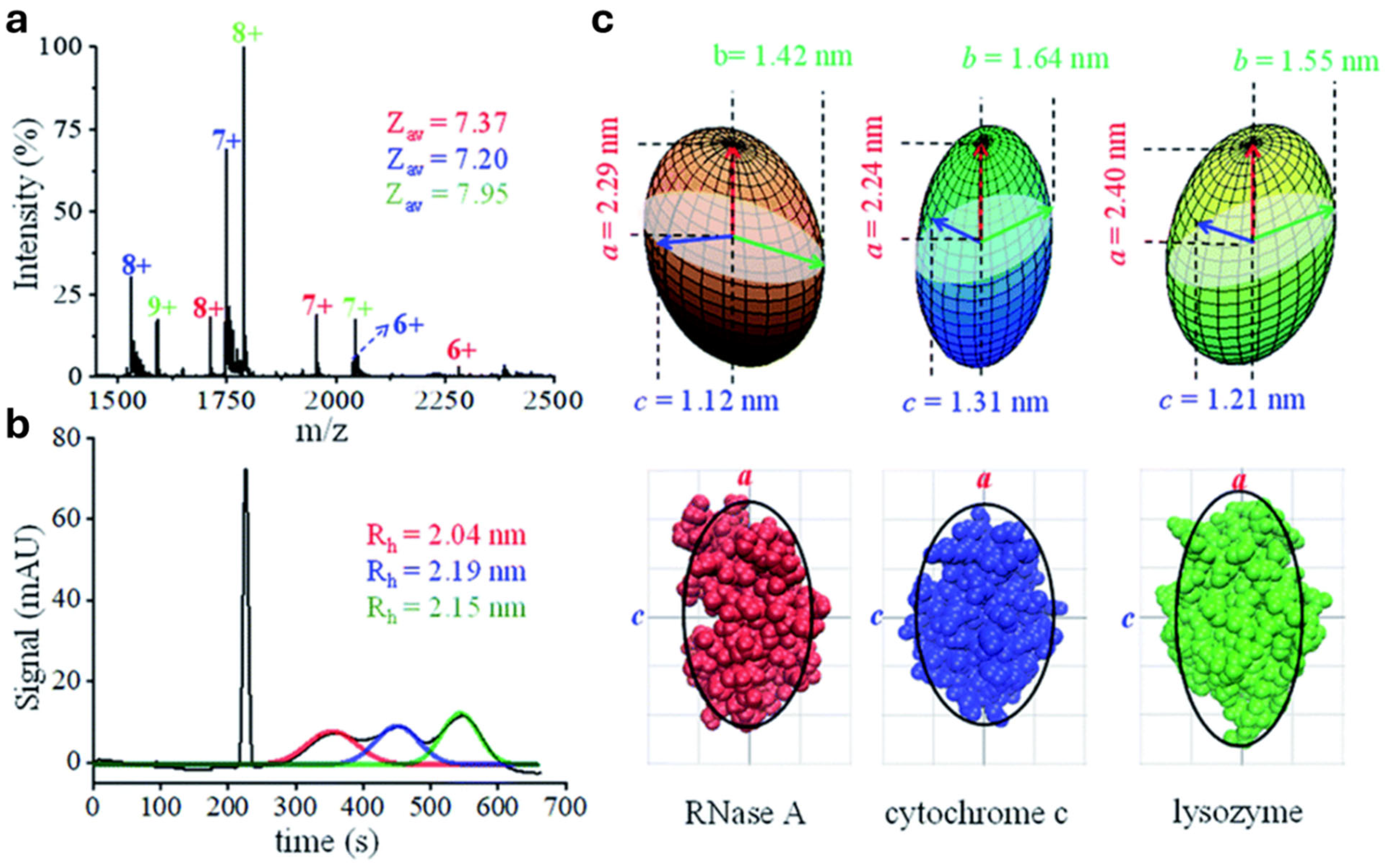
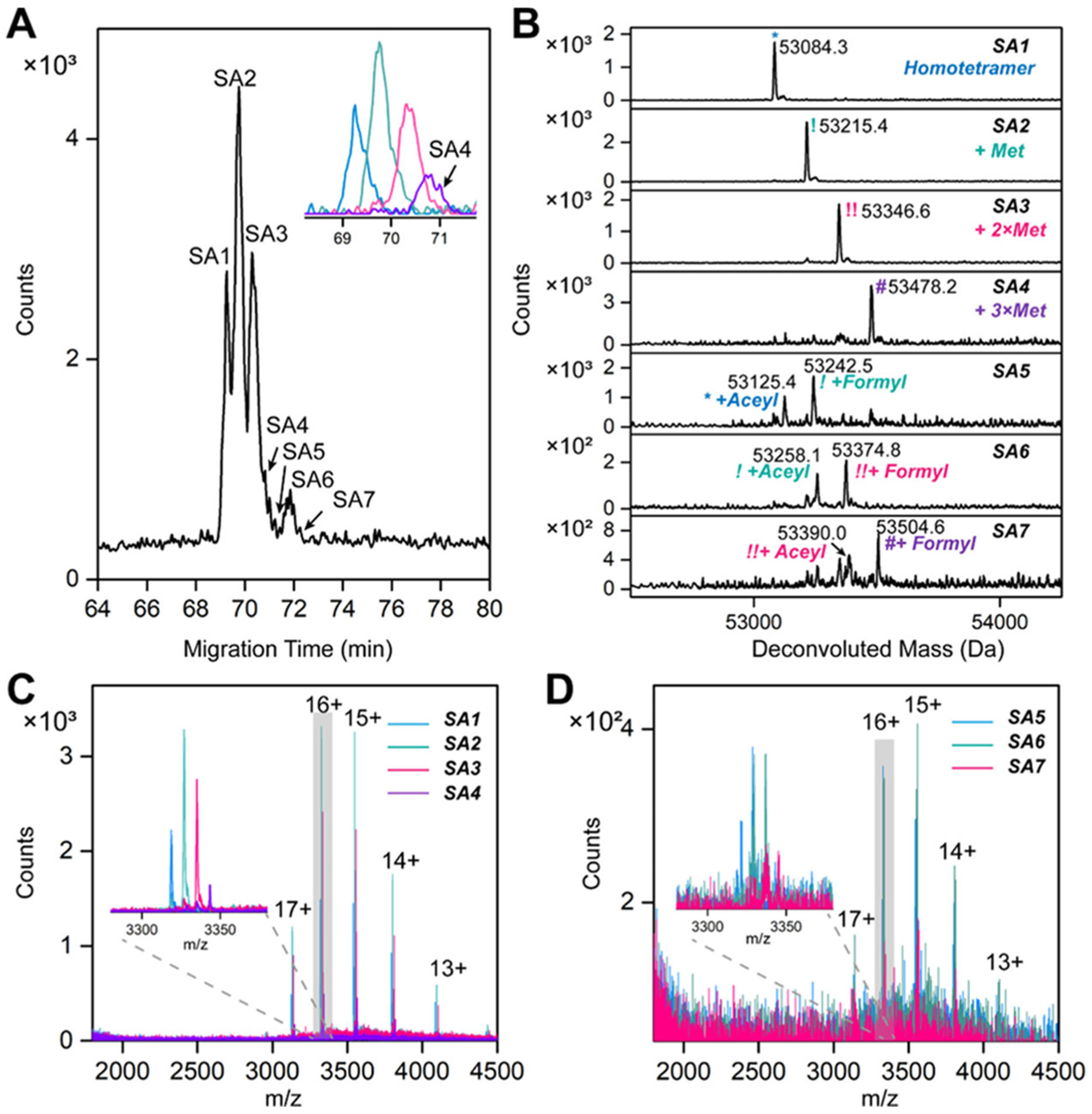
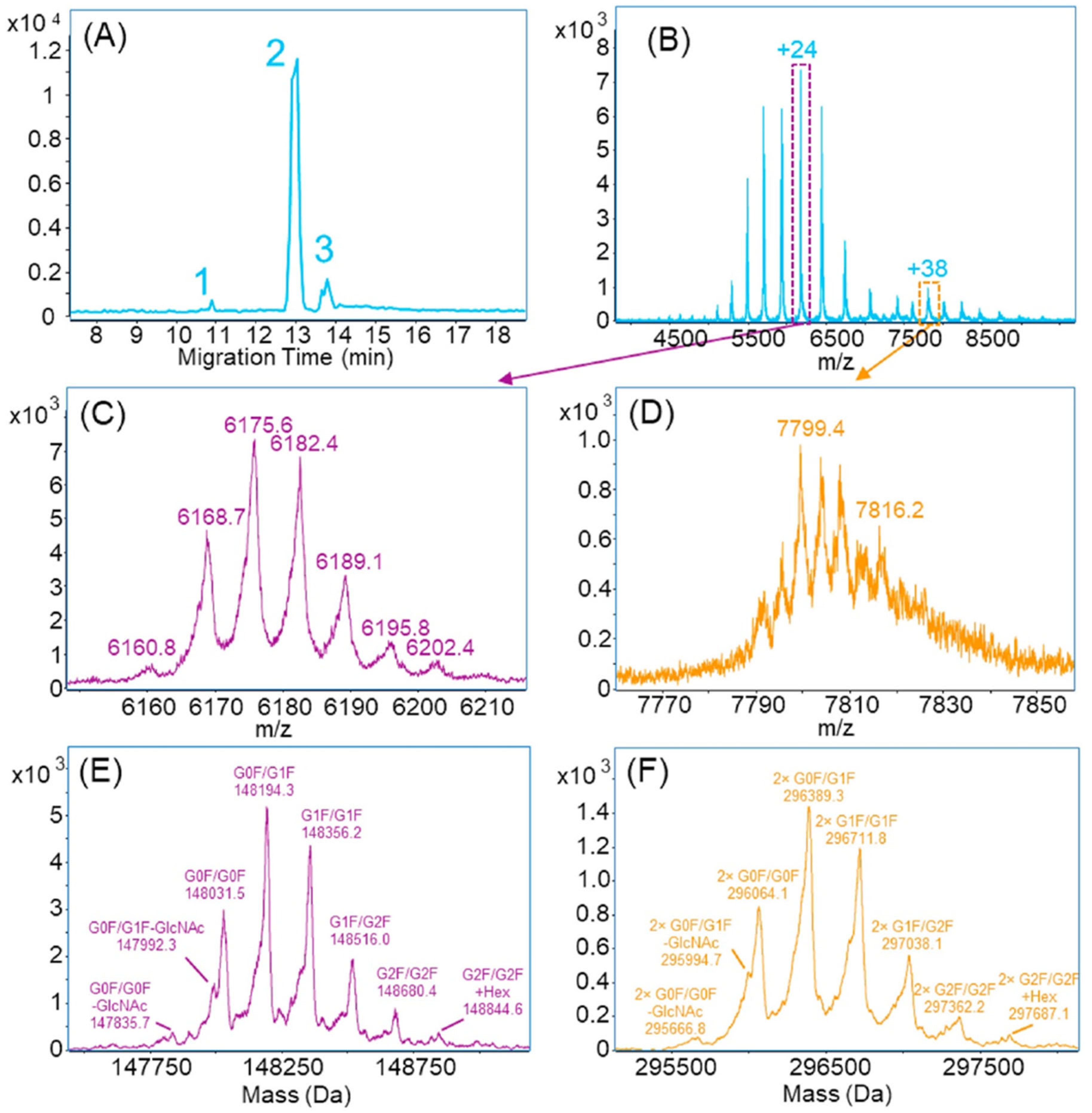
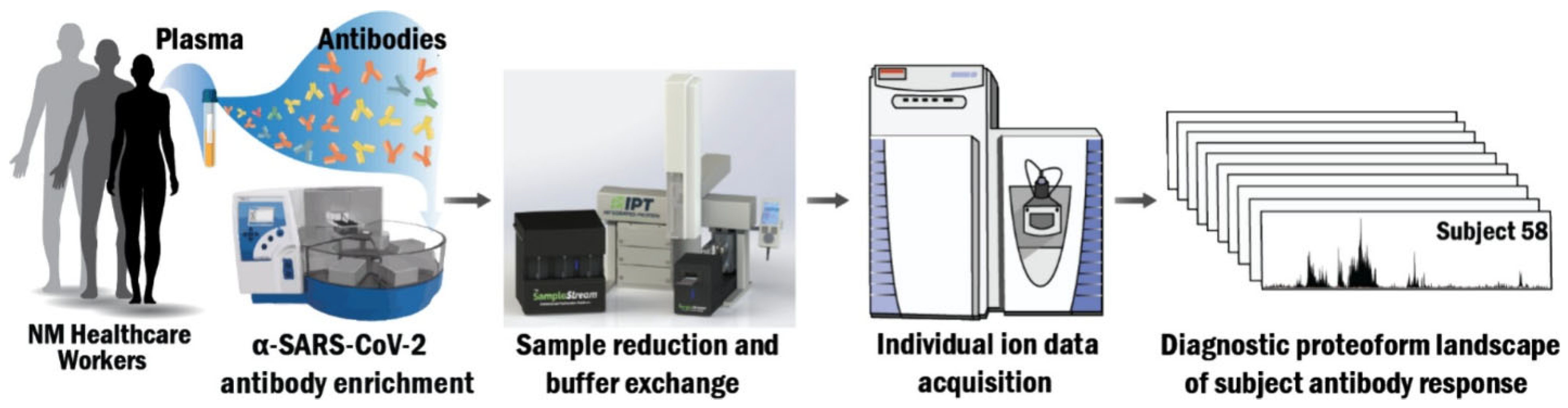
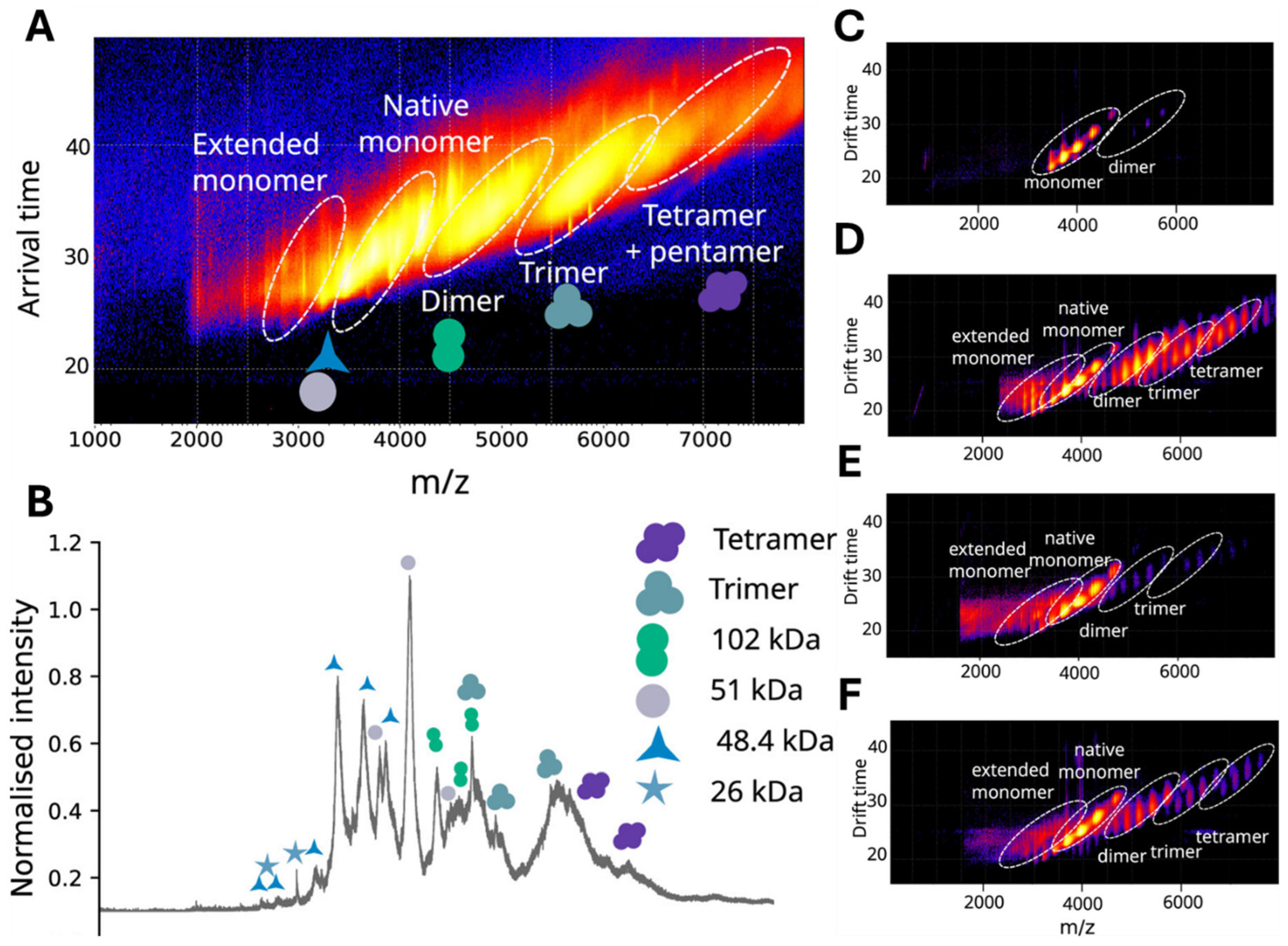
| Instrument | Parameter Settings | Analyzed Protein Species, Their Concentration, and Sample Introduction Approach | Reference |
|---|---|---|---|
| Bruker 15-Tesla SolarisX FT-ICR MS | Capillary voltage: 0.7–0.95 kV | Membrane protein complex, E. coli AquaporinZ homotetramer (97 kDa) (15–30 µM), acquired via direct nanospray-ESI | [96] |
| Dry gas temperature: 100 °C | |||
| Dry gas flow rate: 3.0 L/min | |||
| Ion Funnel RF amplitude: 300 Vapp | |||
| Ion Funnel 1 voltage: 150 V | |||
| Ion Funnel 2 voltage: 6 V | |||
| Skimmer 1: 50–125 V | |||
| Skimmer 2: 5 V | |||
| Multipole 1 RF Frequency: 2 MHz | |||
| Quadrupole RF frequency: 1.4 MHz | |||
| Transfer Hexapole RF Frequency: 1 MHz | |||
| Ion accumulation time: 500 ms | |||
| Waters Synapt G2-HDMS Q-TOF MS | Capillary voltage: 0.5–1 kV 1, 0.8–1.2 kV 2 | 1 Membrane protein complex, E. coli AquaporinZ homotetramer (97 kDa) (15–30 µM), acquired via direct nanospray-ESI 2 Noncovalent protein–ligand complex: Lysozyme and tri-N-acetylchitotriose (NAG3), Trypsin/Pefabloc, Carbonic Anhydrase II/Chlorothiazide, β-Lactoglobulin A/Lauric Acid (5 µM concentration for each protein with 5–25 µM of ligand), acquired via direct nano-ESI | 1 [96] 2 [65] |
| Sample cone: 40 V | |||
| Source temperature: 30 °C | |||
| Trap CE: 4–110 V 1 (High trap CE used for unfolding), 4–102 | |||
| Transfer CE: 3 V 1, 2 V 2 | |||
| Trap pressure: 3 × 10−3 mbar 1, 7 × 10−3 mbar 2 | |||
| Transfer pressure: 3 × 10−3 mbar 1, 6.7 × 10−3 mbar 2 | |||
| Trap direct current (DC) bias: −2 V 1, 3 V 2 | |||
| Thermo Fisher Scientific Q Exactive HF Plus Orbitrap EMR MS | Source voltage: 1.5 kV | Proteolysis-targeting chimeras (PROTACs)-ternary complex (5 µM), acquired via direct nanospray-ESI | [66] |
| Capillary temperature: 100 °C and 50 °C | |||
| FT resolution: 140,000 (at 200 m/z) | |||
| In-source CID voltage: 10 V | |||
| HCD CE: 10 V | |||
| Automatic gain control (AGC) target: 5 × 106 | |||
| SCIEX ZenoTOF 7600 | Spray voltage: 3500 V | NIST mAb (0.7–7 µM, converted from published concentration), ADC (Enhertu) (0.6–6 µM, converted from published concentration), acquired via microflow SEC-ESI-MS | [97] |
| Curtain gas: 40 psi | |||
| CAD gas: 9 | |||
| Ion source gas 1: 60 psi | |||
| Ion source gas 2: 60 psi | |||
| Source temperature: 250–300 °C | |||
| Declustering potential (DP): 120 V | |||
| CE: 12 V | |||
| Accumulation time: 0.25 s | |||
| Time bins to sum: 120 | |||
| Agilent 6545XT AdvanceBio LC/Q-TOF | Dry gas temperature: 365 °C 3, 150 °C 4 | 3 Intact protein complex: Pyruvate kinase tetramer (232 kDa), glutamate dehydrogenase hexamer (335 kDa), and β-galactosidase tetramer (466 kDa) (2–20 µM), acquired via microflow SEC-ESI-MS 4 Intact protein: Myoglobin (Concentration/amount injected not reported), acquired via microflow SEC-ESI-MS | [94] |
| Dry gas flow: 12 L/min 3, 10 L/min 4 | |||
| Nebulizer: 35 psig 3, 30 psig 4 | |||
| Sheath gas temperature: 300 °C 3, 150 °C 4 | |||
| Sheath gas flow: 12 L/min 3, 10 L/min 4 | |||
| Capillary voltage: 5500 V 3, 5000 V 4 | |||
| Nozzle voltage: 2000 V | |||
| Fragmentor: 300 V 3, 250 V 4 | |||
| Skimmer: 220 V 3, 100 V 4 | |||
| Acquisition rate: 1 spectrum/s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayon, N.J. Preanalytical Strategies for Native Mass Spectrometry Analysis of Protein Modifications, Complexes, and Higher-Order Structures. AppliedChem 2025, 5, 35. https://doi.org/10.3390/appliedchem5040035
Ayon NJ. Preanalytical Strategies for Native Mass Spectrometry Analysis of Protein Modifications, Complexes, and Higher-Order Structures. AppliedChem. 2025; 5(4):35. https://doi.org/10.3390/appliedchem5040035
Chicago/Turabian StyleAyon, Navid J. 2025. "Preanalytical Strategies for Native Mass Spectrometry Analysis of Protein Modifications, Complexes, and Higher-Order Structures" AppliedChem 5, no. 4: 35. https://doi.org/10.3390/appliedchem5040035
APA StyleAyon, N. J. (2025). Preanalytical Strategies for Native Mass Spectrometry Analysis of Protein Modifications, Complexes, and Higher-Order Structures. AppliedChem, 5(4), 35. https://doi.org/10.3390/appliedchem5040035






