A Thorough Insight into the Biofilm Properties of Potential Fish Pathogen Tenacibaculum discolor Strain FMCC B487
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Biofilm Assays
2.2.1. Effect of Environmental Factors on Biofilm Formation
2.2.2. Effect of Monosaccharides on the Growth and Biofilm Formation of T. discolor Strain FMCC B487
2.3. Chemical Composition of Biofilm and EPS
2.3.1. Recovery of Biofilm Fractions
2.3.2. Extracellular Polymeric Substances Production in Medium Supplemented with Carbohydrate
2.3.3. Preparation of Extracts
2.3.4. Chemical and Electrophoretic Assays and Molecular Weight Determination
2.3.5. Characterization of Carbohydrates
2.3.6. Molecular Weight Determination
3. Results
3.1. Biofilm Assays
3.1.1. Effects of Environmental Factors on Biofilm Formation of T. discolor Strain FMCC B487
3.1.2. Effects of Monosaccharides on Growth and Biofilm Development of T. discolor Strain FMCC B487
3.2. Chemical Composition of Planktonic, Adhered Biofilm and EPS Extracts
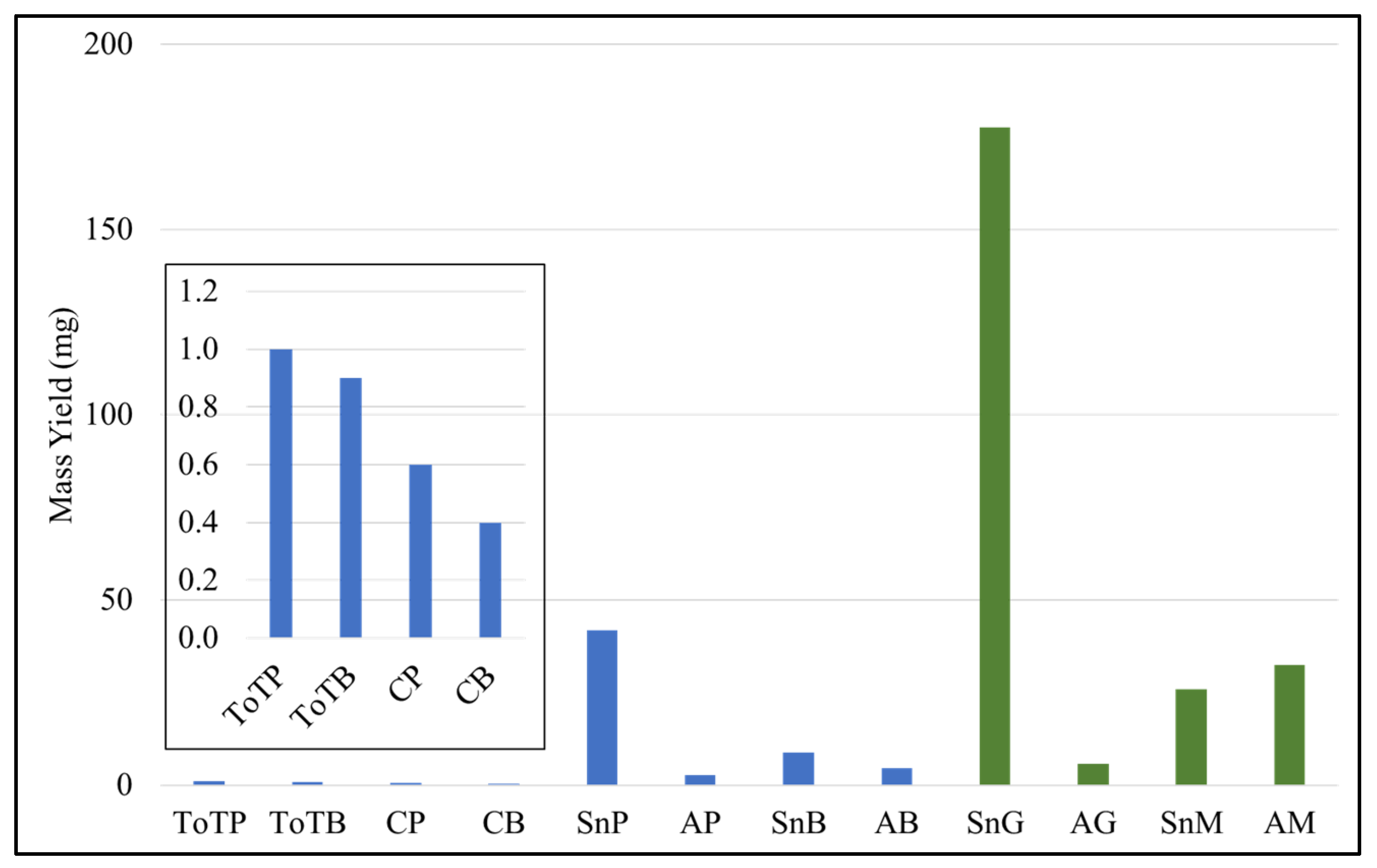
3.2.1. Yield of Cell Extracts
3.2.2. Chemical Composition
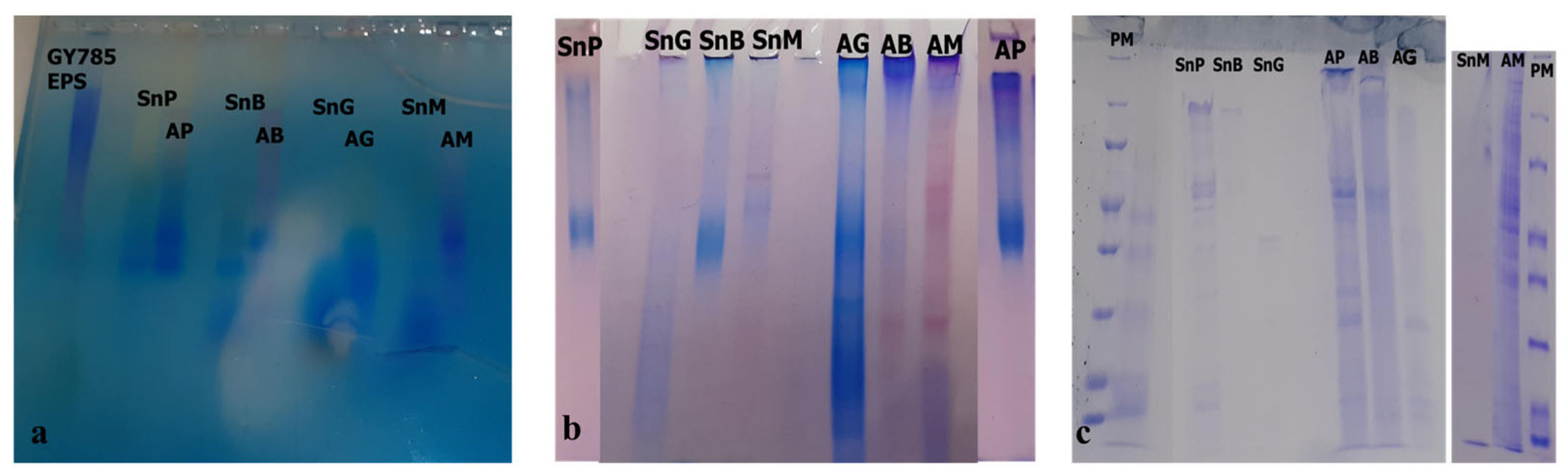
3.2.3. Electrophoretic Analyses
3.2.4. Osidic Composition
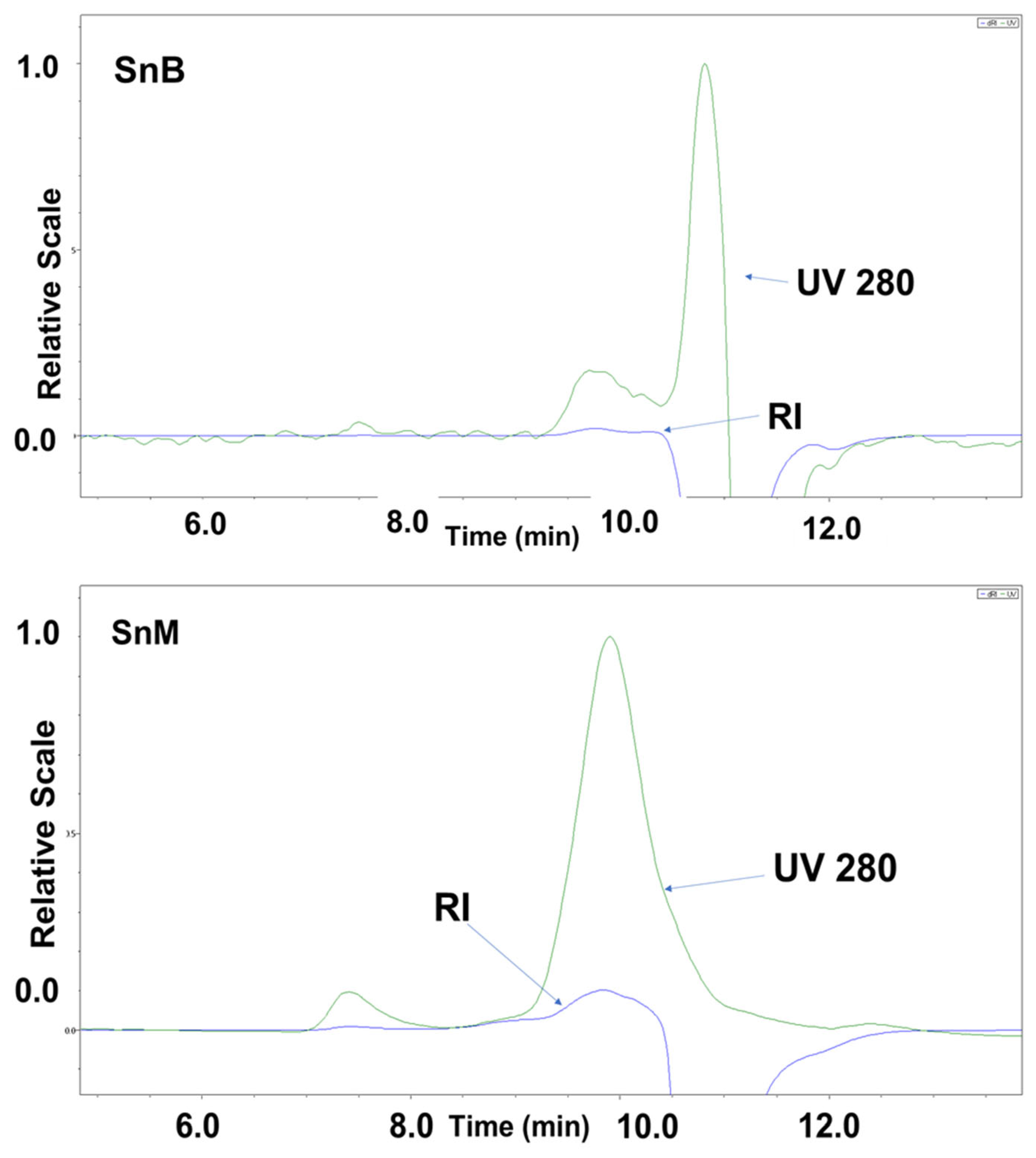
3.2.5. Molecular Weight Analysis
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. In Brief to the State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Jahid, I.K.; Ha, S.D. Microbial biofilms in seafood: A food-hygiene challenge. Food Microbiol. 2015, 49, 41–55. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Irgang, R.; Sandoval, C.; Moreno-Lira, P.; Houel, A.; Duchaud, E.; Poblete-Morales, M.; Ilardi, P. Isolation, Characterization and Virulence Potential of Tenacibaculum dicentrarchi in Salmonid Cultures in Chile. Transbound. Emerg. Dis. 2016, 63, 121–126. [Google Scholar] [CrossRef]
- Fernández-Álvarez, C.; Santos, Y. Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biotechnol. 2018, 102, 9973–9989. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Collarte, C.; Saldarriaga-Córdoba, M.; Irgang, R. New salmonid hosts for Tenacibaculum species: Expansion of tenacibaculosis in Chilean aquaculture. J. Fish Dis. 2020, 43, 1077–1085. [Google Scholar] [CrossRef]
- Irgang, R.; González-Luna, R.; Gutiérrez, J.; Poblete-Morales, M.; Rojas, V.; Tapia-Cammas, D.; Avendaño-Herrera, R. First identification and characterization of Tenacibaculum dicentrarchi isolated from Chilean red conger eel (Genypterus chilensis, Guichenot 1848). J. Fish Dis. 2017, 40, 1915–1920. [Google Scholar] [CrossRef]
- Burioli, E.A.V.; Varello, K.; Trancart, S.; Bozzetta, E.; Gorla, A.; Prearo, M.; Houssin, M. First description of a mortality event in adult Pacific oysters in Italy associated with infection by a Tenacibaculum soleae strain. J. Fish Dis. 2018, 41, 215–221. [Google Scholar] [CrossRef]
- Sakatoku, A.; Fujimura, T.; Ito, M.; Takashima, S.; Isshiki, T. Newly isolated bacterium Tenacibaculum sp. strain Pbs-1 from diseased pearl oysters is associated with black-spot shell disease. Aquaculture 2018, 493, 61–67. [Google Scholar] [CrossRef]
- Suzuki, M.; Nakagawa, Y.; Harayama, S.; Yamamoto, S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: Proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Ten. Int. J. Syst. Evol. Microbiol. 2001, 51, 1639–1652. [Google Scholar] [CrossRef]
- Khalil, R.H.; Diab, A.M.; Shakweer, M.S.; Ghetas, H.A.; Khallaf, M.M.; Omar, A.A. New perspective to control of tenacibaculosis in sea bass Dicentrarchus labrax L. Aquac. Res. 2018, 49, 2357–2365. [Google Scholar] [CrossRef]
- Kolygas, M.N.; Gourzioti, E.; Vatsos, I.N.; Athanassopoulou, F. Identification of Tenacibaculum maritimum strains from marine farmed fish in Greece. Vet. Rec. 2018, 170, 623. [Google Scholar] [CrossRef]
- Muñoz-Atienza, E.; Araújo, C.; Magadán, S.; Hernández, P.E.; Herranz, C.; Santos, Y.; Cintas, L.M. In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus L.) farming. Fish Shellfish Immunol. 2014, 41, 570–580. [Google Scholar] [CrossRef]
- Småge, S.B.; Frisch, K.; Brevik, Ø.J.; Watanabe, K.; Nylund, A. First isolation, identification and characterisation of Tenacibaculum maritimum in Norway, isolated from diseased farmed sea lice cleaner fish Cyclopterus lumpus L. Aquaculture 2016, 464, 178–184. [Google Scholar] [CrossRef]
- van Gelderen, R.; Carson, J.; Nowak, B. Effect of extracellular products of Tenacibaculum maritimum in Atlantic salmon, Salmo salar L. J. Fish Dis. 2009, 32, 727–731. [Google Scholar] [CrossRef]
- Watanabe, K.I.; Nishioka, T. Antibacterial effect of chemical reagents against Tenacibaculum maritimum. Fish Pathol. 2010, 45, 66–68. [Google Scholar] [CrossRef]
- Wynne, J.W.; Thakur, K.K.; Slinger, J.; Samsing, F.; Milligan, B.; Powell, J.F.F.; McKinnon, A.; Nekouei, O.; New, D.; Cichmond, Z.; et al. Microbiome Profiling Reveals a Microbial Dysbiosis During a Natural Outbreak of Tenacibaculosis (Yellow Mouth) in Atlantic Salmon. Front. Microbiol. 2020, 11, 586387. [Google Scholar] [CrossRef]
- Nowlan, J.P.; Lumsden, J.S.; Russell, S. Advancements in characterizing Tenacibaculum infections in Canada. Pathogens 2020, 9, 1029. [Google Scholar] [CrossRef]
- Levipan, H.A.; Tapia-Cammas, D.; Molina, V.; Irgang, R.; Toranzo, A.E.; Magariños, B.; Avendaño-Herrera, R. Biofilm development and cell viability: An undervalued mechanism in the persistence of the fish pathogen Tenacibaculum maritimum. Aquaculture 2019, 511, 734267. [Google Scholar] [CrossRef]
- Levipan, H.A.; Irgang, R.; Tapia-Cammas, D.; Avendaño-Herrera, R. A high-throughput analysis of biofilm formation by the fish pathogen Tenacibaculum dicentrarchi. J. Fish Dis. 2019, 42, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Feng, D.; Song, G.L.; Su, P.; Zheng, D. The effect of a biofilm-forming bacterium Tenacibaculum mesophilum D-6 on the passive film of stainless steel in the marine environment. Sci. Total Environ. 2022, 815, 152909. [Google Scholar] [CrossRef] [PubMed]
- Kee, H.L.; Mikheyeva, I.V.; Mickol, R.L.; Dawson, S.C.; Newman, D.K.; Leadbetter, J.R. Draft Genome Sequence of the Iridescent Marine Bacterium Tenacibaculum discolor Strain IMLK18. Microbiol. Resour. Announc. 2019, 8, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.; Cleary, D.F.R.; Pires, A.C.C.; Rodrigues, A.M.; Quintino, V.; Calado, R.; Gomes, N.C.M. Molecular analysis of bacterial communities and detection of potential pathogens in a recirculating aquaculture system for Scophthalmus maximus and Solea senegalensis. PLoS ONE 2013, 8, e0080847. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro-Vidal, M.; Riaza, A.; Santos, Y. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int. J. Syst. Evol. Microbiol. 2008, 58, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Schoina, E.; Doulgeraki, A.I.; Miliou, H.; Nychas, G.-J.E. Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System. Aquac. J. 2022, 2, 164–179. [Google Scholar] [CrossRef]
- Wang, L.; Linares-Otoya, V.; Liu, Y.; Mettal, U.; Marner, M.; Armas-Mantilla, L.; Willbold, S.; Kurtán, T.; Linares-Otoya, L.; Schäberle, T.F. Discovery and Biosynthesis of Antimicrobial Phenethylamine Alkaloids from the Marine Flavobacterium Tenacibaculum discolor sv11. J. Nat. Prod. 2022, 85, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Marner, M.; Mettal, U.; Liu, Y.; Schäberle, T.F. Seven New Alkaloids Isolated from Marine Flavobacterium Tenacibaculum discolor sv11. Mar. Drugs 2022, 20, 620. [Google Scholar] [CrossRef] [PubMed]
- Lianou, A.; Koutsoumanis, K.P. Strain variability of the biofilm-forming ability of Salmonella enterica under various environmental conditions. Int. J. Food Microbiol. 2012, 160, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Ladero, V.; del Rio, B.; Redruello, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. Biofilm-Forming Capacity in Biogenic Amine-Producing Bacteria Isolated from Dairy Products. Front. Microbiol. 2016, 7, 591. [Google Scholar] [CrossRef]
- Yuan, L.; Burmølle, M.; Sadiq, F.A.; Wang, N.; He, G. Interspecies variation in biofilm-forming capacity of psychrotrophic bacterial isolates from Chinese raw milk. Food Control 2018, 91, 47–57. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Passerini, D.; Fécamp, F.; Marchand, L.; Kolypczuk, L.; Bonnetot, S.; Sinquin, C.; Verrez-Bagnis, V.; Hervio-Heath, D.; Colliec-Jouault, S.; Delbarre-Ladrat, C. Characterization of Biofilm Extracts from Two Marine Bacteria. Appl. Sci. 2019, 9, 4971. [Google Scholar] [CrossRef]
- Raguénès, G.H.C.; Peres, A.; Ruimy, R.; Pignet, P.; Christen, R.; Loaec, M.; Rougeaux, H.; Barbier, G.; Guezennec, J.G. Alteromonas infernus sp. Nov., a new polysaccharideproducing bacterium isolated from a deep-sea hydrothermal vent. J. Appl. Microbiol. 1997, 82, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Zykwinska, A.; Le Berre, L.T.; Sinquin, C.; Ropartz, D.; Rogniaux, H.; Colliec-Jouault, S.; Delbarre-Ladrat, C. Enzymatic depolymerization of the GY785 exopolysaccharide produced by the deep-sea hydrothermal bacterium Alteromonas infernus: Structural study and enzyme activity assessment. Carbohydr. Polym. 2018, 188, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Kamerling, J.P.; Gerwig, G.J.; Vliegenthart, J.F.G.; Clamp, J.R. Characterization by gas-liquid chromatography mass spectrometry of permethylsilylglycosides obtained in the methanolysis of glycoproteins and glycolipids. Biochem. J. 1975, 151, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Montreuil, J.; Bouquelet, S.; Debray, H.; Fournet, B.; Spik, G.; Strecker, G. Glycoproteins. In Carbohydrate Analysis: A Practical Approach, 2nd ed.; Chaplin, M.F., Kennedy, J.K., Eds.; IRL Press: Oxford, UK, 1986; pp. 143–204. [Google Scholar]
- Almeida, D.B.; Magalhães, C.; Sousa, Z.; Borges, M.T.; Silva, E.; Blanquet, I.; Mucha, A.P. Microbial Community Dynamics in a Hatchery Recirculating Aquaculture System (RAS) of Sole (Solea senegalensis). Aquaculture 2021, 539, 736592. [Google Scholar] [CrossRef]
- Bourne, D.G.; Høj, L.; Webster, N.S.; Swan, J.; Hall, M.R. Biofilm Development within a Larval Rearing Tank of the Tropical Rock Lobster, Panulirus ornatus. Aquaculture 2006, 260, 27–38. [Google Scholar] [CrossRef]
- Fernández-Álvarez, C.; Torres-Corral, Y.; Santos, Y. Comparison of serological and molecular typing methods for epidemiological investigation of Tenacibaculum species pathogenic for fish. Appl. Microbiol. Biotechnol. 2018, 102, 2779–2789. [Google Scholar] [CrossRef]
- Jahid, I.K.; Mizan, M.F.R.; Ha, A.J.; Ha, S.D. Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 2015, 49, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Shikuma, N.J.; Yildiz, F.H. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J. Bacteriol. 2009, 191, 4082–4096. [Google Scholar] [CrossRef]
- Li, X.; Sun, J.; Zhang, M.; Xue, X.; Wu, Q.; Yang, W.; Yin, Z.; Zhou, D.; Lu, R.; Zhang, Y. The Effect of Salinity on Biofilm Formation and c-di-GMP Production in Vibrio parahaemolyticus. Curr. Microbiol. 2022, 79, 25. [Google Scholar] [CrossRef]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Brown, M.R. The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 1991, 145, 79–99. [Google Scholar] [CrossRef]
- Whyte, J.N.C.; Nagata, W.D. Carbohydrate and fatty acid composition of the rotifer, Brachionus plicatilis, fed monospecific diets of yeast or phytoplankton. Aquaculture 1990, 89, 263–272. [Google Scholar] [CrossRef]
- Chai, Y.; Beauregard, P.B.; Vlamakis, H.; Losick, R.; Kolter, R. Galactose Metabolism Plays a Crucial Role in Biofilm Formation by B. subtilis. mBio 2012, 3, e00184-12. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Ashrafudoulla, M.; Sadekuzzaman, M.; Kang, I.; Ha, S.D. Effects of NaCl, glucose, and their combinations on biofilm formation on black tiger shrimp (Penaeus monodon) surfaces by Vibrio parahaemolyticus. Food Control 2018, 89, 203–209. [Google Scholar] [CrossRef]
- Piotrowski, M.; Wultańska, D.; Obuch-Woszczatyński, P.; Pituch, H. Fructooligosaccharides and mannose affect Clostridium difficile adhesion and biofilm formation in a concentration-dependent manner. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1975–1984. [Google Scholar] [CrossRef]
- Ryu, E.J.; An, S.J.; Sim, J.; Sim, J.; Lee, J.; Choi, B.K. Use of D-galactose to regulate biofilm growth of oral streptococci. Arch. Oral. Biol. 2020, 111, 104666. [Google Scholar] [CrossRef]
- Ryu, E.J.; Sim, J.; Sim, J.; Lee, J.; Choi, B.K. D-Galactose as an autoinducer 2 inhibitor to control the biofilm formation of periodontopathogens. J. Microbiol. 2016, 54, 632–637. [Google Scholar] [CrossRef]
- Nadesalingam, J.; Dodds, A.W.; Reid, K.B.M.; Palaniyar, N. Mannose-Binding Lectin Recognizes Peptidoglycan via the N -Acetyl Glucosamine Moiety, and Inhibits Ligand-Induced Proinflammatory Effect and Promotes Chemokine Production by Macrophages. J. Immunol. 2005, 175, 1785–1794. [Google Scholar] [CrossRef]
- Oyofo, B.A.; Droleskey, R.E.; Norman, J.O.; Mollenhauer, H.H.; Ziprin, R.L.; Corrier, D.E.; DeLoach, J.R. Inhibition by mannose of in vitro colonization of chicken small intestine by Salmonella typhimurium. Poult. Sci. 1989, 68, 1351–1356. [Google Scholar] [CrossRef]
- Poosarla, V.G.; Wood, T.L.; Zhu, L.; Miller, D.S.; Yin, B.; Wood, T.K. Dispersal and inhibitory roles of mannose, 2-deoxy-d-glucose and N-acetylgalactosaminidase on the biofilm of Desulfovibrio vulgaris. Environ. Microbiol. Rep. 2017, 9, 779–787. [Google Scholar] [CrossRef]
- Sharon, N.; Eshdat, Y.; Silverblatt, F.J.; Ofek, I. Bacterial adherence to cell surface sugars. Ciba Found. Symp. 1981, 80, 119–141. [Google Scholar] [CrossRef]
- de Alexandre Sebastião, F.; Pilarski, F.; Lemos, M.V.F. Composition of extracellular polymeric substances (EPS) produced by Flavobacterium columnare isolated from tropical fish in Brazil. Braz. J. Microbiol. 2013, 44, 861–864. [Google Scholar] [CrossRef]
- Finore, I.; Di Donato, P.; Mastascusa, V.; Nicolaus, B.; Poli, A. Fermentation technologies for the optimization of marine microbial exopolysaccharide production. Mar. Drugs 2014, 12, 3005–3024. [Google Scholar] [CrossRef]
- Escribano, M.P.; Balado, M.; Toranzo, A.E.; Lemos, M.L.; Magariños, B. The secretome of the fish pathogen Tenacibaculum maritimum includes soluble virulence-related proteins and outer membrane vesicles. Front. Cell. Infect. Microbiol. 2023, 13, 1197290. [Google Scholar] [CrossRef]
- Tonetti, M.; Sturla, L.; Bisso, A.; Zanardi, D.; Benatti, U.; De Flora, A. The metabolism of 6-deoxyhexoses in bacterial and animal cells. Biochimie 1998, 80, 923–931. [Google Scholar] [CrossRef]
- Webb, N.A.; Mulichak, A.M.; Lam, J.S.; Rocchetta, H.L.; Garavito, R.M. Crystal structure of a tetrameric GDP-d-mannose 4,6-dehydratase from a bacterial GDP-d-rhamnose biosynthetic pathway. Protein Sci. 2004, 13, 529–539. [Google Scholar] [CrossRef]
- Kisiela, D.I.; Magala, P.; Interlandi, G.; Carlucci, L.A.; Ramos, A.; Tchesnokova, V.; Basanta, B.; Yarov-Yaroyoy, V.; Avagyan, H.; Hovhannisyan, A.; et al. Toggle switch residues control allosteric transitions in bacterial adhesins by participating in a concerted repacking of the protein core. PLoS Pathog. 2021, 17, e1009440. [Google Scholar] [CrossRef]
- Ohashi, K.; Hataya, S.; Nakata, A.; Matsumoto, K.; Kato, N.; Sato, W.; Carlos-Shanley, C.; Beebe, E.T.; Currie, C.R.; Fox, B.G.; et al. Mannose- and mannobiose-specific responses of the insect-associated cellulolytic bacterium Streptomyces sp. strain SirexAA-E. Appl. Environ. Microbiol. 2021, 87, e02719-20. [Google Scholar] [CrossRef]




| Extract | Origin | Fraction | Extraction Procedure | Target |
|---|---|---|---|---|
| ToT-P | Biofilm | Total planktonic cell broth | Ethyl acetate | Total compounds |
| ToT-B | Biofilm | Total adhered cells | Ethyl acetate | |
| SnP | Biofilm | Supernatant of planktonic cell broth | Aqueous solution | Extracellular compounds |
| SnB | Biofilm | Supernatant of biofilm adhered cells | Aqueous solution | |
| SnG | Shake flask cultures supplemented with glucose 30% w/v | Supernatant (Glucose supplemented) | Aqueous solution | |
| SnM | Shake flask cultures supplemented with mannose 15% w/v | Supernatant (Mannose supplemented) | Aqueous solution | |
| CP | Biofilm | Planktonic cells | Ethyl acetate | Total compounds of cells |
| CB | Biofilm | Biofilm adhered cells | Ethyl acetate | |
| AP | Biofilm | Planktonic cells | NaOH | Cell membrane associated compounds |
| AB | Biofilm | Biofilm adhered cells | NaOH | |
| AG | Shake flask cultures supplemented with glucose 30% w/v | Cell pellet | NaOH | |
| AM | Shake flask cultures supplemented with mannose 15% w/v | Cell pellet | NaOH |
| Carbohydrate | Concentration (g/L) | Initial OD 600 | Max OD 600 | Lag Time (hours) | μ Max (h−1) |
|---|---|---|---|---|---|
| Control | - | 0.125 ± 0.028 | 0.908 ± 0.081 | 0.0 ± 0.0 | 0.05 ± 0.00 |
| Glc | 10 | 0.044 ± 0.024 | 0.926 ± 0.077 | 0.4 ± 0.6 | 0.05 ± 0.01 |
| 20 | 0.039 ± 0.023 | 0.929 ± 0.077 | 0.9 ± 1.3 * | 0.04 ± 0.01 | |
| 30 | 0.097 ± 0.033 | 0.870 ± 0.087 | 4.1 ± 1.9 * | 0.04 ± 0.01 | |
| Man | 10 | 0.061 ± 0.029 | 1.093 ± 0.084 * | 0.3 ± 0.5 * | 0.05 ± 0.01 |
| 20 | 0.050 ± 0.025 | 1.131 ± 0.078 * | 0.5 ± 0.7 * | 0.05 ± 0.01 | |
| 30 | 0.054 ± 0.039 | 1.106 ± 0.076 * | 1.7 ± 1.7 * | 0.04 ± 0.01 * | |
| Gal | 10 | 0.071 ± 0.028 | 1.099 ± 0.072 * | 0.1 ± 0.3 | 0.05 ± 0.01 |
| 20 | 0.054 ± 0.027 | 1.122 ± 0.059 * | 0.2 ± 0.7 | 0.05 ± 0.01 | |
| 30 | 0.062 ± 0.019 | 0.995 ± 0.083 * | 1.3 ± 1.4 * | 0.04 ± 0.01 * | |
| GlcN | 10 | 0.007 ± 0.017 | 1.228 ± 0.065 * | 0.3 ± 0.0 * | 0.07 ± 0.00 * |
| 20 | 0.037 ± 0.019 | 1.157 ± 0.073 * | 1.7 ± 0.8 * | 0.06 ± 0.01 * | |
| 30 | 0.090 ± 0.009 | 0.299 ± 0.092 * | 2.0 ± 4.6 | 0.01 ± 0.00 * | |
| Fru | 10 | 0.084 ± 0.020 | 0.956 ± 0.069 | 2.9 ± 1.2 * | 0.05 ± 0.01 |
| 20 | 0.156 ± 0.018 | 0.903 ± 0.061 | 17.7 ± 5.0 * | 0.05 ± 0.01 | |
| 30 | 0.103 ± 0.015 | 0.024 ± 0.041 * | 9.0 ± 10.8 * | 0.01 ± 0.02 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schoina, E.; Marchand, L.; Zykwinska, A.; Sinquin, C.; Leroi, F.; Delbarre-Ladrat, C.; Nychas, G.-J.E. A Thorough Insight into the Biofilm Properties of Potential Fish Pathogen Tenacibaculum discolor Strain FMCC B487. Aquac. J. 2024, 4, 76-91. https://doi.org/10.3390/aquacj4020006
Schoina E, Marchand L, Zykwinska A, Sinquin C, Leroi F, Delbarre-Ladrat C, Nychas G-JE. A Thorough Insight into the Biofilm Properties of Potential Fish Pathogen Tenacibaculum discolor Strain FMCC B487. Aquaculture Journal. 2024; 4(2):76-91. https://doi.org/10.3390/aquacj4020006
Chicago/Turabian StyleSchoina, Eirini, Laetitia Marchand, Agata Zykwinska, Corinne Sinquin, Françoise Leroi, Christine Delbarre-Ladrat, and George-John E. Nychas. 2024. "A Thorough Insight into the Biofilm Properties of Potential Fish Pathogen Tenacibaculum discolor Strain FMCC B487" Aquaculture Journal 4, no. 2: 76-91. https://doi.org/10.3390/aquacj4020006
APA StyleSchoina, E., Marchand, L., Zykwinska, A., Sinquin, C., Leroi, F., Delbarre-Ladrat, C., & Nychas, G.-J. E. (2024). A Thorough Insight into the Biofilm Properties of Potential Fish Pathogen Tenacibaculum discolor Strain FMCC B487. Aquaculture Journal, 4(2), 76-91. https://doi.org/10.3390/aquacj4020006





