Incorporating Uncertainty Quantification for the Performance Improvement of Academic Recommenders
Abstract
1. Introduction
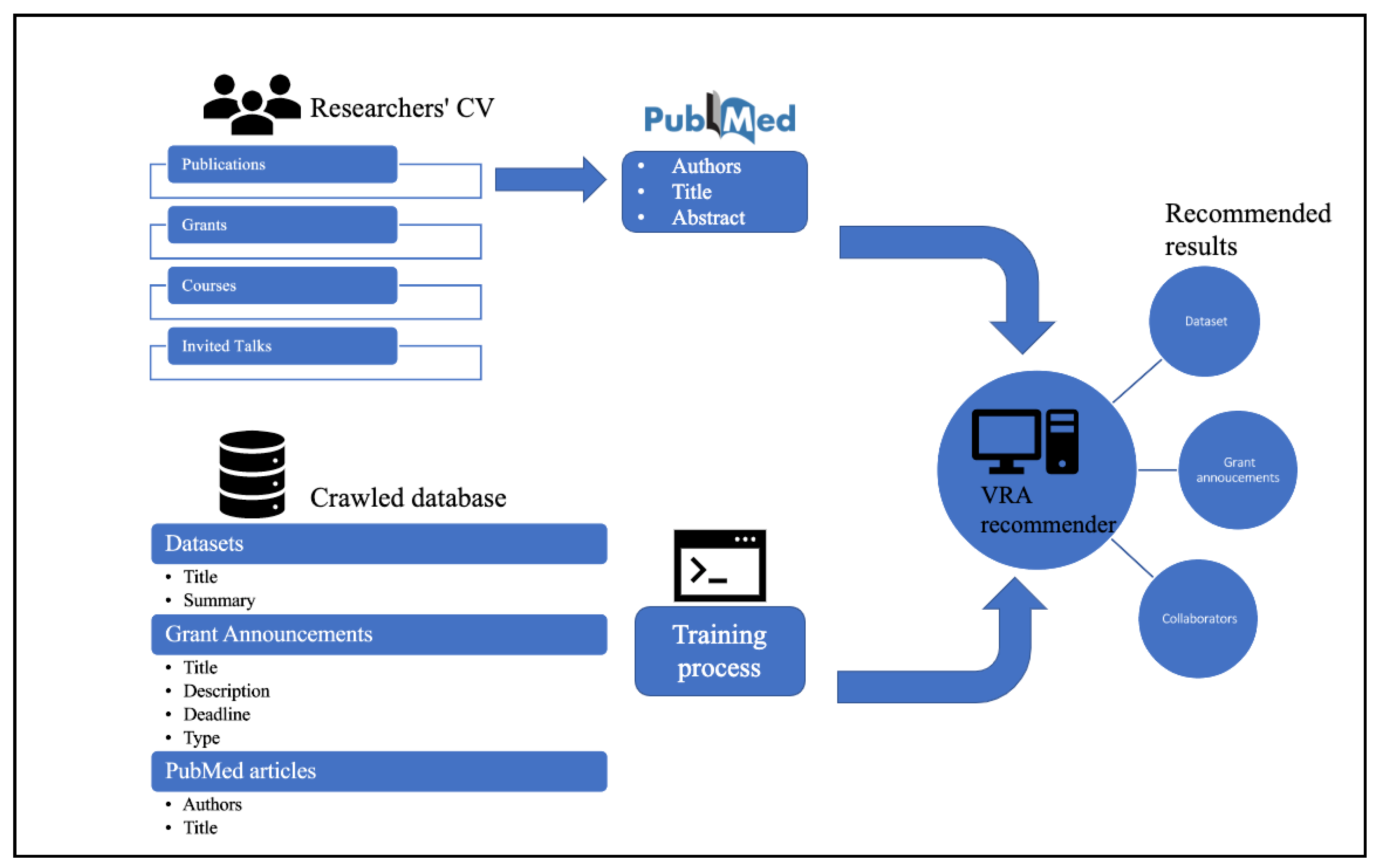
- First, we carried out UQ experiments on different deep learning components of our virtual research assistant (VRA) platform, a web-based recommender for population health professionals that recommends datasets, grants, and collaborators. (The service platform is available at http://genestudy.org/recommends/#/ (accessed on 6 April 2023). We utilized two widely accepted UQ methods: MC dropout and ensemble. Using UQ, we were able to better understand the behaviors of our recommender outputs, while our grant recommender (BERT-based) tends to be overconfident with its outputs, and our collaborator recommender (TGN-based) tends to be underconfident in producing matching probabilities.
- Secondly, we introduced a new metric to incorporate the UQ information into our ranking scores. With this information, we were able to down-rank recommendations that the models were less sure about, thus reducing the risks associated with uncertain recommendations for better user experience. Moreover, the evaluations revealed that our proposed method with ensemble was able to produce consistently better results in a variety of metrics including calibration.
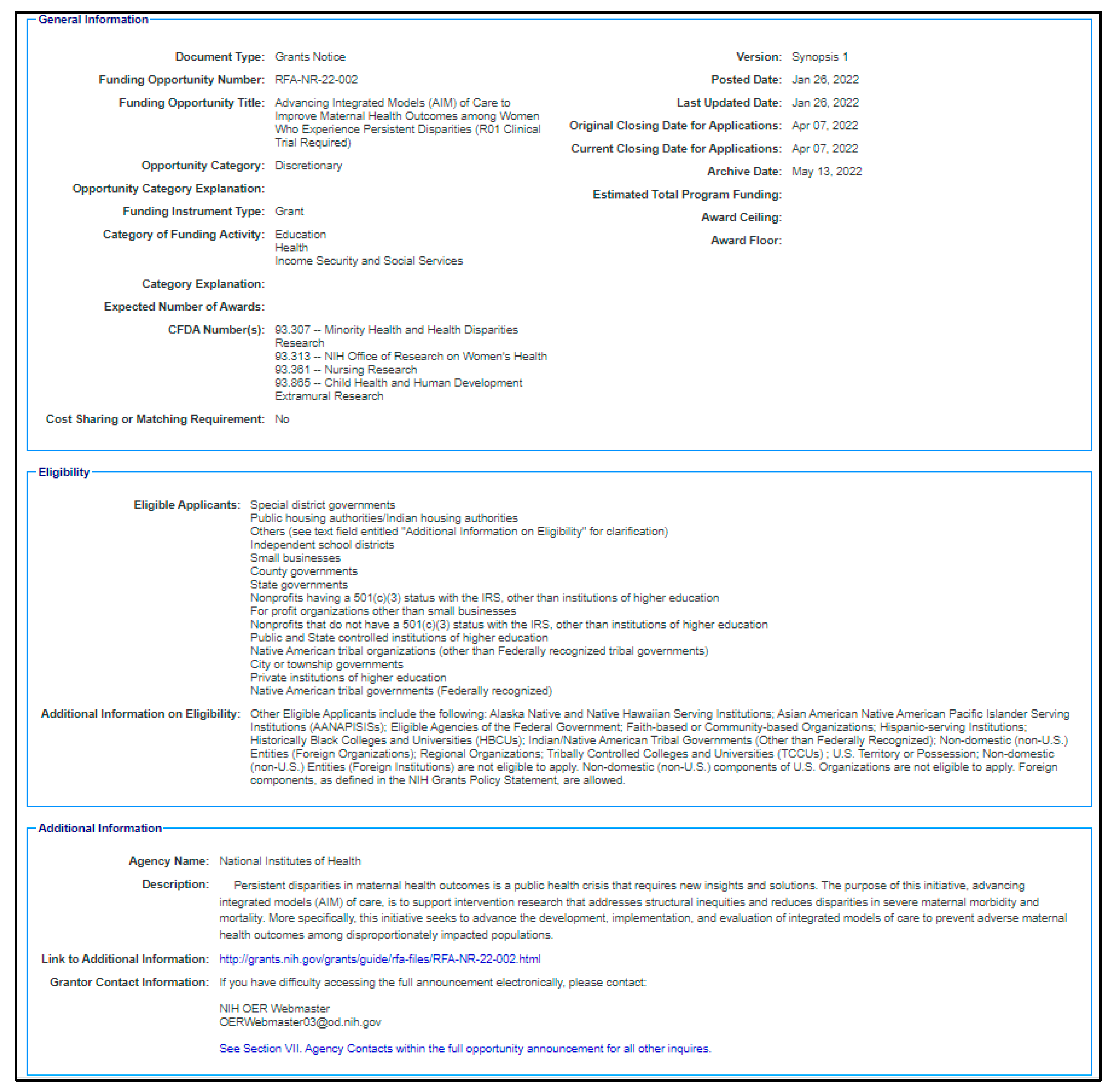
2. Data
3. Methods
3.1. Our Virtual Research Assistant (VRA) Architecture
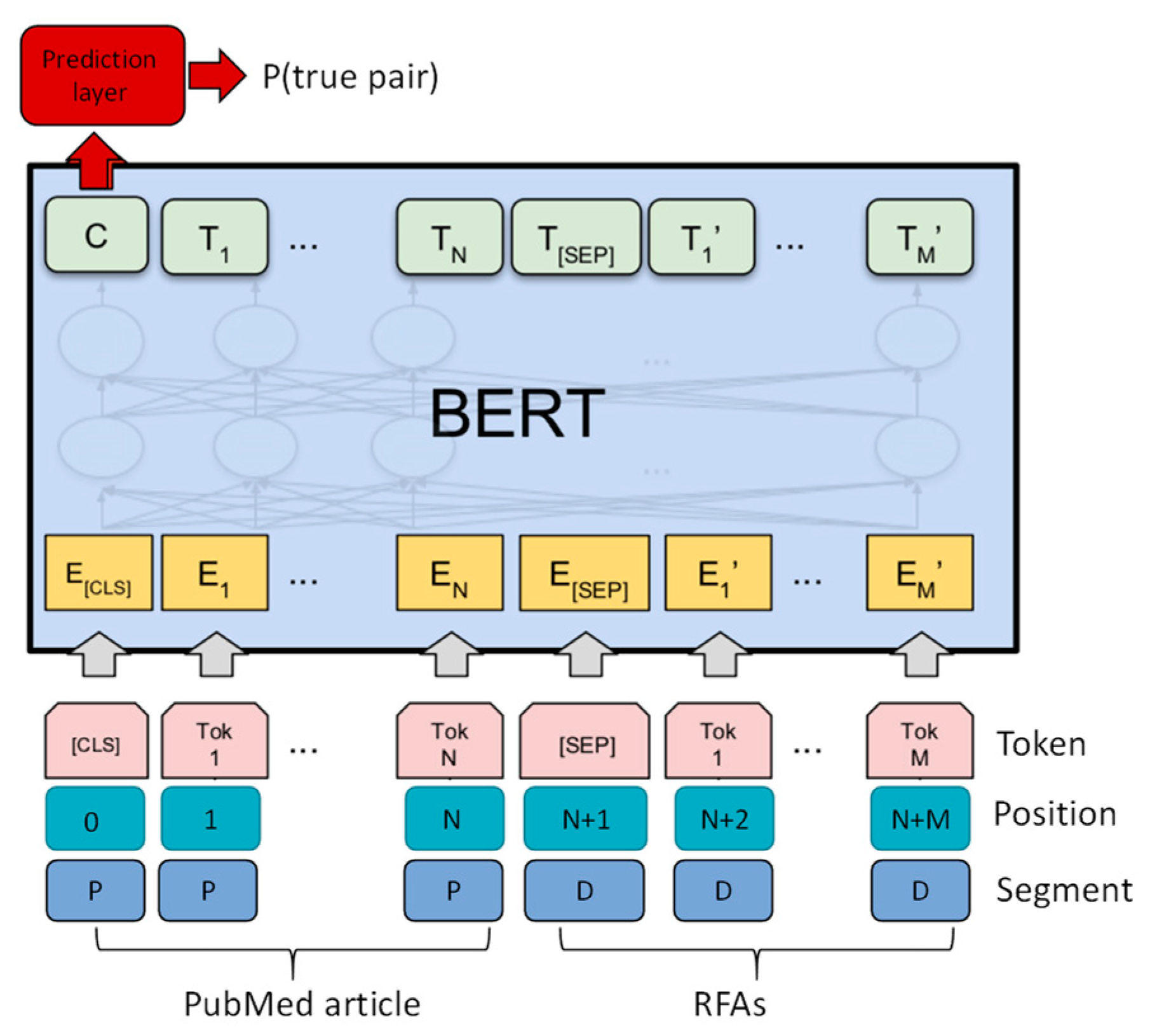
3.1.1. Grant Recommender: BERT-Based
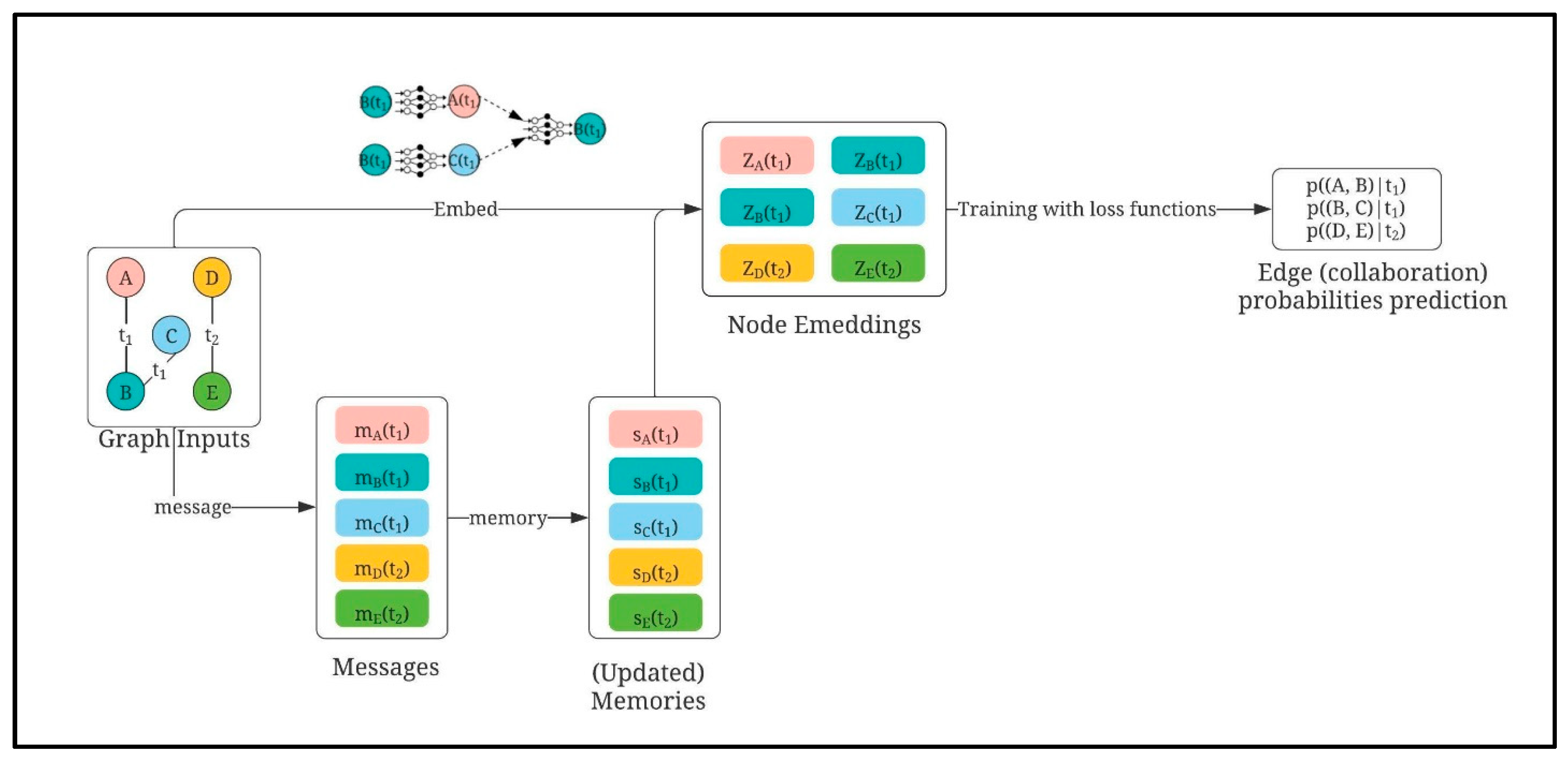
3.1.2. Collaborator Recommender: TGN-Based
3.2. Uncertainty Quantification Methods
3.2.1. Monte Carlo (MC) Dropout
3.2.2. Ensemble
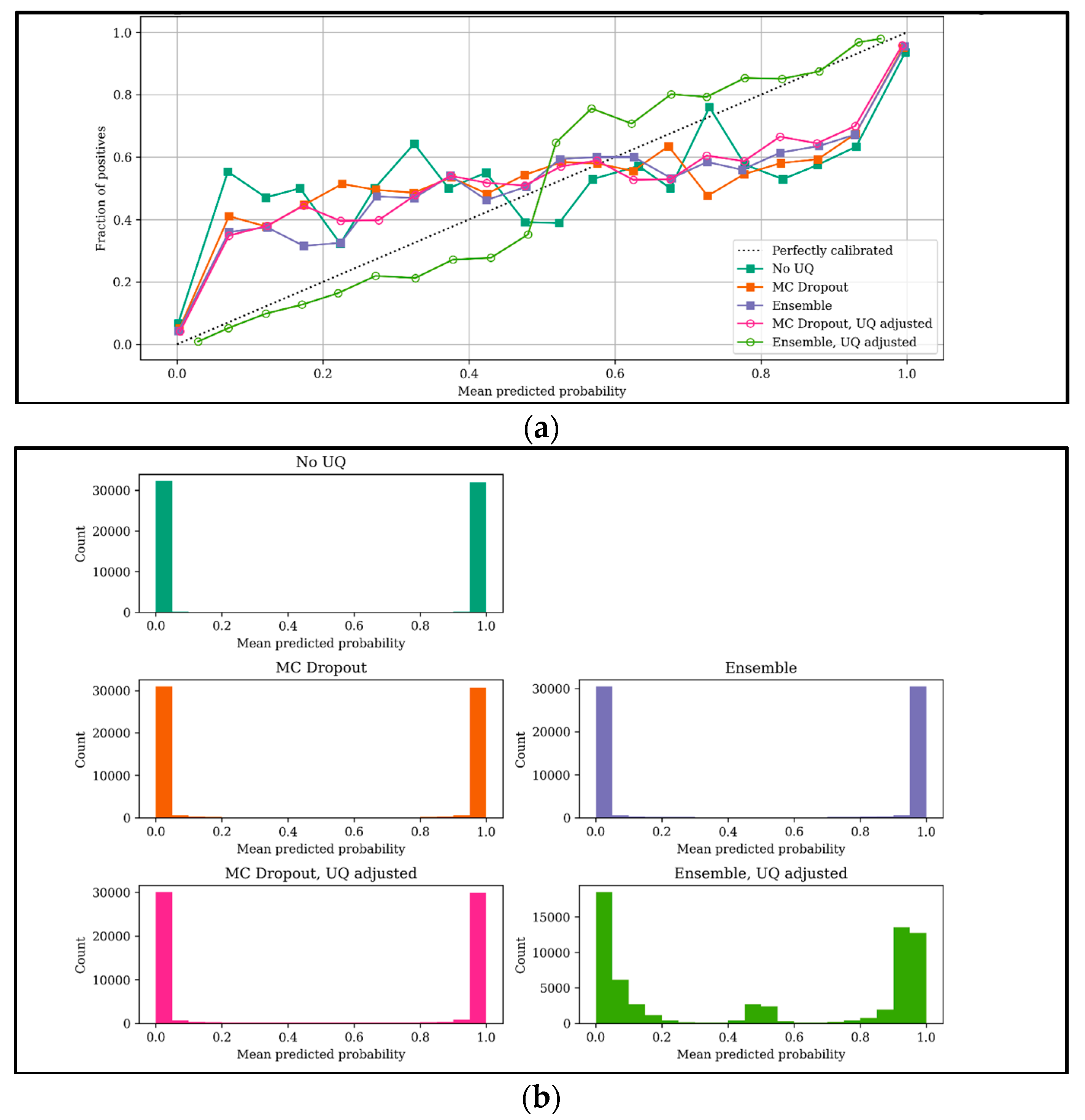
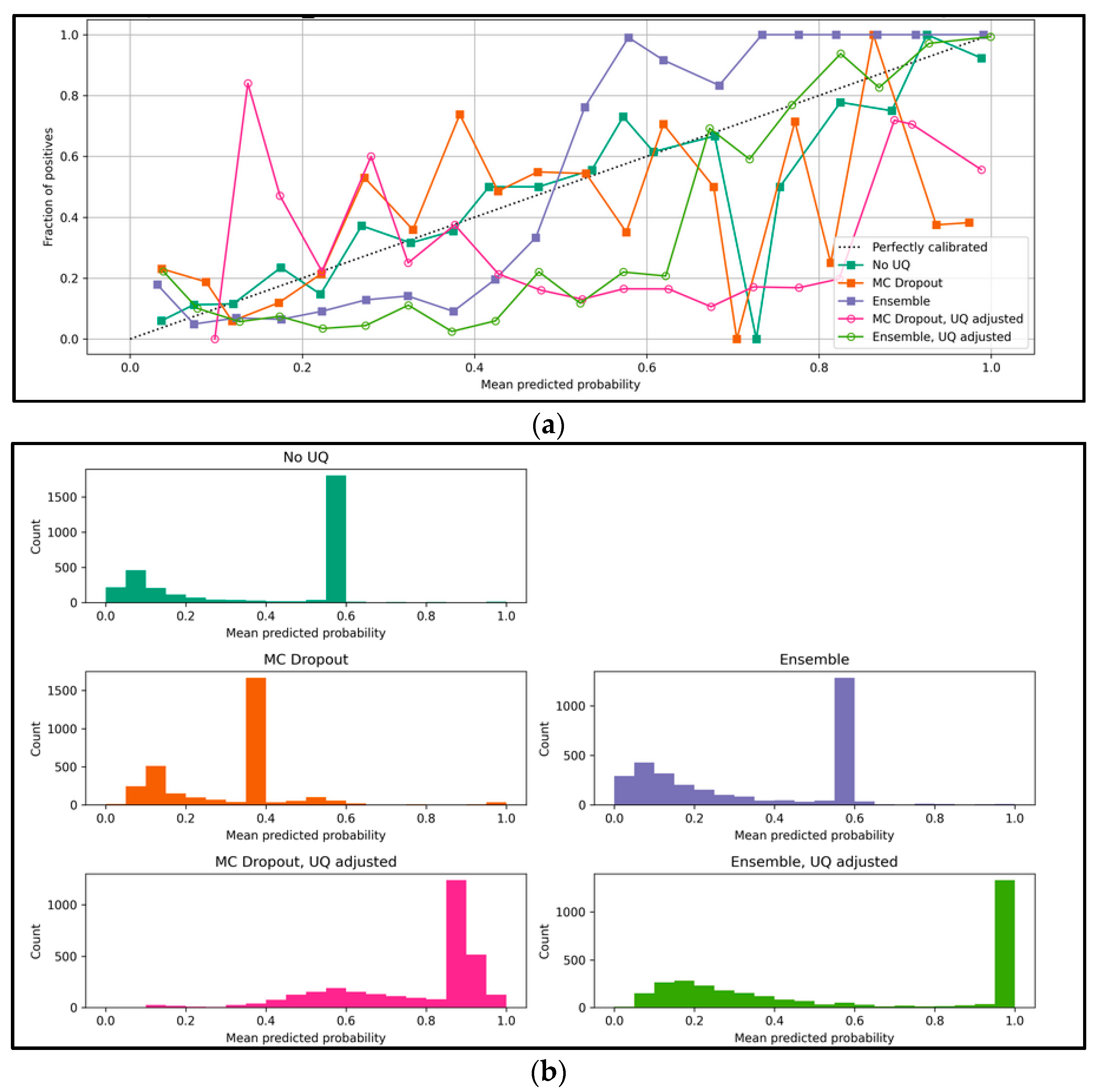
3.3. Proposed UQ Adjusted Results
3.4. Evaluation Metrics
- AUC: A receiver operating characteristic curve, or ROC curve, is a graphical plot that illustrates the diagnostic ability of a binary classifier system as its discrimination threshold is varied. The AUC is the area under a ROC curve, which provides an aggregated measure of performance across all possible classification thresholds [37].
- Average precision (AP): This summarizes precision-recall curve as the weighted mean of precisions achieved at each threshold, with the increase in recall from the previous threshold used as the weight.
- Mean reciprocal rank (MRR): The reciprocal rank (RR) measures the reciprocal of the rank at which the first relevant document was retrieved. RR is 1 if the relevant document was retrieved at rank 1, RR is 0.5 if the document is retrieved at rank 2, and so on. When we average the retrieved items across the queries , the measure is called the MRR.
- Recall@1 (R@1): At the k-th retrieved item, this metric measures the proportion of relevant items that are retrieved. We evaluated recall@1.
- Precision@1 (P@1): At the k-th retrieved item, this metric measures the proportion of the retrieved items that are relevant. In our case, we are interested in precision@1.
- Expected calibration error (ECE): Calibration measures the discrepancy between long-run frequencies and subjective forecasts [38,39]. To make use of uncertainty quantification methods, we need to make sure the (binary) classifier estimates are as close to perfect calibration as possible, meaning that if we discretize our model predictions in interval bins, then we expect that the fraction of positives and predicted probabilities of each bin should agree. Mathematically, let be the set of samples whose predicted probabilities fall into interval ; the fraction of positives for is:where is the true class label for sample . The predicted probability within bin is:where is the predicted probability for sample
4. Results
4.1. Grant Recommender: BERT-Based Results
4.2. Collaborator Recommender: TGN-Based Results
5. Conclusions and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, H.; Kim, B.; Guan, M.; Gupta, M. To Trust or Not to Trust a Classifier. In Advances in Neural Information Processing Systems, Montréal, Canada; Curran Associates, Inc.: Red Hook, NY, USA, 2018; Volume 31, pp. 5546–5557. Available online: https://papers.nips.cc/paper/2018/hash/7180cffd6a8e829dacfc2a31b3f72ece-Abstract.htm (accessed on 20 May 2022).
- Environmental Protection Agency (EPA). Uncertainty and Variability: The Recurring and Recalcitrant Elements of Risk Assessment; National Academies Press (US): Washington, DC, USA, 2009. Available online: https://www.ncbi.nlm.nih.gov/books/NBK214636/ (accessed on 20 May 2022).
- Gawlikowski, J.; Tassi, C.R.N.; Ali, M.; Lee, J.; Humt, M.; Feng, J.; Kruspe, A.; Triebel, R.; Jung, P.; Roscher, R.; et al. A Survey of Uncertainty in Deep Neural Networks. arXiv 2021, arXiv:2107.03342. [Google Scholar]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamzadeh, M.; Fieguth, P.; Cao, X.; Khosravi, A.; Acharya, U.R.; et al. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Loquercio, A.; Segu, M.; Scaramuzza, D. A General Framework for Uncertainty Estimation in Deep Learning. IEEE Robot. Autom. Lett. 2020, 5, 3153–3160. [Google Scholar] [CrossRef]
- Hüllermeier, E.; Waegeman, W. Aleatoric and Epistemic Uncertainty in Machine Learning: An Introduction to Concepts and Methods. Mach. Learn. 2021, 110, 457–506. [Google Scholar] [CrossRef]
- Guo, C.; Pleiss, G.; Sun, Y.; Weinberger, K.Q. On Calibration of Modern Neural Networks. arXiv 2017, arXiv:1706.04599. [Google Scholar]
- Yao, J.; Pan, W.; Ghosh, S.; Doshi-Velez, F. Quality of Uncertainty Quantification for Bayesian Neural Network Inference. arXiv 2019, arXiv:1906.09686. [Google Scholar]
- Ryu, S.; Kwon, Y.; Kim, W.Y. A Bayesian graph convolutional network for reliable prediction of molecular properties with uncertainty quantification. Chem. Sci. 2019, 10, 8438–8446. [Google Scholar] [CrossRef]
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. arXiv 2016, arXiv:1506.02142. [Google Scholar]
- Hasanzadeh, A.; Hajiramezanali, E.; Boluki, S.; Zhou, M.; Duffield, N.; Narayanan, K.; Qian, X. Bayesian Graph Neural Networks with Adaptive Connection Sampling. arXiv 2020, arXiv:2006.04064. [Google Scholar]
- Rong, Y.; Huang, W.; Xu, T.; Huang, J. DropEdge: Towards Deep Graph Convolutional Networks on Node Classification. arXiv 2020, arXiv:1907.10903. [Google Scholar]
- Mobiny, A.; Nguyen, H.V.; Moulik, S.; Garg, N.; Wu, C.C. DropConnect Is Effective in Modeling Uncertainty of Bayesian Deep Networks. arXiv 2019, arXiv:1906.04569. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Salimans, T.; Welling, M. Variational Dropout and the Local Reparameterization Trick. In Advances in Neural Information Processing Systems 28 (NIPS 2015); MIT Press: Cambridge, MA, USA, 2015; Volume 28, pp. 2575–2583. Available online: https://dl.acm.org/doi/10.5555/2969442.2969527 (accessed on 21 May 2022).
- Lakshminarayanan, B.; Pritzel, A.; Blundell, C. Simple and Scalable Predictive Uncertainty Estimation using Deep Ensembles. Adv. Neural Inf. Process. Syst. 2017, 30. Available online: https://proceedings.neurips.cc/paper/2017/hash/9ef2ed4b7fd2c810847ffa5fa85bce38-Abstract.html (accessed on 3 January 2022).
- Valdenegro-Toro, M. Deep Sub-Ensembles for Fast Uncertainty Estimation in Image Classification. arXiv 2019, arXiv:1910.08168. [Google Scholar]
- Wen, Y.; Tran, D.; Ba, J. BatchEnsemble: An Alternative Approach to Efficient Ensemble and Lifelong Learning, Presented at the International Conference on Learning Representations. 2019. Available online: https://openreview.net/forum?id=Sklf1yrYDr (accessed on 21 May 2022).
- Kennamer, N.; Ihler, A.; Kirkby, D. Empirical Study of MC-Dropout in Various Astronomical Observing Conditions. In CVPR Workshops, Long Beach, California; 2019; pp. 17–20. Available online: https://openaccess.thecvf.com/content_CVPRW_2019/papers/Uncertainty%20and%20Robustness%20in%20Deep%20Visual%20Learning/Kennamer_Empirical_Study_of_MC-Dropout_in_Various_Astronomical_Observing_Conditions_CVPRW_2019_paper.pdf (accessed on 20 May 2022).
- Ng, M.; Guo, F.; Biswas, L.; Petersen, S.E.; Piechnik, S.K.; Neubauer, S.; Wright, G. Estimating Uncertainty in Neural Networks for Cardiac MRI Segmentation: A Benchmark Study. IEEE Trans. Biomed. Eng. 2022, 1–12. [Google Scholar] [CrossRef]
- Ovadia, Y.; Fertig, E.; Ren, J.; Nado, Z.; Sculley, D.; Nowozin, S.; Dillon, J.V.; Lakshminarayanan, B.; Snoek, J. Can You Trust Your Model’s Uncertainty? Evaluating Predictive Uncertainty Under Dataset Shift. arXiv 2019, arXiv:1906.02530. [Google Scholar]
- Fan, X.; Zhang, X.; Yu, X. (Bill) Uncertainty quantification of a deep learning model for failure rate prediction of water distribution networks. Reliab. Eng. Syst. Saf. 2023, 109088. [Google Scholar] [CrossRef]
- Zeldes, Y.; Theodorakis, S.; Solodnik, E.; Rotman, A.; Chamiel, G.; Friedman, D. Deep density networks and uncertainty in recommender systems. arXiv 2018, arXiv:1711.02487. [Google Scholar]
- Shelmanov, A.; Tsymbalov, E.; Puzyrev, D.; Fedyanin, K.; Panchenko, A.; Panov, M. How Certain is Your Transformer? In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, Online, 19–23 April 2021; Association for Computational Linguistics: Cedarville, OH, USA, 2021; pp. 1833–1840. [Google Scholar]
- Penha, G.; Hauff, C. On the Calibration and Uncertainty of Neural Learning to Rank Models for Conversational Search. In Proceedings of the 16th Conference of the European Chapter of the Association for Computational Linguistics: Main Volume, Online, 19–23 April 2021; Association for Computational Linguistics: Cedarville, OH, USA, 2021; pp. 160–170. [Google Scholar]
- Siddhant, A.; Lipton, Z.C. Deep Bayesian Active Learning for Natural Language Processing: Results of a Large-Scale Empirical Study. arXiv 2018, arXiv:1808.05697. [Google Scholar]
- Wang, A.; Singh, A.; Michael, J.; Hill, F.; Levy, O.; Bowman, S. GLUE: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding. In Proceedings of the 2018 EMNLP Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP, Brussels, Belgium, 31 October–1 November 2018; Association for Computational Linguistics: Cedarville, OH, USA, 2018; pp. 353–355. [Google Scholar]
- Zhu, J.; Patra, B.; Yaseen, A. Recommender systems of scholarly papers using public datasets. AMIA Jt. Summits Transl. Sci. Proc. 2021, 2021, 672–679. [Google Scholar]
- Zhu, J.; Patra, B.G.; Wu, H.; Yaseen, A. A novel NIH research grant recommender using BERT. PLoS ONE 2023, 18, e0278636. [Google Scholar] [CrossRef]
- Zhu, J.; Yaseen, A. A Recommender for Research Collaborators Using Graph Neural Networks. Front. Artif. Intell. 2022, 5. Available online: https://www.frontiersin.org/articles/10.3389/frai.2022.881704 (accessed on 2 October 2022). [CrossRef]
- Zhu, J.; Wu, H.; Yaseen, A. Sensitivity Analysis of a BERT-based scholarly recommendation system. In Proceedings of the International FLAIRS Conference Proceedings, Hutchinson Island, Jensen Beach, FL, USA, 15–18 May 2022; Volume 35. [Google Scholar]
- Rossi, E.; Chamberlain, B.; Frasca, F.; Eynard, D.; Monti, F.; Bronstein, M. Temporal Graph Networks for Deep Learning on Dynamic Graphs. arXiv 2020, arXiv:2006.10637. [Google Scholar]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; p. 19. [Google Scholar]
- Redford, M. Neal Monte Carlo Implementation. In Bayesian Learning for Neural Networks; Lecture Notes in Statistics; Springer: New York, NY, USA, 1996; Volume 118. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zar, J.H. Biostatistical Analysis; Prentice Hall: Upper Saddle River, NJ, USA, 1999; ISBN 978-0-13-081542-2. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Dawid, A.P. The Well-Calibrated Bayesian. J. Am. Stat. Assoc. 1982, 77, 605–610. [Google Scholar] [CrossRef]
- DeGroot, M.H.; Fienberg, S.E. The Comparison and Evaluation of Forecasters. J. R. Stat. Soc. Ser. D (Stat.) 1983, 32, 12–22. [Google Scholar] [CrossRef]
- Naeini, M.P.; Cooper, G.F.; Hauskrecht, M. Obtaining Well Calibrated Probabilities Using Bayesian Binning. Proc. Conf. AAAI Artif. Intell. 2015, 2015, 2901–2907. [Google Scholar]
- Niculescu-Mizil, A.; Caruana, R. Predicting good probabilities with supervised learning. In Proceedings of the 22nd International Conference on Machine Learning—ICML ’05, Bonn, Germany, 7–11 August 2005; ACM Press: New York, NY, USA; pp. 625–632. [Google Scholar] [CrossRef]

| Splits | # of Unique Publications | # of Records |
|---|---|---|
| Training (7) | 135,766 | 216,766 |
| Validation (1) | 17,456 | 28,056 |
| Testing (2) | 40,730 | 65,104 |
| Splits | # of Links | # of Nodes |
|---|---|---|
| Training (7) | 9589 | 9358 |
| Validation (1.5) | 1754 | 1796 |
| Testing (1.5) | 1559 | 1574 |
| Recommended | Not Recommended | |
|---|---|---|
| Relevant | True positives (TP) | False negatives (FN) |
| Not relevant | False positives (FP) | True negatives (TN) |
| Models | AUC | AP | MRR | R@1 | P@1 | ECE |
|---|---|---|---|---|---|---|
| Regular | 0.977 | 0.975 | 0.933 | 0.810 | 0.871 | 0.073 |
| MC dropout | 0.978 | 0.977 | 0.947 | 0.816 | 0.882 | 0.067 |
| MC dropout, UQ adjusted | 0.979 | 0.978 | 0.939 | 0.816 | 0.882 | 0.064 |
| Ensemble | 0.981 | 0.980 | 0.941 | 0.818 | 0.884 | 0.067 |
| Ensemble, UQ adjusted | 0.975 | 0.967 | 0.938 | 0.815 | 0.879 | 0.030 |
| Models | AUC | AP | ECE |
|---|---|---|---|
| Regular | 0.792 | 0.727 | 0.165 |
| MC dropout | 0.796 | 0.711 | 0.184 |
| MC dropout, UQ adjusted | 0.817 | 0.744 | 0.162 |
| Ensemble | 0.938 | 0.960 | 0.282 |
| Ensemble, UQ adjusted | 0.960 | 0.972 | 0.154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Novelo, L.L.; Yaseen, A. Incorporating Uncertainty Quantification for the Performance Improvement of Academic Recommenders. Knowledge 2023, 3, 293-306. https://doi.org/10.3390/knowledge3030020
Zhu J, Novelo LL, Yaseen A. Incorporating Uncertainty Quantification for the Performance Improvement of Academic Recommenders. Knowledge. 2023; 3(3):293-306. https://doi.org/10.3390/knowledge3030020
Chicago/Turabian StyleZhu, Jie, Luis Leon Novelo, and Ashraf Yaseen. 2023. "Incorporating Uncertainty Quantification for the Performance Improvement of Academic Recommenders" Knowledge 3, no. 3: 293-306. https://doi.org/10.3390/knowledge3030020
APA StyleZhu, J., Novelo, L. L., & Yaseen, A. (2023). Incorporating Uncertainty Quantification for the Performance Improvement of Academic Recommenders. Knowledge, 3(3), 293-306. https://doi.org/10.3390/knowledge3030020






