Text Mining to Understand Disease-Causing Gene Variants
Abstract
1. Introduction
1.1. Background
1.2. Motivation
1.3. Objectives
2. Literature Review
2.1. Online Resources
2.1.1. Databases
2.1.2. Variant Classification Tools
2.1.3. Pathway Analysis Tools
2.2. Text Mining
2.3. Large Language Models
3. Examples and Case Studies
3.1. Autism Spectrum Disorder as an Example of the Implications of Genetic Variants
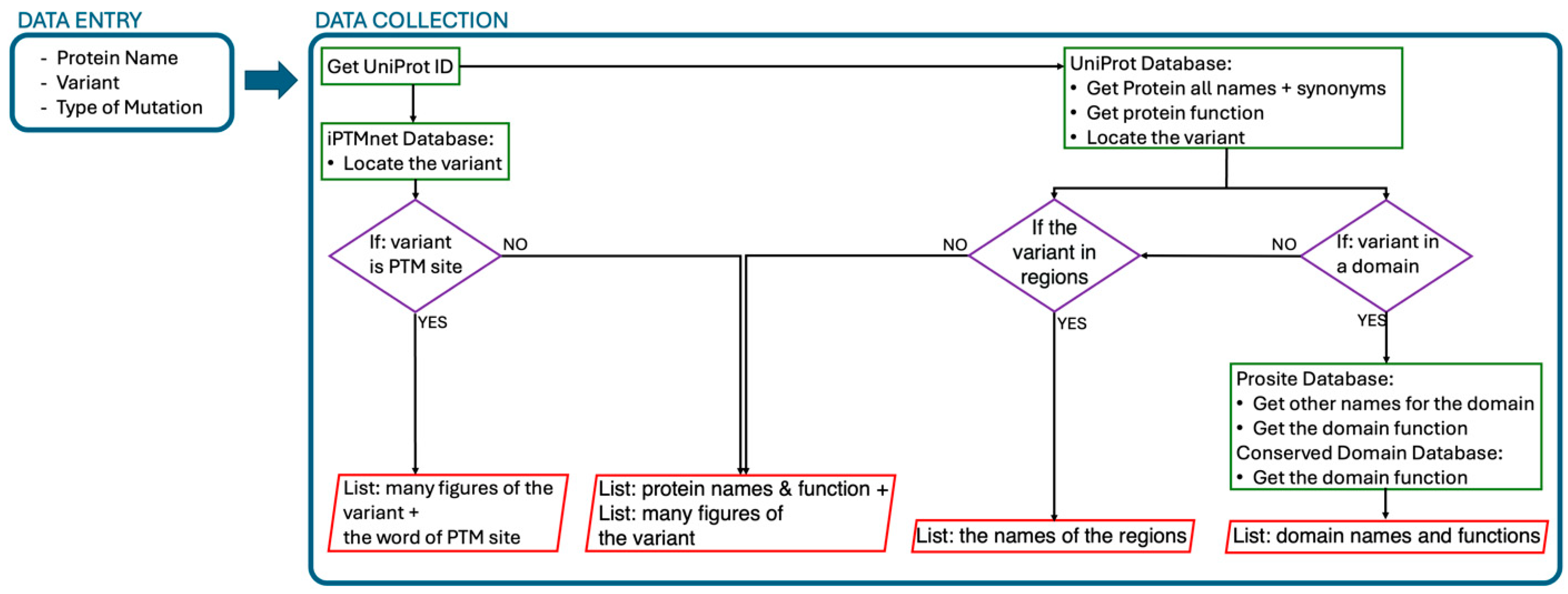
3.2. The Automated Curation
3.3. UniProt Database Curation
3.4. iPTMnet Database Curation
3.5. Arranging the Information
3.6. Knowledge Retrieval
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
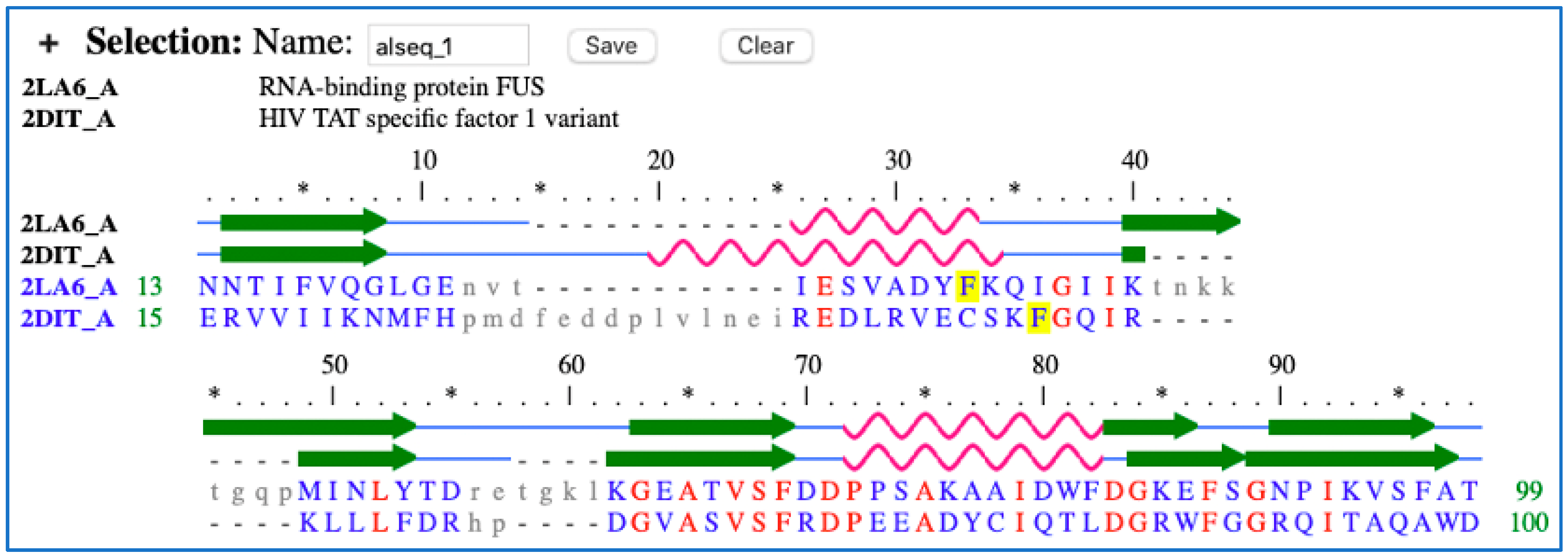
Appendix A.1. HTATSF1
Appendix A.2. OR6C65
Appendix A.3. ITIH6
Appendix A.4. DDX26B
References
- Goh, G.; Choi, M. Application of whole exome sequencing to identify disease-causing variants in inherited human diseases. Genom. Inform. 2012, 10, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Kereszturi, É. Diversity and Classification of Genetic Variations in Autism Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 16768. [Google Scholar] [CrossRef]
- Le Guen, Y.; Belloy, M.E.; Grenier-Boley, B.; de Rojas, I.; Castillo-Morales, A.; Jansen, I.; Nicolas, A.; Bellenguez, C.; Dalmasso, C.; Küçükali, F.; et al. Association of Rare APOE Missense Variants V236E and R251G With Risk of Alzheimer Disease. JAMA Neurol. 2022, 79, 652–663. [Google Scholar] [CrossRef]
- Feliciano, P.; Zhou, X.; Astrovskaya, I.; Turner, T.N.; Wang, T.; Brueggeman, L.; Barnard, R.; Hsieh, A.; Snyder, L.G.; Muzny, D.M.; et al. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom. Med. 2019, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Husson, T.; Lecoquierre, F.; Cassinari, K.; Charbonnier, C.; Quenez, O.; Goldenberg, A.; Guerrot, A.M.; Richard, A.C.; Drouin-Garraud, V.; Brehin, A.C.; et al. Rare genetic susceptibility variants assessment in autism spectrum disorder: Detection rate and practical use. Transl. Psychiatry 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Jiang, Q.; Lin, J.; Wei, Q.; Li, C.; Hou, Y.; Cao, B.; Zhang, L.; Ou, R.; Liu, K.; et al. TBK1 variants in Chinese patients with amyotrophic lateral sclerosis: Genetic analysis and clinical features. Eur. J. Neurol. 2023, 30, 3079–3089. [Google Scholar] [CrossRef]
- Vihinen, M. Functional effects of protein variants. Biochimie 2021, 180, 104–120. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Liu, Z.; Qian, W.; Cai, W.; Song, W.; Wang, W.; Maharjan, D.T.; Cheng, W.; Chen, J.; Wang, H.; Xu, D.; et al. Inferring the Effects of Protein Variants on Protein-Protein Interactions with Interpretable Transformer Representations. Research 2023, 6, 0219. [Google Scholar] [CrossRef]
- Ali, S.; Ali, U.; Qamar, A.; Zafar, I.; Yaqoob, M.; Ain, Q.U.; Rashid, S.; Sharma, R.; Nafidi, H.A.; Bin Jardan, Y.A.; et al. Predicting the effects of rare genetic variants on oncogenic signaling pathways: A computational analysis of HRAS protein function. Front. Chem. 2023, 11, 1173624. [Google Scholar] [CrossRef]
- Sun, Y.V. Integration of biological networks and pathways with genetic association studies. Hum. Genet. 2012, 131, 1677–1686. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ahmed, F.; Samantasinghar, A.; Soomro, A.M.; Kim, S.; Choi, K.H. A systematic review of computational approaches to understand cancer biology for informed drug repurposing. J. Biomed. Inform. 2023, 142, 104373. [Google Scholar] [CrossRef] [PubMed]
- Nezamuldeen, L.; Jafri, M.S. Protein–Protein Interaction Network Extraction Using Text Mining Methods Adds Insight into Autism Spectrum Disorder. Biology 2023, 12, 1344. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Allot, A.; Peng, Y.; Wei, C.-H.; Lee, K.; Phan, L.; Lu, Z. LitVar: A semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic Acids Res. 2018, 46, W530–W536. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- McCoy, M.D.; Hamre, J., 3rd; Klimov, D.K.; Jafri, M.S. Predicting Genetic Variation Severity Using Machine Learning to Interpret Molecular Simulations. Biophys. J. 2021, 120, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Hamre, J.R., 3rd; Klimov, D.K.; McCoy, M.D.; Jafri, M.S. Machine learning-based prediction of drug and ligand binding in BCL-2 variants through molecular dynamics. Comput. Biol. Med. 2022, 140, 105060. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Hombach, D.; Köhler, S.; Cooper, D.N.; Schuelke, M.; Seelow, D. RegulationSpotter: Annotation and interpretation of extratranscriptic DNA variants. Nucleic Acids Res. 2019, 47, W106–W113. [Google Scholar] [CrossRef] [PubMed]
- Parthiban, V.; Gromiha, M.M.; Schomburg, D. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Res. 2006, 34, W239–W242. [Google Scholar] [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M. MutPred2: Inferring the molecular and phenotypic impact of amino acid variants. bioRxiv 2017, 134981. [Google Scholar] [CrossRef] [PubMed]
- López-Ferrando, V.; Gazzo, A.; De La Cruz, X.; Orozco, M.; Gelpí, J.L. PMut: A web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res. 2017, 45, W222–W228. [Google Scholar] [CrossRef] [PubMed]
- Masso, M.; Vaisman, I.I. AUTO-MUTE: Web-based tools for predicting stability changes in proteins due to single amino acid replacements. Protein Eng. Des. Sel. 2010, 23, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Benedix, A.; Becker, C.M.; de Groot, B.L.; Caflisch, A.; Böckmann, R.A. Predicting free energy changes using structural ensembles. Nat. Methods 2009, 6, 3. [Google Scholar] [CrossRef]
- Song, Y.; Di Maio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Potapov, V.; Cohen, M.; Schreiber, G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng. Des. Sel. 2009, 22, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Palli, R.; Palshikar, M.G.; Thakar, J. Executable pathway analysis using ensemble discrete-state modeling for large-scale data. PLoS Comput. Biol. 2019, 15, e1007317. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, Ö.; Anwar, N.; Schultz, N.; Bader, G.D.; Sander, C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2010, 39, D685–D690. [Google Scholar] [CrossRef] [PubMed]
- Demir, E.; Cary, M.P.; Paley, S.; Fukuda, K.; Lemer, C.; Vastrik, I.; Wu, G.; D’eustachio, P.; Schaefer, C.; Luciano, J. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010, 28, 935. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Schwab, J.D.; Kühlwein, S.D.; Ikonomi, N.; Kühl, M.; Kestler, H.A. Concepts in Boolean network modeling: What do they all mean? Comput. Struct. Biotechnol. J. 2020, 18, 571–582. [Google Scholar] [CrossRef]
- Veliz-Cuba, A.; Aguilar, B.; Hinkelmann, F.; Laubenbacher, R. Steady state analysis of Boolean molecular network models via model reduction and computational algebra. BMC Bioinform. 2014, 15, 221. [Google Scholar] [CrossRef]
- Irurzun-Arana, I.; Pastor, J.M.; Trocóniz, I.F.; Gómez-Mantilla, J.D. Advanced Boolean modeling of biological networks applied to systems pharmacology. Bioinformatics 2017, 33, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Nezamuldeen, L.; Jafri, M.S. Boolean Modeling of Biological Network Applied to Protein-Protein Interaction Network of Autism Patients. Biology 2024, 13, 606. [Google Scholar] [CrossRef]
- Przybyła, P.; Shardlow, M.; Aubin, S.; Bossy, R.; Eckart de Castilho, R.; Piperidis, S.; McNaught, J.; Ananiadou, S. Text mining resources for the life sciences. Database 2016, 2016, baw145. [Google Scholar] [CrossRef] [PubMed]
- Verspoor, K.M.; Cohn, J.D.; Ravikumar, K.E.; Wall, M.E. Text mining improves prediction of protein functional sites. PLoS ONE 2012, 7, e32171. [Google Scholar] [CrossRef] [PubMed]
- Samandari Bahraseman, M.R.; Khorsand, B.; Esmaeilzadeh-Salestani, K.; Sarhadi, S.; Hatami, N.; Khaleghdoust, B.; Loit, E. The use of integrated text mining and protein-protein interaction approach to evaluate the effects of combined chemotherapeutic and chemopreventive agents in cancer therapy. PLoS ONE 2022, 17, e0276458. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.H.; Harris, B.R.; Kao, H.Y.; Lu, Z. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics 2013, 29, 1433–1439. [Google Scholar] [CrossRef]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Salekin, S.; Zhang, J.M.; Huang, Y. A deep learning model for predicting transcription factor binding location at single nucleotide resolution. In Proceedings of the 2017 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Orland, FL, USA, 16–19 February 2017; pp. 57–60. [Google Scholar]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rush, A.M. Dilated convolutions for modeling long-distance genomic dependencies. arXiv 2017, arXiv:1710.01278. [Google Scholar]
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016, 44, e107. [Google Scholar] [CrossRef]
- Yang, B.; Liu, F.; Ren, C.; Ouyang, Z.; Xie, Z.; Bo, X.; Shu, W. BiRen: Predicting enhancers with a deep-learning-based model using the DNA sequence alone. Bioinformatics 2017, 33, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Bao, W.; Huang, D.S. Recurrent Neural Network for Predicting Transcription Factor Binding Sites. Sci. Rep. 2018, 8, 15270. [Google Scholar] [CrossRef]
- Pan, X.; Rijnbeek, P.; Yan, J.; Shen, H.B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Genom. 2018, 19, 511. [Google Scholar] [CrossRef]
- He, Y.; Shen, Z.; Zhang, Q.; Wang, S.; Huang, D.S. A survey on deep learning in DNA/RNA motif mining. Brief. Bioinform. 2021, 22, bbaa229. [Google Scholar] [CrossRef]
- Kaddour, J.; Harris, J.; Mozes, M.; Bradley, H.; Raileanu, R.; McHardy, R. Challenges and applications of large language models. arXiv 2023, arXiv:2307.10169. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Luo, R.; Sun, L.; Xia, Y.; Qin, T.; Zhang, S.; Poon, H.; Liu, T.-Y. BioGPT: Generative Pre-trained Transformer for Biomedical Text Generation and Mining. Brief. Bioinform. 2022, 23, bbac409. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Zagirova, D.; Pushkov, S.; Leung, G.H.D.; Liu, B.H.M.; Urban, A.; Sidorenko, D.; Kalashnikov, A.; Kozlova, E.; Naumov, V.; Pun, F.W.; et al. Biomedical generative pre-trained based transformer language model for age-related disease target discovery. Aging 2023, 15, 9293–9309. [Google Scholar] [CrossRef]
- Huang, L.; Lin, J.; Li, X.; Song, L.; Zheng, Z.; Wong, K.C. EGFI: Drug-drug interaction extraction and generation with fusion of enriched entity and sentence information. Brief. Bioinform. 2022, 23, bbab451. [Google Scholar] [CrossRef] [PubMed]
- Karkera, N.; Acharya, S.; Palaniappan, S.K. Leveraging pre-trained language models for mining microbiome-disease relationships. BMC Bioinform. 2023, 24, 290. [Google Scholar] [CrossRef] [PubMed]
- Das Baksi, K.; Pokhrel, V.; Pudavar, A.E.; Mande, S.S.; Kuntal, B.K. BactInt: A domain driven transfer learning approach for extracting inter-bacterial associations from biomedical text. Comput. Biol. Chem. 2024, 109, 108012. [Google Scholar] [CrossRef] [PubMed]
- Philippidis, A. Nvidia Looks to Genentech for Its Next Leap in AI Drug Discovery: Roche subsidiary becomes newest biopharma partner for Silicon Valley giant as it grows life sciences footprint. GEN Edge 2023, 5, 828–833. [Google Scholar] [CrossRef]
- Sevgen, E.; Moller, J.; Lange, A.; Parker, J.; Quigley, S.; Mayer, J.; Srivastava, P.; Gayatri, S.; Hosfield, D.; Korshunova, M. ProT-VAE: Protein transformer variational autoencoder for functional protein design. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wong, F.; de la Fuente-Nunez, C.; Collins, J.J. Leveraging artificial intelligence in the fight against infectious diseases. Science 2023, 381, 164–170. [Google Scholar] [CrossRef]
- Roberts, J.B.; Nava, A.A.; Pearson, A.N.; Incha, M.R.; Valencia, L.E.; Ma, M.; Rao, A.; Keasling, J.D. Foldy: A web application for interactive protein structure analysis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Al-Mubarak, B.; Abouelhoda, M.; Omar, A.; AlDhalaan, H.; Aldosari, M.; Nester, M.; Alshamrani, H.A.; El-Kalioby, M.; Goljan, E.; Albar, R. Whole exome sequencing reveals inherited and de novo variants in autism spectrum disorder: A trio study from Saudi families. Sci. Rep. 2017, 7, 5679. [Google Scholar] [CrossRef]
- Coudert, E.; Gehant, S.; de Castro, E.; Pozzato, M.; Baratin, D.; Neto, T.; Sigrist, C.J.A.; Redaschi, N.; Bridge, A.; Consortium, U. Annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics 2023, 39, btac793. [Google Scholar] [CrossRef]
- Requests: HTTP for Humans™. Available online: https://requests.readthedocs.io/en/latest/ (accessed on 1 July 2024).
- Richardson, L. Beautiful Soup Documentation; April, 2007. Available online: https://tedboy.github.io/bs4_doc/ (accessed on 1 July 2024).
- Huang, H.; Arighi, C.N.; Ross, K.E.; Ren, J.; Li, G.; Chen, S.C.; Wang, Q.; Cowart, J.; Vijay-Shanker, K.; Wu, C.H. iPTMnet: An integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 2018, 46, D542–D550. [Google Scholar] [CrossRef]
- Ahmad, R.M.; Ali, B.R.; Al-Jasmi, F.; Sinnott, R.O.; Al Dhaheri, N.; Mohamad, M.S. A review of genetic variant databases and machine learning tools for predicting the pathogenicity of breast cancer. Brief. Bioinform. 2023, 25, bbad479. [Google Scholar] [CrossRef]
- Berger, S.M.; Appelbaum, P.S.; Siegel, K.; Wynn, J.; Saami, A.M.; Brokamp, E.; O’Connor, B.C.; Hamid, R.; Martin, D.M.; Chung, W.K. Challenges of variant reinterpretation: Opinions of stakeholders and need for guidelines. Genet. Med. 2022, 24, 1878–1887. [Google Scholar] [CrossRef]
- Garcia, F.A.O.; de Andrade, E.S.; Palmero, E.I. Insights on variant analysis. Front. Genet. 2022, 13, 1010327. [Google Scholar] [CrossRef]
- Daigle, J.G.; Lanson, N.A., Jr.; Smith, R.B.; Casci, I.; Maltare, A.; Monaghan, J.; Nichols, C.D.; Kryndushkin, D.; Shewmaker, F.; Pandey, U.B. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum. Mol. Genet. 2012, 22, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Cléry, A.; Allain, F.H.T. From structure to function of RNA binding domains. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2000–2013. [Google Scholar]
- Madej, T.; Lanczycki, C.J.; Zhang, D.; Thiessen, P.A.; Geer, R.C.; Marchler-Bauer, A.; Bryant, S.H. MMDB and VAST+: Tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014, 42, D297–D303. [Google Scholar] [CrossRef] [PubMed]
- Corsini, N.S.; Peer, A.M.; Moeseneder, P.; Roiuk, M.; Burkard, T.R.; Theussl, H.C.; Moll, I.; Knoblich, J.A. Coordinated Control of mRNA and rRNA Processing Controls Embryonic Stem Cell Pluripotency and Differentiation. Cell Stem Cell 2018, 22, 543–558.e512. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics 2020, 36, 131–135. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Lu, S.; Madej, T.; Marchler, G.H.; Cheng, T.; Chong, L.C.; et al. iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode. Front. Mol. Biosci. 2022, 9, 831740. [Google Scholar] [CrossRef] [PubMed]
- Olender, T.; Lancet, D.; Nebert, D.W. Update on the olfactory receptor (OR) gene superfamily. Hum. Genom. 2008, 3, 87. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Ronnett, G.V.; Moon, C. G proteins and olfactory signal transduction. Annu. Rev. Physiol. 2002, 64, 189–222. [Google Scholar] [CrossRef]
- Sarafoleanu, C.; Mella, C.; Georgescu, M.; Perederco, C. The importance of the olfactory sense in the human behavior and evolution. J. Med. Life 2009, 2, 196–198. [Google Scholar] [PubMed]
- Rinaldi, A. The scent of life. The exquisite complexity of the sense of smell in animals and humans. EMBO Rep. 2007, 8, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, B.; Masukawa, L.M.; Shepherd, G.M. Excitable properties of olfactory receptor neurons. J. Neurosci. 1987, 7, 2338–2343. [Google Scholar]
- Tonacci, A.; Sansone, F.; Pala, A.P.; Centrone, A.; Napoli, F.; Domenici, C.; Conte, R. Effect of feeding on neurovegetative response to olfactory stimuli. In Proceedings of the 2017 E-Health and Bioengineering Conference (EHB), Sinaia, Romania, 22–24 June 2017; pp. 9–12. [Google Scholar]
- Ashwin, C.; Chapman, E.; Howells, J.; Rhydderch, D.; Walker, I.; Baron-Cohen, S. Enhanced olfactory sensitivity in autism spectrum conditions. Mol. Autism 2014, 5, 53. [Google Scholar] [CrossRef]
- Rozenkrantz, L.; Zachor, D.; Heller, I.; Plotkin, A.; Weissbrod, A.; Snitz, K.; Secundo, L.; Sobel, N. A Mechanistic Link between Olfaction and Autism Spectrum Disorder. Curr. Biol. 2015, 25, 1904–1910. [Google Scholar] [CrossRef]
- Wicker, B.; Monfardini, E.; Royet, J.P. Olfactory processing in adults with autism spectrum disorders. Mol. Autism 2016, 7, 4. [Google Scholar] [CrossRef]
- Zhuo, L.; Kimata, K. Structure and function of inter-α-trypsin inhibitor heavy chains. Connect. Tissue Res. 2008, 49, 311–320. [Google Scholar] [CrossRef]
- Morikis, D.; Lambris, J.D. Structural Biology of the Complement System; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Whittaker, C.A.; Hynes, R.O. Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 2002, 13, 3369–3387. [Google Scholar] [CrossRef] [PubMed]
- Bost, F.; Diarra-Mehrpour, M.; Martin, J.P. Inter-alpha-trypsin inhibitor proteoglycan family—A group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 1998, 252, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yoneda, M.; Kimata, K. A serum-derived hyaluronan-associated protein (SHAP) is the heavy chain of the inter alpha-trypsin inhibitor. J. Biol. Chem. 1993, 268, 26725–26730. [Google Scholar] [CrossRef]
- Zhao, M.; Yoneda, M.; Ohashi, Y.; Kurono, S.; Iwata, H.; Ohnuki, Y.; Kimata, K. Evidence for the covalent binding of SHAP, heavy chains of inter-alpha-trypsin inhibitor, to hyaluronan. J. Biol. Chem. 1995, 270, 26657–26663. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Popovich, P.G. Extracellular matrix regulation of inflammation in the healthy and injured spinal cord. Exp. Neurol. 2014, 258, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Bonneh-Barkay, D.; Wiley, C.A. Brain extracellular matrix in neurodegeneration. Brain Pathol. 2009, 19, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Warren, P.M.; Dickens, S.M.; Gigout, S.; Fawcett, J.W.; Kwok, J.C.F. Regulation of CNS Plasticity Through the Extracellular Matrix. In The Oxford Handbook of Developmental Neural Plasticity; Chao, M.V., Ed.; Oxford University Press: New York, NY, USA, 2018. [Google Scholar]
- Camargo, A.A.; Nunes, D.N.; Samaia, H.B.; Liu, L.; Collins, V.P.; Simpson, A.J.; Dias-Neto, E. Molecular characterization of DDX26, a human DEAD-box RNA helicase, located on chromosome 7p12. Braz. J. Med. Biol. Res. 2001, 34, 1237–1245. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baillat, D.; Hakimi, M.A.; Näär, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Baillat, D.; Wagner, E.J. Integrator: Surprisingly diverse functions in gene expression. Trends Biochem. Sci. 2015, 40, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41, D348–D352. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, T.; Yu, X. A core hSSB1-INTS complex participates in the DNA damage response. J. Cell Sci. 2013, 126, 4850–4855. [Google Scholar] [CrossRef]
- Jodoin, J.N.; Sitaram, P.; Albrecht, T.R.; May, S.B.; Shboul, M.; Lee, E.; Reversade, B.; Wagner, E.J.; Lee, L.A. Nuclear-localized Asunder regulates cytoplasmic dynein localization via its role in the integrator complex. Mol. Biol. Cell 2013, 24, 2954–2965. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wagner, E.J. snRNA 3′ end formation: The dawn of the Integrator complex. Biochem. Soc. Trans. 2010, 38, 1082–1087. [Google Scholar] [CrossRef]
- Kapp, L.D.; Abrams, E.W.; Marlow, F.L.; Mullins, M.C. The integrator complex subunit 6 (Ints6) confines the dorsal organizer in vertebrate embryogenesis. PLoS Genet. 2013, 9, e1003822. [Google Scholar] [CrossRef] [PubMed]
- Otani, Y.; Nakatsu, Y.; Sakoda, H.; Fukushima, T.; Fujishiro, M.; Kushiyama, A.; Okubo, H.; Tsuchiya, Y.; Ohno, H.; Takahashi, S.; et al. Integrator complex plays an essential role in adipose differentiation. Biochem. Biophys. Res. Commun. 2013, 434, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Ferris, A.L.; Wu, X.; Saraf, A.; Khanna, K.K.; Florens, L.; Washburn, M.P.; Hughes, S.H.; Pagano, M. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015, 25, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Lui, K.Y.; Zhao, H.; Qiu, C.; Li, C.; Zhang, Z.; Peng, H.; Fu, R.; Chen, H.A.; Lu, M.Q. Integrator complex subunit 6 (INTS6) inhibits hepatocellular carcinoma growth by Wnt pathway and serve as a prognostic marker. BMC Cancer 2017, 17, 644. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.N.; Heyer, W.D.; LaSalle, J.M. Autism and Cancer Share Risk Genes, Pathways, and Drug Targets. Trends Genet. 2016, 32, 139–146. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nezamuldeen, L.; Jafri, M.S. Text Mining to Understand Disease-Causing Gene Variants. Knowledge 2024, 4, 422-443. https://doi.org/10.3390/knowledge4030023
Nezamuldeen L, Jafri MS. Text Mining to Understand Disease-Causing Gene Variants. Knowledge. 2024; 4(3):422-443. https://doi.org/10.3390/knowledge4030023
Chicago/Turabian StyleNezamuldeen, Leena, and Mohsin Saleet Jafri. 2024. "Text Mining to Understand Disease-Causing Gene Variants" Knowledge 4, no. 3: 422-443. https://doi.org/10.3390/knowledge4030023
APA StyleNezamuldeen, L., & Jafri, M. S. (2024). Text Mining to Understand Disease-Causing Gene Variants. Knowledge, 4(3), 422-443. https://doi.org/10.3390/knowledge4030023






