1. Introduction
Myoglobin is a small, monomeric heme protein that facilitates oxygen storage and transport within muscle tissues [
1,
2]. Beyond this classical role, myoglobin exhibits pseudo-peroxidase activity by catalyzing the reduction of hydrogen peroxide through redox cycling at its heme iron center [
3,
4]. This peroxidase-like activity is associated with antioxidant defense in muscle tissues, especially under oxidative stress [
4], and is enhanced under acidic conditions [
5]. While some reports suggest that hydrogen peroxide exposure increases this activity [
6], the mechanisms and limits of its function remain complex.
The catalytic behavior of myoglobin resembles that of classical peroxidases, including the transient formation of high-valent iron-oxo intermediates [
7], but unlike true peroxidases, myoglobin lacks a built-in electron donor system. Attempts to restore its activity with vitamin C have shown mixed results; while it can donate electrons [
8], its ability to reverse activity loss is questionable; while some oxidative modifications can be reversed with reductants such as vitamin C, others lead to irreversible damage [
6]. Myoglobin has been implicated in lipid oxidation and redox regulation in hypoxic tissues [
9], and its heme group is structurally identical to that of peroxidases [
10]. However, oxidative damage such as dimerization and aggregation may hinder its regeneration.
Substrate inhibition is a kinetic phenomenon in which the reaction velocity increases with substrate concentration up to an optimum but then declines at higher concentrations due to the formation of non-productive enzyme–substrate complexes or other inhibitory interactions. This behavior, observed in a variety of enzymes, produces a characteristic bell-shaped activity profile and can result from mechanisms such as secondary site binding, conformational changes, or interference with product release [
11,
12,
13,
14]. In the case of myoglobin, repeated redox cycling in the presence of hydrogen peroxide may similarly lead to decreased activity, either through classical substrate inhibition or through oxidative structural degradation, such as crosslinking or modification of the heme environment [
6,
15,
16]. Some oxidative modifications, such as ditryptophan dimer formation or porphyrin ring damage via hydroxyl radicals, can alter the protein irreversibly [
5]. While 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) has been used as an external electron donor in some studies, resulting in pseudo-first-order kinetics and radical generation [
10], myoglobin’s damage from peroxide exposure is not consistently reversible.
Mechanistically, substrate inhibition can occur when excess substrate binds to secondary or non-productive sites on the enzyme or enzyme–substrate complex, altering its conformation or blocking product release. Depending on the binding mode, this effect can resemble competitive, uncompetitive, noncompetitive, or allosteric inhibition, each producing distinct kinetic profiles.
In multi-protein systems, such as co-reactions with lactoperoxidase, crosslinking between heme proteins has been observed [
17]. These aggregates may form homodimers or heterodimers and often resist dissociation unless the native protein structure is disrupted by heat. These structural alterations emphasize the vulnerability of myoglobin under oxidative conditions and its potential shift from protective antioxidant to pro-oxidant. While activity enhancement has been observed in acidic pH [
10], it remains unclear whether elevated hydrogen peroxide concentrations in vivo result in classical substrate inhibition or simply irreversible catalyst damage. Thus, distinguishing true substrate inhibition from oxidative inactivation is a central challenge in characterizing myoglobin’s peroxidase function.
Preliminary data indicated that myoglobin activity may decline at high hydrogen peroxide concentrations, a hallmark of substrate inhibition. This observation served as a major motivation for the present study. One of our goals is to gain insight into the mechanism of this inhibition, specifically, whether it reflects a general protein-damaging effect of hydrogen peroxide or a phenomenon more specific to the structure and reactivity of myoglobin and hemoglobin.
Despite these complexities, systematic kinetic studies of myoglobin under varying hydrogen peroxide concentrations are limited. Many prior investigations have not fully explored how ionic strength, substrate accumulation, and time-dependent degradation converge to influence activity. Furthermore, comparisons between myoglobin and hemoglobin under matched oxidative conditions remain underexplored, despite evidence suggesting similar pseudoperoxidase behavior. In this study, we use ABTS as an electron donor to examine the time-dependent peroxidase-like activity of myoglobin and hemoglobin across a range of hydrogen peroxide concentrations. Our analysis includes varying ionic strengths and employs kinetic modeling to distinguish between classical substrate inhibition and irreversible inactivation. These findings help clarify the mechanistic underpinnings of globin activity loss under oxidative stress and provide new insight into the physiological relevance of substrate inhibition in heme proteins.
2. Materials and Methods
Equine skeletal muscle myoglobin, bovine blood hemoglobin, and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Hydrogen peroxide (30% w/v), magnesium sulfate (MgSO4), and all buffer components, 2-(N-morpholino)ethanesulfonic acid (MES) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), were obtained from Fisher Scientific (Waltham, MA, USA) and used without further purification.
Lyophilized myoglobin and hemoglobin were reconstituted in deionized water. Protein concentrations were determined spectrophotometrically using Soret band absorbances at 409 nm (ε409 = 154 mM−1·cm−1 for myoglobin and ε409 = 188 mM−1·cm−1 for hemoglobin). Proteins were used in their ferric form, confirmed by characteristic absorption spectra, and maintained in buffered solutions at the desired pH prior to use.
All reactions were performed at 25 °C in UV-transparent 1 cm cuvettes using a Vernier visible spectrophotometer from Vernier Science Education, Beaverton, OR, USA. Catalytic activity was monitored by measuring ABTS oxidation at 730 nm (ε730 = 15,000 M
−1·cm
−1), reflecting ABTS radical cation formation, as previously described for monitoring pseudo-peroxidase activity [
10]. Standard reaction mixtures contained 1 mM ABTS, 13 mM MgSO
4, and either 20 mM MES (pH 5) or 20 mM HEPES (pH 7). Protein concentrations (1–5 µM) were kept constant within each kinetic dataset but varied slightly between experimental sets to optimize signal-to-noise ratios and maintain initial rate measurements within the linear detection range. All reported rates were normalized to enzyme concentration to allow for direct comparison across conditions.
To maintain steady-state conditions, hydrogen peroxide and ABTS were used at concentrations substantially higher than that of the catalyst. For example, 100 µL of 1 mg/mL hemoglobin (15.5 µM) was diluted to 3 mL, yielding ∼500 nM final concentration. This ensured a >200-fold molar excess of substrate over enzyme, avoiding appreciable substrate depletion during initial velocity measurements and minimizing transient effects.
To monitor reaction rates, the oxidation of ABTS by hydrogen peroxide was tracked spectrophotometrically at 730 nm, where the ABTS radical cation exhibits a strong absorbance (ε730 = 15,000 M
−1·cm
−1) [
10]. This colorimetric change reflects electron transfer from ABTS to hydrogen peroxide and was used as a proxy for pseudo-peroxidase activity. All reactions were performed using MES buffer at pH 5 or HEPES/Tris at pH 7, and ionic strength was standardized with 13 mM MgSO
4. pH 5 was selected to ensure optimal conditions for peroxide reactivity, while pH 7 assays were included to evaluate behavior under near-physiological conditions.
Proteins were maintained in the ferric oxidation state, confirmed by absorbance at the Soret band (409 nm) and visible spectra between 450 and 650 nm (
Figures S1 and S2). Upon exposure to hydrogen peroxide, spectral shifts were observed, consistent with the formation of ferryl species or oxy-like intermediates, as previously reported for myoglobin [
18,
19]. These spectral changes indicate that hydrogen peroxide alters the redox state of the heme center, likely forming reactive intermediates responsible for catalysis. All rate measurements were based on initial absorbance changes and performed under pseudo–first-order conditions, with substrate concentrations in large excess relative to enzyme.
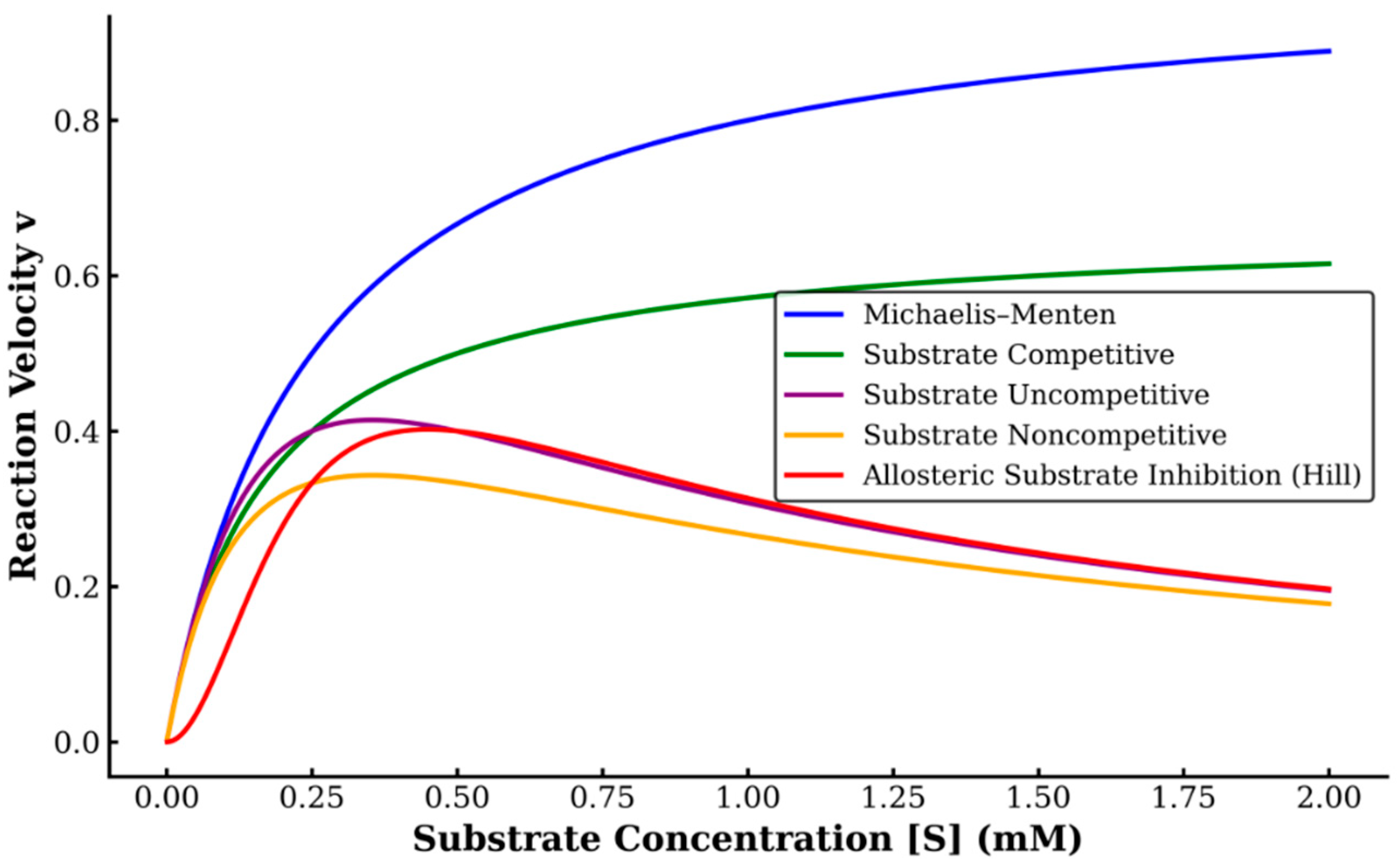
Initial velocities were recorded across various hydrogen peroxide concentrations and fit to the following kinetic models including the Michaelis–Menten equation
and various substrate inhibition models. Specifically, fits were attempted to the classic substrate competitive inhibition model
the uncompetitive
noncompetitive
and the allosteric substrate inhibition model (Hill variation).
where K
i is the inhibition constant, and n is the Hill coefficient.
Curve fitting was performed using KaleidaGraph (v3.6) and Python (v3.11.8). Values for these variables were found by nonlinear regression of the experimental data to the respective model equations, with V
max, K
m, K
i, and n, (where applicable) treated as free parameters. All curve fitting was performed using KaleidaGraph (Synergy Software) and custom Python scripts utilizing nonlinear regression tools [
20]. Plots were generated using the Matplotlib visualization library (v3.6.3) in Python [
21]. A sample of the type of code used is provided in the
Supplementary Materials (Code S1).
Figure 1 illustrates the theoretical behavior of each kinetic model used for curve fitting, highlighting their distinct responses to increasing substrate concentrations. The classic Michaelis–Menten model shows saturation without inhibition, while competitive, uncompetitive, noncompetitive, and Hill-type allosteric inhibition models demonstrate decreasing activity at higher substrate concentrations to varying degrees. For time-course measurements, ABTS was added after a fixed 90 s delay following pre-incubation of protein and hydrogen peroxide. Additional experiments tested hydrogen peroxide pre-incubation up to 50 min. Declines in initial rates were interpreted as indicators of time-dependent inactivation.
To evaluate ionic strength effects, assays were repeated with and without 13 mM MgSO
4. Supplementation with 0.1% bovine serum albumin (BSA) was included to assess whether alternative electron donors influenced inhibition profiles or protected proteins from oxidative inactivation as has been suggested [
22].
3. Results
3.1. Substrate Inhibition of Myoglobin and Hemoglobin
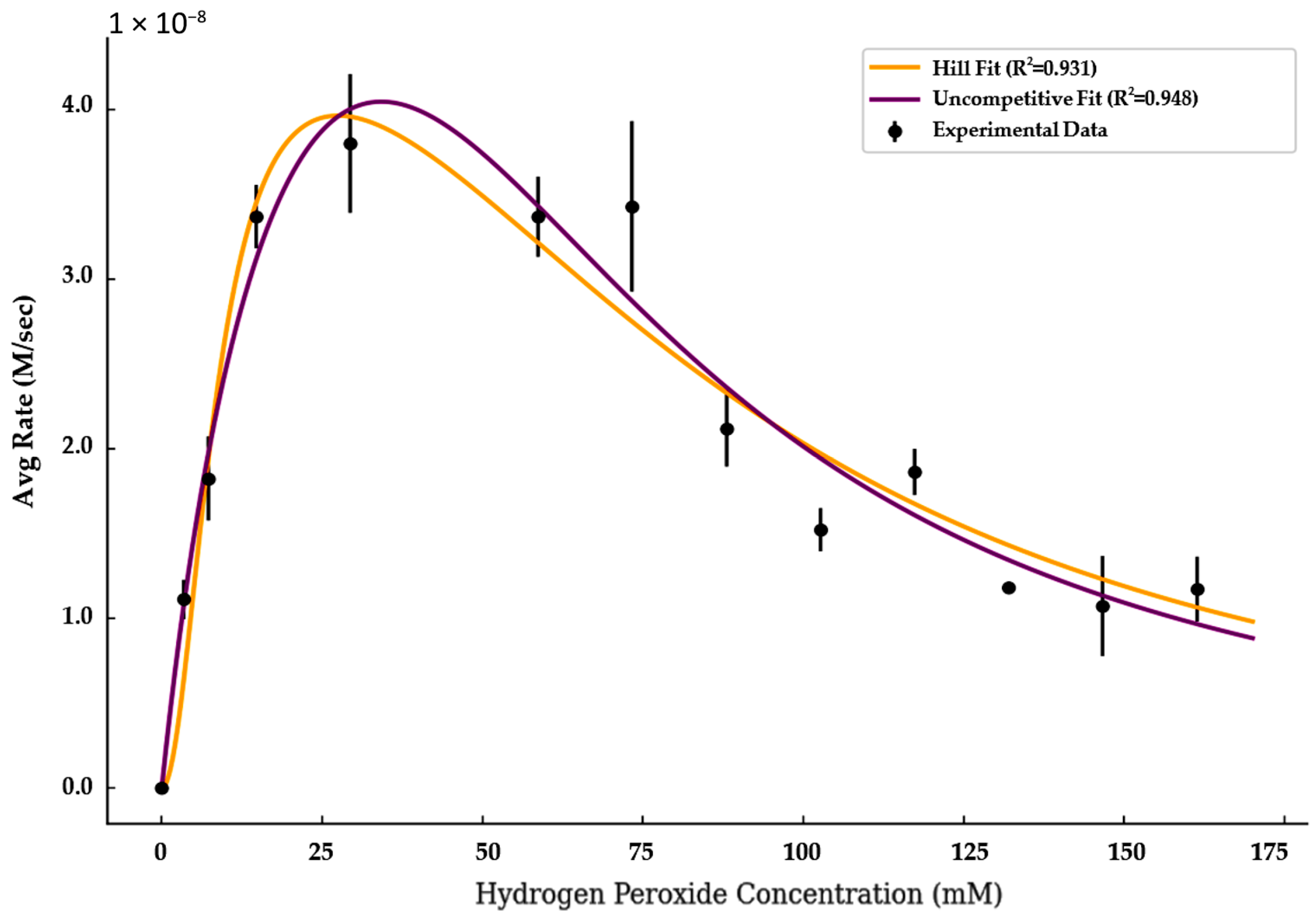
Kinetic analysis of myoglobin and hemoglobin’s pseudo-peroxidase activity revealed a pronounced decline in activity at elevated hydrogen peroxide concentrations. While some of this decline may result from general oxidative damage, the shape of the response curves and the ability of inhibition models to capture these trends suggest that true substrate inhibition also plays a role (
Figure 1).
To quantify this behavior, myoglobin activity was analyzed using classical Michaelis–Menten kinetics and substrate inhibition models (Equations (1)–(5)). The Michaelis–Menten model failed to account for the observed activity decline at high peroxide concentrations in both enzymes. In contrast, the Hill-type and uncompetitive inhibition models yielded the strongest fits to the experimental data.
The Hill model captured cooperative activation at low peroxide levels and inhibition at higher concentrations, with a Hill coefficient of 2.00, a V
max of 4.77 × 10
−8 M/s, a K
m of 8.74 mM, and a K
i of 86.38 mM (R
2 = 0.931). The uncompetitive model achieved an even stronger fit (R
2 = 0.949), with a V
max of 7.89 × 10
−8 M/s, a K
m of 21.6 mM, and a K
i of 3700 mM. These results suggest that both cooperative and non-cooperative substrate inhibition mechanisms may contribute to myoglobin’s catalytic behavior under oxidative conditions (
Figure 2).
The Hill coefficient of 2.00 indicates that myoglobin’s catalytic activity increases cooperatively with rising hydrogen peroxide concentrations at low substrate levels. This steep activation profile may reflect multivalent substrate interactions or peroxide-induced aggregation, which can enhance activity before inhibitory effects emerge at higher concentrations
The large inhibition constant (Ki ≈ 3700 mM) in the uncompetitive model suggests that inhibition requires very high hydrogen peroxide concentrations, implying weak binding of peroxide to the enzyme–substrate complex. This may indicate that the decline in activity at elevated peroxide levels arises not solely from classical kinetic inhibition but potentially from secondary oxidative effects.
To complement the findings from myoglobin, a similar kinetic modeling approach was applied to hemoglobin. To evaluate the kinetic behavior of hemoglobin in the presence of hydrogen peroxide, all the substrate inhibition models were explored and the two with the best fit were compared. The uncompetitive inhibition model provided a substantially better fit to the experimental data, with an R
2 value of 0.995, compared to 0.963 for the Hill-type inhibition model. The uncompetitive model also yielded a higher estimated Vmax and a much larger Ki, suggesting weaker inhibition at high substrate concentrations. These results support the conclusion that hemoglobin undergoes inhibition through a mechanism more consistent with uncompetitive substrate binding, rather than cooperative inhibition. A selection of the raw data for these experiments is available in the
Supplementary Materials (Tables S1 and S2, Figures S3 and S4).
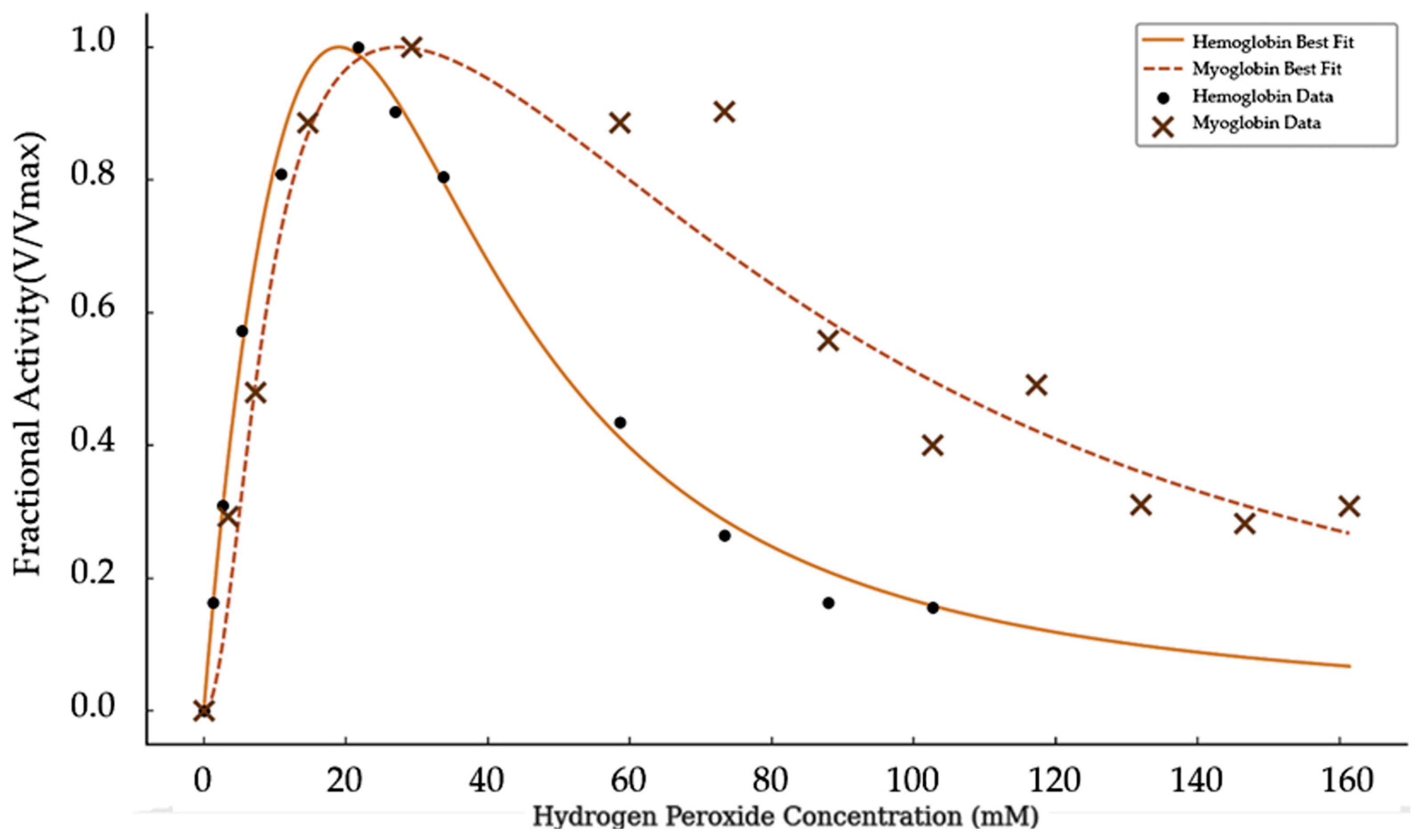
Comparing
Figure 2 and
Figure 3 it is clear that hemoglobin and myoglobin respond differently to increasing concentrations of hydrogen peroxide, as shown in
Figure 4.
Figure 4 presents normalized activity for the two catalysts as a function of substrate concentration. Hemoglobin activity rises quickly at low peroxide levels but declines sharply at moderate concentrations. In contrast, myoglobin maintains higher activity across a broader range of peroxide concentrations and shows a more gradual decrease.
These distinct inhibition patterns suggest that hydrogen peroxide does not simply cause nonspecific oxidative damage. If that were the case, greater or equal inhibition would be expected in myoglobin, which was tested at lower concentrations. Instead, the steeper decline in hemoglobin activity points to a protein-specific mechanism by which hydrogen peroxide interferes with catalytic function.
Although myoglobin is monomeric and therefore cannot exhibit classical multi-subunit cooperativity like hemoglobin in its substrate binding, the Hill coefficient obtained here reflects apparent cooperativity within its substrate inhibition kinetics. In this context, “cooperativity” describes the steep initial increase in activity with rising hydrogen peroxide concentrations, which may result from peroxide-induced conformational changes, transient aggregation, or other intermolecular effects that enhance turnover before inhibitory binding dominates. This apparent cooperativity is thus a feature of the kinetic inhibition mechanism rather than a product of traditional allosteric subunit interactions.
The normalized activity curves (
Figure 4) reveal additional mechanistic differences between the two proteins. Hemoglobin reaches peak activity at lower peroxide concentrations and declines rapidly, suggesting a sharp transition into an inhibited state. In contrast, myoglobin exhibits a broader peak and retains substantial activity even at high peroxide levels, indicating a more gradual and possibly less cooperative inhibitory process.
3.2. Time-Dependent Loss of Activity
Beyond initial kinetic responses, the stability of catalytic activity over time was also explored to better understand inactivation mechanisms. Spectrophotometric time-course experiments performed at 1.2 mM hydrogen peroxide demonstrated that both proteins exhibited time-dependent loss of activity in the presence of hydrogen peroxide, but with markedly different sensitivities. Hemoglobin activity declined almost immediately and was nearly abolished within 5 min, whereas myoglobin retained partial activity for over 20 min before gradually tapering off. These results suggest that while both proteins undergo oxidative inactivation, hemoglobin is more rapidly and severely affected, supporting a model in which peroxide induces protein-specific damage or aggregation (
Figure 5).
These results may suggest that the inhibition of pseudo-peroxidase activity by hydrogen peroxide may involve a two-step mechanism. Initially, myoglobin retains its activity longer than hemoglobin, possibly due to its monomeric structure. Over time, however, both proteins show a secondary, more rapid decline in activity, possibly relating to a subsequent oxidative process.
This pattern may point to a two-step inhibition mechanism including the initial aggregation of the catalyst, which would be more pronounced for monomeric myoglobin, followed by oxidation of residues close enough to impact the heme and further compromise enzyme activity. Aggregation for the myoglobin is likely different than the aggregation step in hemoglobin due to the tetrameric nature of hemoglobin.
Control experiments confirmed that the observed loss of activity was specific to hydrogen peroxide exposure. When hydrogen peroxide was added last, just before measurement, catalytic activity was preserved. In contrast, pre-incubation of the protein with hydrogen peroxide led to significant loss of function, indicating that peroxide exposure time is a critical factor (
Figure 6a).
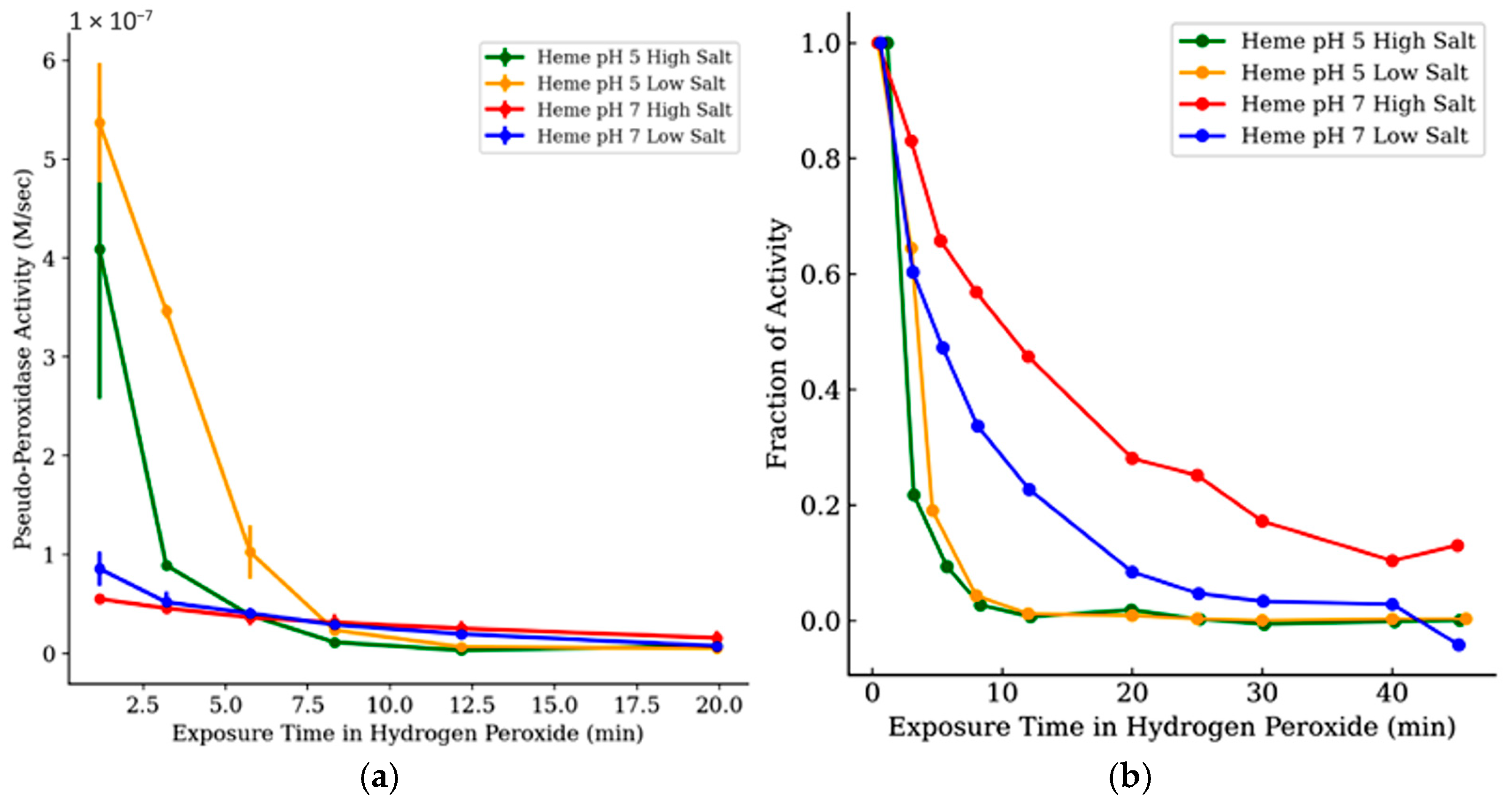
3.3. Influence of Ionic Strength and pH
The effect of pH and ionic strength on hemoglobin’s pseudo-peroxidase activity was evaluated by tracking both absolute and comparing relative activity over time under different buffer conditions (
Figure 6b and
Figure 7a,b). At pH 5, hemoglobin showed the highest initial activity in low-salt conditions, but this activity declined rapidly. High-salt conditions at pH 5 resulted in slightly lower initial rates and an even steeper loss of activity. No visible precipitation or turbidity was observed at 13 mM MgSO
4 under any pH condition, which is far below concentrations typically associated with protein salting-out effects. In contrast, pH 7 conditions, especially with high ionic strength, preserved activity more effectively over time, despite lower initial rates. These results suggest that while acidic, low-salt conditions may enhance early turnover, they also promote rapid oxidative inactivation. Neutral pH buffers appear to mitigate this decline, likely by stabilizing the protein structure or reducing peroxide-mediated damage.
Figure 7b shows that when activities are normalized, pH 7 conditions consistently maintain a greater fraction of hemoglobin’s activity across all time points. This indicates that buffer pH plays a critical role not just in catalytic performance but in protecting the enzyme from peroxide-induced degradation during extended exposures.
In control experiments testing reagent addition order, the catalytic activity was consistently higher when hydrogen peroxide was added last, suggesting that prolonged peroxide exposure is the cause of catalyst inactivation prior to measurement (
Figure 6a). In reactions monitored over the first 100 s (
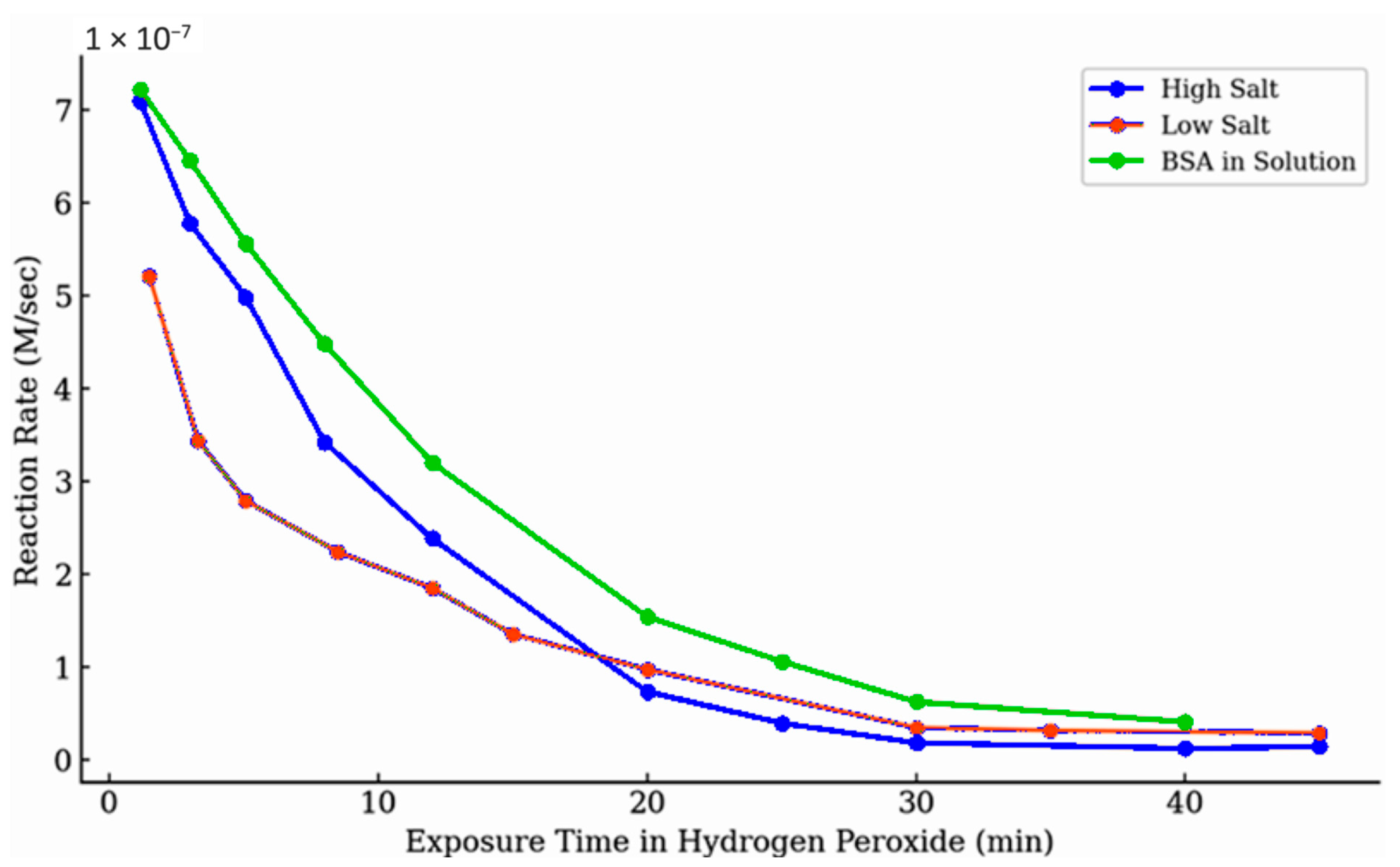
Figure 6b), myoglobin activity showed distinct trends based on salt concentration. Under high-salt conditions, activity declined only slightly, by less than 20%, suggesting limited early inhibition. In contrast, low-salt conditions resulted in a noticeably flatter trajectory, indicating a faster onset of activity loss.
Figure 8 shows a sample of the impact of exposure time on the fraction of activity maintained at different concentrations of hydrogen peroxide at different time points. These observations are consistent with the broader trend seen in
Figure 9, where activity decreases more rapidly under low-salt conditions during prolonged exposure to hydrogen peroxide. This may be further evidence of a two-step mechanism in which the first step is heavily dependent on ionic strength, but this was only a quantitative observation and not clearly identified as a significant factor. It is not unreasonable to expect the aggregation of the enzymes to be impacted by the available ions in solution that help maintain the proteins’ three-dimensional structures.
The presence of a strong electron donor during these short-term measurements may have also played a role in sustaining activity. In the conditions used for
Figure 6b, ABTS was included from the beginning of the reaction, allowing for rapid electron transfer and preventing accumulation of the oxidized heme species. This may have limited the potential for secondary oxidative processes, such as aggregation or protein surface modification. In contrast, the experiments shown in
Figure 7,
Figure 8,
Figure 9 and
Figure 10 involved preincubation of the catalyst in elevated hydrogen peroxide concentrations without ABTS present. Under those conditions, the oxidized form of the catalyst may have persisted longer, increasing the likelihood of inactivation or structural changes before the reaction was initiated.
To further assess time-dependent inactivation, hemoglobin was pre-incubated in various hydrogen peroxide concentrations for either 3 or 40 min prior to catalysis (
Figure 8). After 3 min, activity remained high across all concentrations tested. However, after 40 min, catalytic function was nearly abolished at most peroxide concentrations. Notably, a measurable fraction of activity persisted at the lowest hydrogen peroxide concentration (0.05 mM), even after extended incubation, indicating that peroxide-induced inactivation is both concentration and time-dependent, and that lower concentrations may allow the enzyme to retain some function over longer durations.
The trends observed in both high- and low-salt conditions were consistent across multiple hemoglobin experiments, as summarized in
Figure 7a,b. In the presence of ABTS, high-ionic strength conditions briefly sustained hemoglobin activity, whereas low-salt solutions saw a faster decline. Control experiments confirmed that the order of reagent addition significantly influenced observed activity decay. When hydrogen peroxide was added last the initial rate was reproducible and showed minimal activity loss related to time before reaction initiation. The same was seen when enzyme was added last, minimal activity decay was observed. This is in contrast to activity loss seen when the reaction was initiated by ABTS, allowing time for the catalyst to interact with elevated hydrogen peroxide concentrations.
3.4. Role of BSA and Oxidative Modification
The addition of bovine serum albumin (BSA) did not substantially preserve myoglobin activity over time (
Figure 9). BSA was included as a potential protective agent, providing alternative proteinaceous sites that might compete with oxidative modification of the globins. Some modest preservation of activity might be argued, but increasing the BSA concentration did not enhance this effect, suggesting that peroxide preferentially targets critical functional regions of myoglobin rather than being diverted to alternative sites.
An additional experiment was run to test whether or not peroxidase activity inhibition was completely time dependent and independent of hydrogen peroxide concentration (
Figure 10). Enzymatic activity was monitored following incubation with either 40 nM or 675 nM hydrogen peroxide for up to 50 min. At low hydrogen peroxide concentration, activity remained relatively stable over time, indicating minimal time-dependent inactivation at this concentration. In contrast, a substantial loss of activity was observed at the higher concentration, supporting a concentration-dependent mechanism of inhibition. These results suggest that inactivation is unlikely due to gradual nonspecific protein oxidation and may involve targeted modifications at higher peroxide levels.
4. Discussion
The decline in catalytic activity for both myoglobin and hemoglobin at elevated hydrogen peroxide concentrations indicates a mechanistic shift beyond simple oxidative degradation. The biphasic activity profiles, an initial increase in rate followed by a decline, are characteristic of substrate-inhibition-like kinetics [
10,
13,
14,
23]. Such behavior is consistent with a reversible, kinetic process rather than immediate oxidative collapse of the heme [
3,
5]. The strong statistical fits to Hill and uncompetitive models further reinforce this view, suggesting that activity loss follows structured inhibition dynamics [
15] instead of reflecting nonspecific damage. Mechanistically, this inhibition may arise from transient peroxide interactions at secondary or non-productive sites, or from conformational adjustments that reduce catalytic efficiency without directly destroying the active site.
The apparent cooperative fit of the Hill model for myoglobin is somewhat unexpected given its monomeric nature. In this context, we interpret the Hill coefficient primarily as an empirical descriptor of curvature rather than evidence of true allosteric behavior [
13,
16]. One possible explanation is that peroxide exposure promotes aggregation or reversible oxidative modifications at peripheral residues, which could generate transient intermolecular interactions that mimic cooperative kinetics [
6,
24]. In contrast, the uncompetitive model, with a relatively high Ki, places inhibition after substrate binding, consistent with a scenario where the enzyme–substrate complex remains intact but progressively less efficient. Such behavior may reflect structural destabilization rather than classical binding inhibition [
13,
15].
The relatively large inhibition constant obtained from the uncompetitive model suggests that high peroxide concentrations are required before substantial activity loss occurs, consistent with a mechanism driven by gradual structural changes rather than direct kinetic competition [
25]. Such changes likely involve progressive conformational disruption or heme modification rather than immediate active-site inactivation [
5,
19]. Differences between myoglobin and hemoglobin reinforce this interpretation: the tetrameric architecture of hemoglobin may help distribute oxidative stress and confer greater structural resilience, whereas monomeric myoglobin appears more vulnerable to early destabilization [
26]. The time-dependent decline in activity further supports a two-phase model for myoglobin: an early stage where peroxide targets peripheral residues or promotes reversible aggregation [
6,
27], followed by later stages where the heme environment itself is altered. This sequence allows partial preservation of function during the initial phase even as structural perturbations accumulate. Taken together, these results indicate that hemoglobin activity is more acutely vulnerable to hydrogen peroxide than myoglobin, undergoing rapid inactivation within minutes, whereas myoglobin retains partial activity for a longer period before gradual loss.
In light of these kinetics, the differing behaviors of hemoglobin and myoglobin may help frame oxidative stress in blood versus muscle. Under our experimental conditions, hemoglobin is inactivated more acutely, whereas myoglobin retains partial activity for longer before a gradual decline (
Figure 4 and
Figure 5). This distinction is consistent with hemoglobin’s ferryl/heme-loss pathways and vulnerability at elevated hydrogen peroxide [
5,
19], and with myoglobin’s monomeric architecture where early, partly reversible oxidative modifications or aggregation can precede later damage [
6,
24,
28]. In vivo, erythrocyte antioxidant systems normally limit peroxide, so hemoglobin damage is most relevant when buffering is overwhelmed or hemoglobin is released (e.g., hemolysis) [
3,
5]; conversely, during muscle injury or inflammation, localized hydrogen peroxide and lipid oxidation chemistry implicate myoglobin in early oxidative events before functional loss [
4,
12]. While the peroxide levels used here are above physiological steady states, our results provide an accelerated model that highlights comparative susceptibilities of blood and muscle globins under oxidative stress.
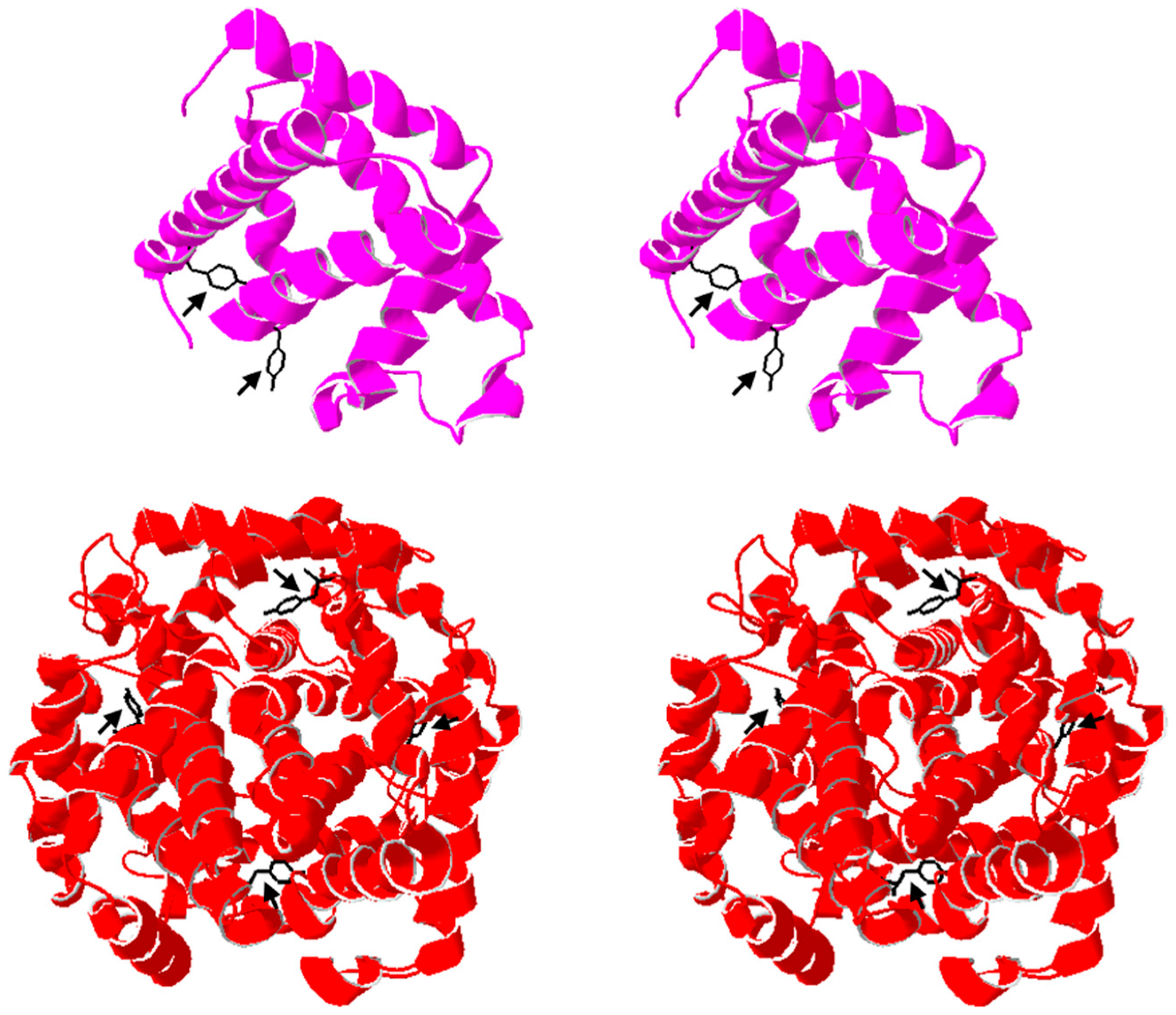
To explore whether structural accessibility may help explain peroxide-induced inactivation, we examined representative crystal structures of myoglobin (PDB 1DWR) and hemoglobin (PDB 2MHB) to visualize tyrosine residues previously implicated in oxidative crosslinking (
Figure 11). In myoglobin, at least one tyrosine residue is highly solvent-exposed and has been associated with peroxide-driven dimerization [
6,
17,
28]. In contrast, hemoglobin contains homologous tyrosines within each subunit, but these are more buried within the tetrameric framework, which could limit their reactivity [
29]. This qualitative comparison suggests that structural positioning and solvent accessibility may contribute to the greater susceptibility of myoglobin to early aggregation, consistent with reports of its oxidative dimerization propensity [
6,
24]. While not experimental data, this visualization provides mechanistic context for our kinetic results and highlights how tertiary and quaternary structural features can modulate vulnerability to oxidative inhibition.
Order-of-addition experiments showed that inactivation depends on cumulative peroxide exposure rather than instantaneous concentration, as adding peroxide last preserved activity. Buffer conditions further modulated stability: acidic, low-salt environments enhanced initial rates but hastened inactivation, whereas neutral pH and high salt prolonged activity [
27,
30,
31]. BSA provided no protection, consistent with targeted rather than nonspecific oxidation [
22,
32]. Together, these findings highlight a trade-off between catalytic performance and structural longevity under oxidative stress.
While these findings give insight into kinetic and structural features of myoglobin and hemoglobin under controlled oxidative stress, the physiological impact must be viewed in context. The peroxide concentrations used here far exceed typical cellular levels, which are tightly regulated by catalase, peroxidases, and glutathione systems. In vivo, these proteins also function in antioxidant-rich and highly crowded environments that limit aggregation and buffer oxidative modifications. Thus, although our data highlight potential mechanisms of peroxide-induced inactivation, they likely represent an accelerated model of damage rather than a direct reflection of normal physiological conditions.
Overall, the data support a two-phase inactivation model for myoglobin: an early, reversible perturbation followed by slower, oxidative damage [
32]. Factors such as ionic strength, exposure time, and protein architecture modulate this process [
27]. These findings clarify the kinetic vulnerabilities of heme proteins and point to future spectroscopic and structural studies as the next step in defining the underlying molecular changes.
5. Conclusions
This study provides compelling evidence that both myoglobin and hemoglobin exhibit substrate inhibition under high-hydrogen peroxide conditions, with activity loss best described by models incorporating uncompetitive or cooperative inhibition. Hemoglobin proved more acutely vulnerable, showing rapid inactivation within minutes, whereas myoglobin retained partial activity over a broader concentration and time range before gradually declining. These differences likely reflect structural features, including myoglobin’s greater solvent-exposed tyrosines and monomeric nature, which may favor early aggregation, compared with hemoglobin’s tetrameric assembly.
Inactivation was both concentration time- and pH-dependent and may be further modulated by ionic strength, with neutral buffers and high salt stabilizing activity relative to acidic, low-salt conditions. Taken together, the results support a multi-step inhibition pathway in which early, partly reversible perturbations give way to slower, more permanent oxidative damage. While the peroxide levels used here exceed physiological conditions, this accelerated model highlights comparative susceptibilities of muscle and blood globins under oxidative stress. These findings refine the mechanistic understanding of pseudo-peroxidase inhibition and relate to other and future work on reversibility and structural characterization to further elucidate the molecular basis of globin inactivation.