Abstract
The specific identification of three major morphotypes of the tropical holopelagic Sargassum species causing massive strandings on the African and Caribbean coastlines was attempted by morphological characterisation as well as quantitative and qualitative analyses of several metabolites. Of the 25 morphological variables studied on 208 samples from the North Atlantic Ocean, 22 were used to establish a dichotomous identification key, allowing without any doubt the identification of each morphotype based on their morphological criteria alone. We also attempted to differentiate morphotypes using chemical fingerprintings (HR-MAS NMR) and markers by analysing pigment level and composition using High Pressure Liquid Chromatography, terpene profiles by Thin Layer Chromatography, phenolic compound levels by the Folin-Ciocalteu assay and structures by 2D Nuclear Magnetic Resonance spectroscopy, and fatty acid composition by Gas Chromatography. While pigment level and composition, terpene profiles, and phenolic contents were not discriminating, quantification of eight fatty acids enabled the differentiation of the three morphotypes. Furthermore, phlorotannin purification permitted their structural characterisation allowing discrimination between the three morphotypes. Our study highlights the potential of the free fatty acid profile and phlorotannin structure as good chemomarkers in order to discriminate between the three morphotypes of holopelagic Sargassum.
1. Introduction
Since 2011, massive biomasses of holopelagic brown macroalgae have been stranding on the South-American, Caribbean, and African shores [1,2]. This concerning phenomenon is growing every year and increasingly impacts more coastlines, with ecological, health, and economic consequences. Rafts responsible for these strandings are associated with two emblematic species of the Sargasso Sea in the tropical North Atlantic Ocean, Sargassum natans (Linnaeus) Gaillon and S. fluitans (Børgensen) Børgensen [3,4]. These species present morphologic and phylogenetic differences [5,6,7]. Offshore, the Sargassum rafts of variable dimensions are part of the marine ecosystem and constitute habitats, food sources, and means of migration for more than a hundred species, including several endemic species [8,9]. However, near the coasts, these rafts can have detrimental effects on benthic communities by blocking sunlight or causing anoxia in the environment [9]. They are also responsible for the introduction of exogenous species, threatening the coastal equilibrium [8,10]. Onshore, strandings have caused health issues induced by the release of hydrogen sulphide (H2S) by the bacteria decomposing the stranded biomasses, as well as economic problems, in particular for the tourism and fishing industries [8,11]. These Sargasso rafts and their consequences are thus major issues for the coastlines of the tropical North Atlantic Ocean.
In recent years, several oceanographic expeditions led in the tropical North Atlantic Ocean [12,13] revealed the presence, in the rafts, of three distinct morphotypes of holopelagic Sargassum (Figure 1). The morphological diversity of both species is already known, as Winge [14] and later Parr [15] described four types of S. natans (I, II, VIII, IX) and two types of S. fluitans (III, X) based on morphological characteristics [14,15]. The preliminary morphological identification of the morphotypes found during the expeditions associated morphotype 1 with S. natans VIII (Figure 1B,E), morphotype 2 with S. natans I (Figure 1C,F), and morphotype 3 with S. fluitans III (Figure 1D,G) according to Parr’s classification [15]. The morphological characteristics allowing this differentiation are overall appearance, smooth or thorny axes, aspects of fronds, and presence or absence of wings on the aerocysts. Sargassum natans VIII Parr is characterised by tough and smooth axes, long and wide fronds, and the presence, although rare, of wings on aerocysts. Sargassum natans I Parr is characterised by thin and smooth axes, long and narrow fronds, and the presence of wings on aerocysts. Sargassum fluitans III Parr is characterised by bushy and thorny axes, short and wide fronds, and the absence of wings on aerocysts [7]. Even though the morphological differentiation of the three morphotypes is indisputable, the molecular differentiation through the genome is challenging. Indeed, Dibner et al. [6] observed low but consistent genetic differences between the three morphotypes, possibly calling for taxonomic reclassification. Most recently, Siuda et al. [16] carried out a morphological and molecular study (6 different markers) on the 3 morphotypes, leading them to carry out a taxonomic revision and propose to change the nomination of the morphotypes of holopelagic Sargassum as follows: S. fluitans var. fluitans for S. fluitans III, S. natans var. natans for S. natans I, and finally S. natans var. wingei for S. natans VIII.
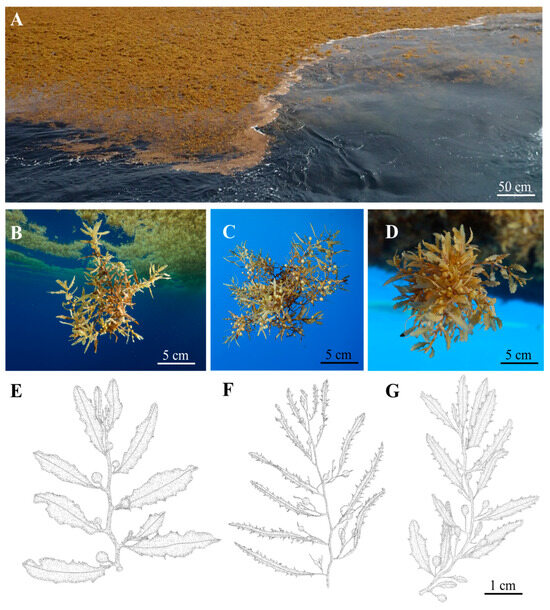
Figure 1.
Sargassum raft observed at station 9 (12°46.160 N; 55°31.040 W) in the Sargasso Sea (A) and in situ specimens of the three Sargassum morphotypes: S. natans var. wingei ((B) known before as S. natans VIII), S. natans var. natans ((C) known before as S. natans I), and S. fluitans var. fluitans ((D) known before as S. fluitans III). (E–G) represent drawings of the three morphotypes commonly found in rafts along expeditions. Pictures: V. Stiger©UBO (A), T. Thibaut©MIO-AMU (B–D). Drawings by M. Helias©UBO.
As molecular discrimination of the three morphotypes is still complicated, without any clear separation of morphotypes into three distinct species, a chemotaxonomic study was proposed to highlight potential differences between the three morphotypes at the scale of the metabolome.
Chemotaxonomy is a useful way to classify and/or identify organisms according to their similar or unique chemical composition, both in quality and/or quantity, which usually encompasses fatty acids, terpenes, proteins, carbohydrates, and secondary metabolites. Chemical fingerprinting can also be used to discriminate specimens from different taxonomic levels, because it highlights the specific presence or absence of a compound which acts as a marker for this level and is therefore referred to a “chemomarker” [17]. Within the Sargassaceae family, terpene fingerprinting by thin-layer chromatography was used to discriminate populations of Bifurcaria bifurcata living in different hydrodynamic conditions [18]. Moreover, liquid chromatography-mass spectrometry (LC-MS) has been proven to be useful for discriminating against Cystoseira spp. sensu lato in Brittany. Indeed, the presence of a single peak on the LC chromatogram highlighted the presence of chemomarkers, which were identified as meroditerpenes in Gongolaria nodicaulis [19]. Amino acid fingerprinting by 1H Nuclear Magnetic Resonance (NMR) also permitted the discrimination of the holopelagic Sargassum morphotypes from benthic Sargassum spp., as well as the discrimination of the morphotypes among them [20].
A chemical fingerprint can also be obtained by running an algal fragment through a NMR spectrometer equipped with a High-Resolution Magic-Angle Spinning (HR-MAS) probe, also called in vivo NMR [21], leading to HR-MAS NMR spectra showing the major metabolites of the organism. This approach proved to be efficient within the Sargassaceae family as it permitted the distinction between specimens from two different species of Turbinaria, T. conoides and T. ornata [22,23], from different species of Cystoseira sensu lato [24], and from different populations of Sargassum muticum [25]. Additionally, it also enables the quantification of metabolites in the alga [26] without the extraction of metabolites and therefore only requiring a careful spectra analysis.
In the present study, we looked at the distinction between the three morphotypes of holopelagic Sargassum spp. through two different approaches: (1) using morphological criteria observed on numerous specimens of each morphotype, i.e., in examining herbarium specimens in order to identify specific and discriminant criteria, and (2) using chemical fingerprinting at the scale of the thallus or from pigments, terpenes, fatty acids, and phlorotannins profiles and in isolating chemomarkers.
2. Materials and Methods
2.1. Morphological Analysis
A total of 208 herbarium samples from the tropical North Atlantic were carefully analysed and measured. They were collected during two expeditions, Caribbean [13] and Transatlantic [12], and from opportunistic sampling that took place on various Atlantic coasts (Caribbean, Brazil, and Africa). A total of 62 herbarium specimens were identified as Sargassum natans var. wingei, 53 as S. natans var. natans, and 93 as S. fluitans var. fluitans by morphological characters.
For the morphometric analysis, 25 variables were measured on each of the 208 herbarium specimens. First, the length and width of the main axis and the number of ramifications were determined, as well as the existence, or not, of an orientation in the plant growth. The texture of the main axis was described as thorny or smooth. A frond index as well as an aerocyst index were determined for each Sargassum specimen. Both indices represent the number of fronds or aerocysts on a 3-cm long axis and, by extension, the frond or aerocyst density. To determine each index, a segment of L = 3 cm was randomly selected on each sample, and the number of fronds and aerocysts was counted. Then, the ratio L/N was computed. The more fronds or aerocysts there were, the lower the ratio. For 6 fronds and 6 aerocysts chosen randomly on each thallus, the length and width were measured. Thereby, the length/width ratio was calculated for each frond and aerocyst. The length of the frond and aerocyst peduncles was also measured.
In addition, several qualitative variables were observed: the indentation of the blades (dentated or densely dentated), their shape (linear or lanceolate), the presence and shape of a midrib (central, symmetric, asymmetric), the shape of aerocysts (spherical or ovoid), the presence of a mucron, a frond, or a wing on these aerocysts, the presence of conceptacles and their shape, as well as the presence of cryptostomata.
2.2. Chemical Investigation
2.2.1. Sampling
Samples of the three morphotypes were collected during two expeditions, Caribbean [13] and Transatlantic [12], which took place from June to October 2017 in the tropical Atlantic Ocean. The collection of specimens was achieved within drifting rafts at eight locations, allowing South-North latitudinal and East-West longitudinal gradients in the North Atlantic Ocean (Table 1, see Gobert et al. [27] for a map of stations). After collection, the samples were separated according to their morphology.

Table 1.
Date and spatial coordinates of the stations sampled for the collection of holopelagic Sargassum species for chemical investigation. See Gobert et al. [27] for the localisation of stations on a map.
For the morphological and fingerprint profiles, vouchers of each morphotype per station have been deposited into the herbarium of the Institut Universitaire Européen de la Mer, Brest, France. Fragments of the laterals of each sample were placed in an Eppendorf® tube, freeze-dried to remove any water, and analysed using High-Resolution Magic-Angle Spinning Nuclear Magnetic Resonance (HR-MAS NMR, Bruker, Wissembourg, France; see further section dedicated to the chemical approach).
For the chemical part, the isolation of metabolites was completed from fine algal powder. For this, thalli were rinsed, cleansed of their epiphytes, and freeze-dried. The dried samples were then ground into a fine powder using a ball mill.
2.2.2. Chemicals
Organic solvents (acetic acid, acetone, acetonitrile, chloroform, ethanol, ethylacetate, dichloromethane, diethyl ether, hexane, methanol, and orthophosphoric acid) were purchased from Carlo Erba reagents (Milano, Italy). Chemicals (ammonium acetate, copper sulphate, and phloroglucinol standard) were purchased from Sigma-Aldrich (Burlington, MA, USA). Folin-Ciocalteu’s reagent was purchased from VWR chemicals (Radnor, PA, USA). Pigment standards were purchased from Sigma (Virginia Beach, VA, USA; chlorophyll a, chlorophyll c, and fucoxanthin) and DHI (Hørsholm, Denmark; β-carotene, zeaxanthin, antheraxanthin, and violaxanthin).
2.2.3. Pigment Extraction and Analysis Using High Pressure Liquid Chromatography
Pigments were extracted from 75 mg dry weight (DW) of algal powder in 750 μL of 90% acetone, as previously described [28,29]. After two successive extractions and centrifugations, the supernatants were combined and filtered for high pressure liquid chromatography (HPLC) analysis (Dionex Ultimate 3000, ThermoScientific, Waltham, MA, USA). Pigments were separated using an ACE C18 column (150 × 4.6 mm, 3 μm) with a guard column, an injection volume of 6 μL, and a run-time of 33 min per sample. Before injection, each sample was automatically diluted to ¾ with ammonium acetate buffer (0.5 M, pH 7.2). Separation was achieved using a solvent gradient (methanol:buffer [80:20; v:v]; acetonitrile:milliQ water [87.5:12.5; v:v]; ethylacetate) delivered at a flow rate of 1.0 mL·min−1. A photo-diode array detector (DAD3000, Thermo-Scientific) was used for the detection of pigments at 435, 470, and 650 nm. The identification and quantification of each pigment were based on spectral comparisons and calibration using commercial standards. Pigments are expressed as mg·g−1 of algal dry weight. Three pigment ratios were also calculated:
Chlorophyll a degradation rate, or CD = [Chl a]/[Chl a + Pheo a]
Accessory pigments/Chlorophyll a or AC= [Chl c + Fuco + β-car]/[Chl a]
Xanthophyll cycle pigments/Chlorophyll a or XC= [Viola + Zea]/[Chl a]
2.2.4. Terpenes Extraction and Analysis Using Thin Layer Chromatography
Terpenes were extracted from 1 g dry weight of algal powder in 20 mL of a dichloromethane:methanol mixture [2:1; v:v] using an adapted protocol from [18]. After two successive extractions and centrifugations, the supernatants were combined and dry-evaporated to determine the dry mass yield of terpenes. Then, the extract was solubilised in a dichloromethane:methanol mixture [2:1; v:v], and 20 µL were deposited on a silica TLC aluminium plate (VWR International, Avantor Group, Rosny-sous-Bois, France). Separation was achieved using a hexane:diethyl ether:glacial acetic acid mixture [80:20:2; v:v:v], with the addition of 500 µL of methanol to obtain clearer spots. After elution, the plate was read under visible light, then revealed using an orthophosphoric acid:copper sulphate:water mixture and left in an oven at 100 °C for 15 min. The plate was again read under visible light, and terpenes were identified as the spots appearing after revelation.
2.2.5. Fatty Acid Extraction and Gas-Chromatography Analysis
Total lipids were extracted from 50 mg dry weight (DW) of algal powder in 6 mL of a chloroform-methanol mixture [2:1; v:v], as previously described by Mathieu Resuge et al. [30] and adapted from Folch et al. [31]. An aliquot of the total lipid extract was then used to obtain fatty acid methyl esters (FAME) by acidic trans-methylation, as described by Sardenne et al. [32]. FAME were analysed by gas chromatography (GC) (CP-3800 Gas Chromatography, Varian, Palo Alto, California, USA), coupled to a splitless auto-injector and a flame ionisation detector (FID), as previously described [33]. FAME were separated using polar (30 m × 0.25 mm, 0.25 µm) and apolar (30 m × 0.25 mm, 0.25 µm) columns with hydrogen as carrier gas. Identification of each fatty acid was achieved through comparisons of the retention with those contained in commercial standards (37 components: FAME, PUFA1, and PUFA3, Sigma) and in-house standard mixtures from marine bivalves, fish, and microalgae that were GC-MS certified. FAME quantification was carried out by an internal standard, tricosanoic acid (23:0). Fatty acid proportions are expressed as percent of total fatty acids.
2.2.6. Phenolic Compounds: Extraction, Purification, and Assay
Phenolic compounds were extracted from 24 g dry weight of algal powder in a 240 mL ethanol-distilled water mixture [70:30; v:v]. After three successive extractions and centrifugations, the supernatants were combined and dry-evaporated to determine the dry mass yield of phenolic compounds. Liquid/liquid purification was then used to remove non-polar compounds, pigments, proteins, and carbohydrates by a succession of solvents (dichloromethane, acetone, ethanol, and distilled water, respectively) and concentrate phenolic compounds in an ethylacetate phase, as previously described [34,35]. Phenolic compounds were then further purified by solid phase extraction (SPE) on a C18-E reverse phase silica column (Strata® C18-E, 55 μm, 70 Å, 5 g/20 mL, Giga Tubes, Phenomenex®, Torrance, CA, USA), assisted by a SPE manifold (Vac Elut SPS 24 Manifold, Agilent Technologies, Santa Clara, CA, USA). Separation was achieved using a solvent gradient (distilled water; distilled water:ethanol [70:30, v:v], and ethanol), using an adapted protocol from [36].
Total henolic content (TPC) was determined by spectrophotometry at 620 nm using the Folin-Ciocalteu assay, with phloroglucinol as a standard [36,37]. TPC is expressed as mg of phloroglucinol equivalent per g of algal dry weight.
2.2.7. Nuclear Magnetic Resonance (NMR) Analyses
NMR is a non-destructive analytical tool used to obtain various information about organic compounds, including structures, concentrations, and interactions [21]. All analyses were performed in the shared service from UBO (Brest, France). Three distinct analyses were performed: HR-MAS NMR on solid samples together with NMR of proton (1H NMR) for soluted samples, and two-dimensional NMR (2D NMR) for purified fractions of phlorotannins.
HR-MAS NMR allows the study of the full algal compounds fingerprinted in solid state without prior extraction and using a HR-MAS probe. A freeze-dried cylindrical piece of algae (of equal mass) was placed in a zirconium oxide rotor together with deuterium oxide, and air bubbles were removed by needle stirring. The rotor was then placed on a magnet for NMR analysis [21]. All solid samples were analysed to obtain a typical fingerprint of each morphotype.
Phenolic compound extracts of the three morphotypes were analysed by 1H NMR. For this, dry extracts (~10 mg) were solubilized in 800 µL of either deuterium oxide (D2O) for aqueous extracts or deuterated methanol (CD3OD) for organic extracts, as previously described [21]. All spectra were acquired on a Bruker Avance 400 using the standard pulse sequences available in the Bruker software (Bruker, France). Chemical shifts are expressed in parts per million (ppm). Peak number, form, and multiplicity on the spectrum provide information about the structural conformation of each proton atom in the molecule. On a 1H NMR spectrum, signals between 5.5 and 6.5 ppm are characteristic of aromatic molecules, which include phenolic compounds.
For 2-D NMR couples of proton (1H) and carbon (13C) NMR, dry extracts (~10 mg) were solubilized in 800 µL of deuterated methanol (CD3OD). All spectra were acquired on a Bruker Avance 500 using the standard pulse sequences available in the Bruker software (Bruker, France). Chemical shifts are expressed in parts per million (ppm). The nature of phlorotannins present in the purified fractions was determined with the aid of distortionless enhancement of polarisation transfer (DEPT), heteronuclear multiple quantum coherence (HMQC), and heteronuclear multiple bond correlation (HMBC) experiments, followed by comparison with chemical shifts of 1H (between 5.5 and 6.5 ppm) and 13C (between 95 and 165 ppm) resonances with literature data (Table 2A) [35,38]. These signals will be useful to identify the class of phlorotannins synthetised by morphotypes following the type of bond (Table 2B).

Table 2.
Identification of signals (A) on the 13C axis by their chemical shifts and permitting the determination of (B) categories of phlorotannins, adapted from [35,38,39].
2.3. Statistical Analyses
All statistical analyses concerning morphological criteria and chemical data (except NMR spectra), were carried out on R software (v.4.2.0), coupled to the RStudio (v.2022.12.0) integrated development environment [40].
For the morphological analysis, the data set was first converted into a distance matrix using the Gower coefficient [41], which can take into account both quantitative and qualitative variables. Then, a hierarchical clustering analysis using Ward linkage was applied [41,42,43]. Finally, a Principal Coordinates Analysis (PCoA) was constructed [44,45,46] from the square root of the distance matrix [41] and compared to the clustering analysis results. The PCoA allows the 2-dimension separation of the sample pool according to their dissimilarities.
Similarity percentages analysis (Simper test) [45] was performed to highlight the fatty acids with the most influence on the differentiation of the three morphotypes, with fatty acids accounting for 75% of the dissimilarity between morphotypes. The fatty acid relative abundance table was analysed through a principal component analysis (PCA) [47,48] to map the samples.
For the statistical analysis of the pigment and phenolic contents, as the data set did not respect the requirements for Analysis of Variance ANOVA (homogeneity of variances and normality tested by Bartlett and Shapiro tests, respectively, at the 95% significance level), the non-parametric Scheirer-Ray-Hare tests were applied [49] to highlight significant differences between the morphotypes and/or the stations at the 95% significance level. When significant differences were found, the Dunn post-hoc test [50] at 95% significance level coupled to the Bonferroni correction allowed their identification. Pigment abundance was also analysed through a Principal Component analysis (PCA).
NMR 1H spectra were processed on the software MesReNova 11. Spectra baselines were automatically corrected, followed by the Whittaker smoother correction. An equal-width bucketing of 0.03 ppm was applied between 0 and 7 ppm to finally obtain the data matrix. Statistical analyses were performed using the on-line MetaboAnalyst 6.0 software. A PCA was used to visualise the metabolome variation according to the three morphotypes of holopelagic Sargassum species.
3. Results
3.1. Morphological Identification of the Three Morphotypes
Out of the 25 morphological variables studied, based on our statistical results, 22 were considered discriminating for the three morphotypes of holopelagic Sargassum. These variables were mainly the length, width, texture, and number of ramifications of the main axis, the frond and aerocyst indices, the length/width ratio and the length of peduncles of fronds and aerocysts, the indentation and shape of the blades, the presence and shape of a midrib, the shape and the presence of a mucron, and finally the presence of a frond or a wing on aerocysts.
The clustering analysis through the Ward method showed the existence of three main groups corresponding to the three morphotypes. The PCoA mapping in Figure 2 was an accurate visual representation of the herbarium specimen pool, as shown by the dimensions explaining 58% of the samples.
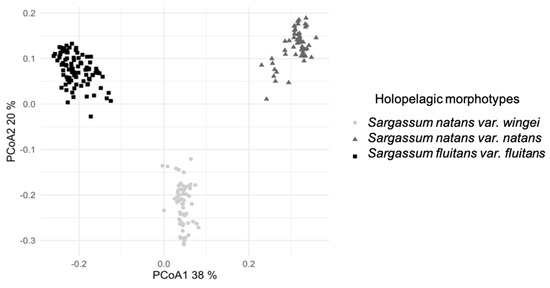
Figure 2.
Principal Coordinates Analysis (PCoA) of the 22 discriminating morphological variables, measured on different samples of the three common morphotypes of holopelagic Sargassum. The base matrix was converted using the Gower coefficient to the square root.
The PCoA showed a clear separation of the Sargassum specimens into three groups, corresponding to S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, according to the selected discriminating criteria. The selected morphological variables were thus suitable to differentiate the three morphotypes of holopelagic Sargassum into distinct groups using pertinent morphological criteria.
3.2. HR-MAS NMR Fingerprinting Profiles
Three spectra per morphotype were selected as the most representative spectra of each morphotype and are illustrated and statistically treated (Figure 3A). The PCA mapping is an accurate visual representation of the chemical fingerprinting pool, as shown by the dimensions explaining 79% of the variability within the samples (Figure 3B).
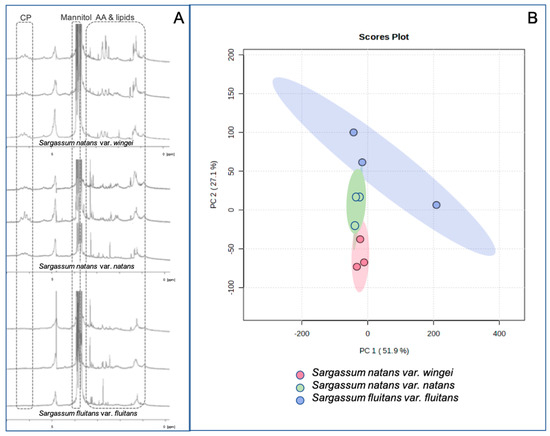
Figure 3.
Comparison of chemical fingerprintings obtained from three morphotypes of holopelagic Sargassum species. (A): in vivo HR-MAS NMR chemical fingerprintings obtained in 3 individuals for each morphotype. (B): Principal Component Analysis (PCA) of the chemical fingerprinting, measured on 9 different samples of the three common morphotypes of holopelagic Sargassum. PC: phenolic compounds; AA: amino acids.
The PCA highlighted that the chemical fingerprinting of the three morphotypes is distinct, especially those from Sargassum natans var. wingei and S. fluitans var. fluitans, with S. natans var. natans positioned in an intermediate position (Figure 3B).
Based on our sampling, Sargassum natans and S. fluitans morphotypes are distinguished by their phlorotannin profiles, with S. fluitans var. fluitans producing fewer phlorotannins than both S. natans morphotypes (Figure 3A). Moreover, distinct signals of amino acids and lipidic compounds were observed in the three morphotypes, permitting us to discriminate between them using these two classes of compounds (Figure 3B). We then used phenolic compounds and fatty acids for our investigation (see further sections).
3.3. Chemical Investigation
3.3.1. Pigment Level and Composition
Total pigment contents were between 0.12 and 2.03 µg.mg−1 algal dry weight (DW), with both observed in the morphotype Sargassum fluitans var. fluitans sampled in two different stations: station 19 for the lowest content and station 9 for the highest. When looking at the different pigments, the variation among the samples was also high, with, for example, the chlorophyll a content between 0.04 and 1.35 µg.mg−1 DW (Table S1). This variation is due to a significant effect of the morphotype and the station on all pigment contents, with the exception of the effect of the station on violaxanthin (Table S2). Thus, S. natans var wingei exhibited the lowest concentration of all pigments, whatever the station, and S. natans var. natans the highest. The same difference among morphotypes was also observed for two ratios (AC and XC), but not for the chlorophyll a degradation rate, for which no effect of the morphotype was observed.
A PCA was then computed with all the pigment levels and ratios (Figure 4), showing strong variation among samples in pigment contents, mainly due to morphotype but also station differences.
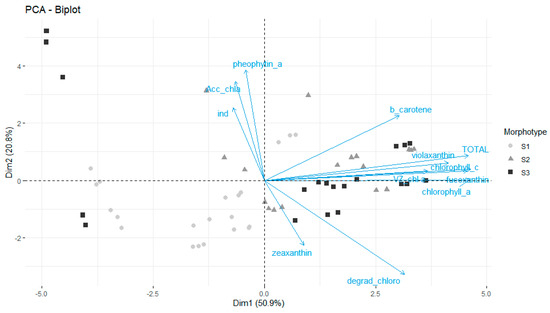
Figure 4.
Principal Component Analysis (PCA) of the holopelagic Sargassum samples using the pigment contents and the pigment ratios. S1: S. natans var. wingei; S2: S. natans var. natans; S3: S. fluitans var. fluitans.
3.3.2. Terpene Profiles
Terpene profiling of the three holopelagic Sargassum morphotypes from different locations in the Central Atlantic Ocean was carried out on Thin Layer Chromatography (TLC). The schematic representation of the TLC plates (Figure S1) displayed the presence of several compounds in the three morphotypes, characterised by the colour and retention factor (Rf) of their respective spots after revelation. The green spot with a front ratio of 0.2 was characteristic of chlorophyll-a. Both grey spots with Rf of 0.4 and 0.9 were characteristic of carotenoids. Lastly, the pink-bluish spot with a Rf of 0.3, which appeared only after the use of a terpene-specific revealer, was characteristic of terpenes. Terpenes were thus present in the three holopelagic Sargassum morphotypes; however, their TLC profiles did not allow differentiation between morphotypes.
3.3.3. Free Fatty Acid Proportions
Gas chromatography of the fatty acid profiles resulted in the identification and quantification of 25 fatty acids in the three holopelagic Sargassum morphotypes (Table 3).

Table 3.
Mass percentage of total fatty acids (%TFA) of the 25 main fatty acids and the three main classes of fatty acids (highlighted in grey), identified in the three holopelagic Sargassum morphotypes. SFA: saturated fatty acids; MUFA: monounsaturated fatty acids; PUFA: polyunsaturated fatty acids.
For all three morphotypes, saturated fatty acids (SFA) were the most abundant group of fatty acids, with 54.64 ± 2.44% TFA, 54.68 ± 1.66% TFA and 58.00 ± 2.31% TFA for Sargassum natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively. This was mainly due to the fact that palmitic acid (16:0) is the most abundant free fatty acid, with 47.91 ± 3.31% TFA, 45.80 ± 3.57% TFA and 47.53 ± 3.90% TFA for S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively. Monounsaturated fatty acids (MUFA) in the three morphotypes had similar but slightly higher contents than polyunsaturated fatty acids (PUFA), with 25.49 ± 2.25% TFA, 22.18 ± 1.50% TFA and 25.03 ± 2.07% TFA of MUFA and 19.08 ± 4.34% TFA, 23.42 ± 3.02% TFA and 16.67 ± 4.15% TFA of PUFA for S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively.
In regard to our study, the first observation was that a qualitative differentiation of the three morphotypes was not feasible on the basis of the presence or absence of a specific fatty acid, as each of the three morphotypes possessed all the fatty acids identified. However, the Simper statistical analysis allowed the selection of the eight fatty acids that contributed the most to the dissimilarity between the fatty acid profiles of the different morphotypes. Indeed, cis-7-hexadecenoic acid (16:1n-9), palmitoleic acid (16:1n-7), and margaric acid (17:0) in Sargassum natans var. wingei (0.43 ± 0.09% TFA, 10.00 ± 1.53% TFA, and 0.38 ± 0.05% TFA, respectively) were significantly higher than in S. natans var. natans (0.26 ± 0.05% TFA, 4.53 ± 0.46% TFA, and 0.14 ± 0.03% TFA, respectively) and S. fluitans var. fluitans (0.28 ± 0.07% TFA and 5.22 ± 0.54% TFA, 0.12 ± 0.04% TFA, respectively). Meanwhile, α-linoleic acid (18:3n-3), eicosatetraenoic acid (20:4n-3), and eicosapentaenoic acid (20:5n-3) in S. natans var. natans (3.69 ± 0.65% TFA, 0.53 ± 0.13% TFA, and 1.55 ± 0.97% TFA, respectively) were significantly higher than in S. natans var. wingei (2.21 ± 0.53% TFA, 0.32 ± 0.16% TFA, and 0.66 ± 0.53% T FA, respectively) and S. fluitans var. fluitans (1.82 ± 0.46% TFA, 0.26 ± 0.12% TFA, and 0.69 ± 0.63% TFA, respectively). Finally, myristic acid (14:0) and cis-11-eicosaenoic acid (20:1n-9) in S3 (4.17 ± 0.32% and 1.60 ± 0.21%, respectively) were significantly higher than in S. natans var. wingei (2.96 ± 0.20%TFA and 0.83 ± 0.14% TFA, respectively) and S. natans var. natans (2.63 ± 0.32% TFA and 1.05 ± 0.27% TFA, respectively). Thus, the proportions of these fatty acids were considered distinctive for the three morphotypes.
A PCA was then computed according to the eight selected fatty acid proportions of the samples. The PCA mapping in Figure 5 was an accurate visual representation of the samples, with the dimensions explaining more than 82% of the sample pool.
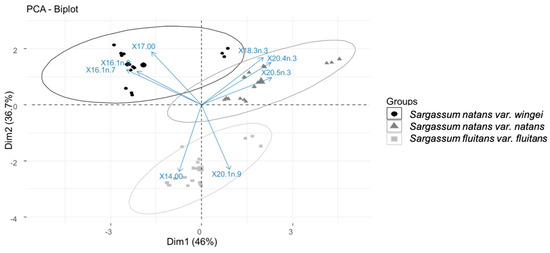
Figure 5.
Principal Component Analysis (PCA) of the holopelagic Sargassum samples using the proportion of the eight fatty acids out of the total fatty acids selected by SIMPER analysis. Ellipses represent a confidence interval of 95%. Cis-7-hexadecenoic acid (16:1n-9), palmitoleic acid (16:1n-7), and margaric acid (17:0) drive the clustering of S. natans var. wingei, α-linoleic acid (18:3n-3), eicosatetraenoic acid (20:4n-3), and eicosapentaenoic acid (20:5n-3) drive the clustering of S. natans var. natans, and myristic acid (14:0) and cis-11-eicosaenoic acid (20:1n-9) drive the clustering of S. fluitans var. fluitans.
The PCA showed a separation of the holopelagic Sargassum samples into three groups, identified as S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, according to the proportions of these eight fatty acids. They were thus suitable to differentiate the three holopelagic Sargassum morphotypes. However, although a clear distinction of S. fluitans var. fluitans from the two other morphotypes was observed at the confidence interval of 95%, a slight overlap of the ellipses of S. natans var. wingei and S. natans var. natans was present.
This overlap was mainly due to one sample, which stretched the ellipses along Dimension 1 for S. natans var. natans and S. fluitans var. fluitans and the two Dimensions for S. natans var. wingei. This sample corresponded to station GP on Table 1, which was on a different longitudinal scale than the rest of the sample pool.
3.3.4. Total Phenolic Content and Structure of Phlorotannins
The mean Total Phenolic Content (TPC) values of the three morphotypes across the stations are presented in Table 4.

Table 4.
Total Phenolic Content (TPC) values in mgPE·g−1 DW of the three morphotypes across the eight stations of the sampling. Each value corresponds to the mean ± SD (n = 3). S1: Sargassum natans var. wingei; S2: S. natans var. natans; S3: S. fluitans var. fluitans.
The statistical analysis of the dataset showed significant differences both at morphotype and station levels (p < 0.05, Scheirer-Ray-Hare test), with stronger differences found at the station level. The effect of the interaction between both parameters was not significant (p > 0.05, Scheirer-Ray-Hare test). The Dunn post-hoc test allowed the identification of significant differences between Sargassum natans var. wingei and S. fluitans var. fluitans, with S. natans var. wingei usually having a higher TPC than S. fluitans var. fluitans. Significant differences were also found between stations, in particular those that can be considered extremes, i.e., Station 19 was the northernmost station, station GP was the easternmost, and mangrove samples were collected stranded on the coast.
The proton Nuclear Magnetic Resonance (1H NMR) analysis of the three morphotypes’ purified fraction enriched in phenolic compounds highlighted the chemical fingerprint of the extracts (Figure 6).
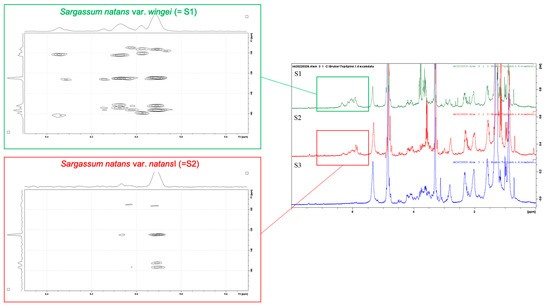
Figure 6.
Structural elucidation of phlorotannins produced by Sargassum natans var. wingei (S1) and S. natans var. natans (S2). 1H Nuclear Magnetic Resonance (NMR) spectra of the three Sargassum morphotypes (S1, S2, and S3 as S. fluitans var. fluitans) (right side). Two-dimensional NMR spectra of S1 S. natans var. wingei (left side up) and S2 S. natans var. natans (left side down). On 2D NMR, chemical shifts of protons are on the X-axis, and chemical shifts of carbons are on the Y-axis. Proton signals between 5.5 and 6.5 ppm are characteristic of aromatic compounds, which encompass phenolic compounds.
The 1H NMR spectra of Sargassum natans var. wingei and S. natans var. natans showed several signals in the aromatic compounds area (5.5 and 6.5 ppm), which encompassed the phenolic compounds. The signals showed several differences between the two morphotypes, both in terms of form and multiplicity of peaks. The 1H NMR spectrum of S. fluitans var. fluitans, however, did not show signals for phenolic compounds, as this morphotype seemed to contain very low amounts of phenolic compounds (see above the TPC obtained for S. fluitans var. fluitans).
The 2-Dimensional Nuclear Magnetic Resonance (2D NMR) spectra were recorded only for Sargassum natans var. wingei and S. natans var. natans (Figure 6), as the purified extract of S. fluitans var. fluitans did not contain sufficient amounts of phenolic compounds to perform the analysis. The 2D NMR spectra of the two morphotypes showed differences in placement and abundance of signals. The spectrum of the purified extract of S. natans var. wingei (Figure 6) showed signals characteristic of methine (95–100 ppm) and phenolic carbons (150–165 ppm), as well as signals for diaryl-ether carbons (125–130 ppm) (Table 2A). They did not show, however, the carbon signal for supplementary hydroxyl functions (145–150 ppm). The spectrum of the purified extract of S. natans var. natans (Figure 6) presented signals characteristic of methine carbons (95–100 ppm), phenolic carbons (150–165 ppm), diaryl-ether carbons (125–130 ppm), and an absence of signals for supplementary hydroxyl functions (145–150 ppm) as well, but differed from S. natans var. wingei by the presence of signals for aryl-aryl carbons (100–105 ppm). There was thus a difference in the structure of phenolic compounds between S. natans var. wingei and S. natans var. natans.
4. Discussion
Since 2011, recurring massive strandings have impacted Western African, South American, and Caribbean shores [1,2,51,52]. The species responsible for these strandings are three common morphotypes of the holopelagic Sargassum species: S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans. They are present in the North Atlantic Ocean as mixed rafts of various sizes, drifting along winds and currents [53,54,55]. The three morphotypes demonstrate clear morphological differences, as described in this present study (Figure 2 and Figure 7) and previous work [7,15]. However, their molecular discrimination, based on classical phylogenetic markers, is still challenging [6,16]. The literature reports examples of Sargassaceae for which different species exist even though no molecular differences could be detected, particularly in the genus Sargassum, subgenus Bactrophycus (S. boreale, S. confusum, and S. microceratium) and subgenus Sargassum (S. mcclurei and S. phyllocystum), for which no difference could be detected for their ITS2 sequences while clear morphological differences exist [56,57]. Nevertheless, Siuda et al. [16] were not able to elevate both morphotypes of Sargassum natans into two distinct species and therefore to create two separate varieties of S. natans. The present study identified additional morphometric differences as well as several chemical markers that allowed the differentiation of these species and varieties. Indeed, in the rafts, the three morphotypes are subjected to the same environmental conditions, which means that any qualitative or quantitative difference in their metabolites is specific to the morphotype, at a given point in the Atlantic Ocean.

Figure 7.
Morphological particularities of each lateral, axis, frond, and floating vesicles selected and isolated from the three holopelagic Sargassum morphotypes (Sargassum natans var. wingei to the left, S. natans var. natans in the middle, S. fluitans var. fluitans right side) present in rafts during the latitudinal expedition. Scale: 1 cm. Herbarium was made by M. Helias©UBO.
4.1. Morphological Discrimination of Morphotypes
The morphological diversity of holopelagic Sargassum has already been described, as Winge and later Parr acknowledged four forms of S. natans (I, II, VIII, and IX) and two forms of S. fluitans (III, X) based on four morphological criteria [14,15]. A plate of selected specimens used in the morphological analysis illustrates particularities found in each holopelagic Sargassum morphotype (Figure 7).
Our study described 18 supplementary discriminating characters to differentiate the three morphotypes. These 22 variables were combined in a dichotomous determination key, with reference to the graphical representation of the three morphotypes (Figure 1, Figure 2, and Figure 7), allowing the identification of a morphotype solely based on morphological criteria.
The main criteria used to isolate holopelagic from benthic Sargassum specimens are the following: absence of cryptostomata and receptacles/conceptacles, no fixing system, generally numerous aerocysts. Moreover, the length/width frond ratio is higher than the 3~5 interval of benthic Sargassum (unpublished data).
We propose the following key in order to morphologically identify the three morphotypes:
1—Linear and dentate fronds present. Length/width frond ratio between 7 and 21 (fronds long and narrow). Fronds inhibit a light brown colour, with a central midrib parallel to the edges and reaching the tip of the frond. Aerocysts are mucronated (mucron sometimes replaced by a small frond). Frond and aerocyst peduncles can reach a size greater than 1 mm, and bear no wing.
.......................................................... S. natans var. natans (Figure 1C,F and Figure 7 in the middle)
1—Lanceolate and dentate fronds present. Length/width frond ratio varies between 3 and 10. Fronds are of a dark brown to a light brown colour, with a central midrib parallel or asymmetrical to the edges and reaching the tip of the fronds. Aerocysts do not have a mucron. Frond and aerocyst peduncles are usually smaller than 1 mm.. Axes are smooth or thorny……………………..………………………………………………….…………….......2
2—Frond index varies between 0.3 and 1 (scarce fronds). Axes are always smooth. Fronds exhibit a parallel midrib. Fronds and axes are usually covered by several species of bryozoans and hydrozoans..........S. natans var. wingei (Figure 1B,E and Figure 7 to the left)
2—Frond index varies between 0.1 and 0.6 (numerous fronds), giving a “leafy” aspect to the axes. Axes are thorny or smooth. Fronds exhibit an asymmetrical midrib.
4.2. Chemical Discrimination of Morphotypes
4.2.1. HR-MAS Fingerprinting
The use of HR-MAS NMR or in vivo NMR made it possible to obtain chemical fingerprints and thus to distinguish between the three morphotypes of holopelagic Sargassum, particularly in the low ppm zone (0–3.5 ppm). This area has already been highlighted as an area allowing the discrimination of various organisms, such as bacterial [58] and microalgal [59] strains, lichen [60], Asteraceae plants [61], and macroalgae, such as Phaeophyceae [23,24]. Our analyses also showed a distinction between the three morphotypes using phlorotannin signals, with both Sargassum natans morphotypes producing more compounds than S. fluitans and different structures of phlorotannins (see Section 4.2.3). The zone presenting the most differences between the three morphotypes is that corresponding to chemical shifts from 0 to 3.5 ppm. This area presents NMR signals from different types of compounds, such as amino acids and lipid compounds, with the potential to find chemomarkers (see further sections).
4.2.2. Apolar/Lipidic Compounds: Pigments, Terpenes, and Fatty Acids
The pigment composition of Sargassum samples in this study is characteristic of brown seaweeds, with chlorophyll a being the main photosynthetic pigment, followed by fucoxanthin, chlorophyll c, violaxanthin, β-carotene, and the photoprotective pigment zeaxanthin in stations where the chlorophyll a content was very low [62]. Moreover, a consistent amount of pheophytin a has been assayed in samples from some stations, suggesting a degradation of the chlorophyll a due to the environment. A significant difference in all pigments among the three Sargassum morphotypes has been identified, with the lowest contents in S. natans var. wingei, the highest in S. natans var. natans, and finally S. fluitans var. fluitans with intermediate contents. However, the effect of the environmental conditions at each station also had a significant impact on the algal pigment content, making it difficult to establish a pigment content or ratio specific to each morphotype.
The qualitative study of holopelagic Sargassum terpenes across several stations showed no differences between the three morphotypes, even across highly geographically different stations (Supplementary Figure S1). However, the technique used in the present study, thin layer chromatography (TLC), only indicated the presence or absence of a class of compounds in an extract without taking into account neither the quantity nor the structure of the compounds. Thus, this technique could not be discriminating enough to separate the three morphotypes and may overlook differences happening within the class of terpenes, either in terms of structure or proportion in the biomass. A finer analysis of the three morphotypes through liquid-chromatography coupled to mass-spectrometry (LC-MS) and nuclear magnetic resonance (NMR) may allow the identification of terpene chemomarkers in the holopelagic Sargassum. Indeed, it has already allowed the discrimination of Gongolaria nodicaulis from four other species of Cystoseira spp. sensu lato in Brittany through the presence of a specific monocyclic meroditerpene [19] and the discrimination of several populations of Bifurcaria through the presence and type of terpenes [18].
The study then moved to the qualitative and quantitative study of 25 fatty acids in holopelagic Sargassum through gas-chromatography (GC). Palmitic acid was found to be the most abundant fatty acid in the three morphotypes, ranging between 46 and 48% of the total fatty acid pool (TFA, Table 3). This result is in agreement with the studies of Van Ginneken et al. [63], who found a proportion of palmitic acid of 41%TFA for S. natans, and the one from Milledge et al. [64], who found a proportion of palmitic acid of 41% TFA for S. natans var. wingei. However, the latter also found proportions of palmitic acid of 24% TFA for both S. natans var. natans and S. fluitans var. fluitans, below the values found in the present study. We can assume that this is due to the fact that the proportions of the fatty acids are modified for one morphotype to adapt to the physical and environmental changes that the macroalga has to face. Concerning the different classes of fatty acids, while the literature and the present study agree that saturated fatty acids (SFA) are the most abundant class, divergences appear in the unsaturated fatty acids. Indeed, the present study found a similar but slightly higher proportion of monounsaturated fatty acids (MUFA, ~24% TFA) compared to polyunsaturated fatty acids (PUFA, ~20% TFA) in the three morphotypes (Table 3), but Milledge et al. [64] found a higher proportion of PUFA (~29% TFA) compared to MUFA (~20% TFA). The proportion of PUFA in the tropical Sargassum can be very variable, as highlighted by Turner and Rooker [65], who found proportions of PUFA ranging from 16 to 62%.
In each morphotype, the proportion of one specific fatty acid was significantly higher than in the other two morphotypes. Indeed, proportions of cis-7-hexadecenoic acid (16:1n-9), palmitoleic acid (16:1n-7), and margaric acid (17:0) were significantly higher in Sargassum natans var. wingei than in S. natans var. natans and S. fluitans var. fluitans. This observation was supported by the study of Milledge et al. [64] for palmitoleic acid (8.3% TFA, 3.5% TFA and 4.1% TFA for S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively), but not for margaric acid (0.13% TFA, 0.88% TFA and 0.76% TFA for S. natans var. wingei, S. natans var. natans and S. fluitans var. fluitans respectively). Proportions of α-linoleic acid (18:3n-3), eicosatetraenoic acid (20:4n-3) and eicosapentaenoic acid (20:5n-3,) were significantly higher in S. natans var. natans than in S. natans var. wingei and S. fluitans var. fluitans, once again in agreement with the study of Milledge et al. [64] for α-linoleic acid (3.5% TFA, 5.9%TFA and 3.5% TFA for S. natans var. wingei, S. natans var. natans and S. fluitans var. fluitans respectively) and eicosapentaenoic acid (<0.05% TFA, 2.77% TFA, and 1.49% TFA for S. natans var. wingei, S. natans var. natans and S. fluitans var. fluitans respectively). Proportions of myristic acid (14:0) and cis-11-eicosaenoic acid (20:1n-9) were significantly higher in S. fluitans var. fluitans than in S. natans var. wingei and S. natans var. natans, which was also observed in the study of Milledge et al. [64] for myristic acid (2.0% TFA, 1.6% TFA, and 2.1% TFA for S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively). However, there are variations in the proportions of fatty acids found in both studies, as well as in the present study, between the various sampled stations [64]. Indeed, the fatty acid profile of macroalgae depends on numerous factors, such as season, geographical location, water temperature, light intensity, contaminants, and growth factors as highlighted by Schmid et al. [29] in several macroalgae. Performing ratios of fatty acid proportions eliminates variations linked to external factors since all the metabolites in the studied specimen will have been subjected to the same environmental conditions, thus allowing a global study of the three morphotypes coming from different geographical locations. For each morphotype, one fatty acid in particular showed the most differences in proportions with the other two morphotypes, namely palmitoleic acid (16:1n-7) for Sargassum natans var. wingei, α-linoleic acid (18:3n-3) for S. natans var. natans, and myristic acid (14:0) for S. fluitans var. fluitans. They were thus selected for the ratio study, which led to the establishment of a dichotomous determination key (Figure 8). The key allows the identification of a morphotype based solely on the analysis of its fatty acid profile and, more specifically, on the quantification of only three fatty acids. However, the determination key is only effective for offshore biomasses, and not beachcast ones, since as soon as the biomass washes up, algal metabolites start to deteriorate at different rates, and fatty acid proportions could be affected.
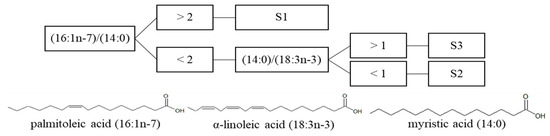
Figure 8.
Dichotomous determination key of the three common morphotypes of holopelagic Sargassum, based on the ratio of three discriminating fatty acids. S1: S. natans var. wingei; S2: S. natans var. natans; S3: S. fluitans var. fluitans.
4.2.3. Phenolic Compounds
The total phenolic content (TPC) of the crude extracts of three morphotypes quantified in this study is higher than that reported in the literature. Indeed, Milledge et al. [64] (2020) and Davis et al. [51] had TPCs ranging from 2.5–3.1, 2.1–6.6, and 1.2–3.7 mgPE.g−1 DW for Sargassum natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively, while the present study found TPCs of 27, 17, and 15 mgPE.g−1 DW for S. natans var. wingei, S. natans var. natans, and S. fluitans var. fluitans, respectively (Table 4). High TPC variations are to be expected, even within the same species, because TPC is greatly influenced by a variety of parameters. Indeed, phenolic compounds are synthesised when the organism is subjected to stresses, both abiotic (such as light quantity and quality, temperature, and salinity) and biotic (such as microfouling and grazing). Depending on the intensity of these stresses, the amount of phenolic compounds synthesised could vary [35,66]. Furthermore, intrinsic parameters such as maturity, stage of development, age of tissue, and algal size also play a role in the phenolic content [35,67]. Drying and extraction techniques can also influence the measured TPC. For example, Davis et al. [51] used sun exposure to dry their samples. Phenolic compounds are highly sensitive to light and temperature, and the phenolic pool could have been degraded during drying.
The phenolic compounds’ chemical structure was then characterised through 2-Dimensional Nuclear Magnetic Resonance (2D NMR), a technique that associates proton (1H) and carbon (13C) NMR analysis. This analysis requires a high degree of purity, so the crude extracts were purified through liquid/liquid separation and analysed through 1H NMR (Figure 6). While the 1H NMR spectra of Sargassum natans var. wingei and S. natans var. natans showed differences in the form and the multiplicity of signals in the aromatic compounds area containing the phenolic compounds (5.5–6.5 ppm), the S. fluitans var. fluitans spectrum did not show identifiable signals, which could be related to a low proportion of compounds in the extract. Thus, only S. natans var. wingei and S. natans var. natans were further purified by solid phase extraction (SPE). The SPE fractions, enriched in phenolic compounds, were analysed through 2D NMR (Figure 6). The 2D NMR spectra of the two morphotypes show different signals between 5.5 and 6.5 ppm, which hints towards a differentiation of the chemical structure of the phenolic compounds. Brown macroalgae’s phenolic compounds, phlorotannins, are aromatic molecules made up of polymerized phloroglucinol units linked by various bonds. Thus, they can be classified into different categories according to the type of linkages present in the molecule (Table 2B).
The 2D NMR spectra of Sargassum natans var. wingei and S. natans var. natans (Figure 6) both show signals characteristic for methine and phenolic carbons, which indicate the presence of phloroglucinol units and thus highlight the presence of phlorotannins. The spectrum of S. natans var. natans displayed carbon signals for diaryl-ether carbons, which are characteristic of ether bonds. The presence of ether bonds in a phloroglucinol polymer results in either phlorethol- or fuhalol-type phlorotannins (Table 2B). However, the absence of carbon signals specific to additional hydroxyl functions on the spectrum excludes the possibility of fuhalol-type units, resulting in the hypothesis that the phlorotannins of S. natans var. natans belong to the phlorethol class. Meanwhile, the spectrum of S. natans var. wingei shows both carbon signals for aryl-aryl and diaryl-ether carbons, which are respectively characteristic of phenyl and ether bonds. The co-occurrence of phenyl and ether bonds in a phloroglucinol polymer typically indicates a phlorotannin belonging to the fucophlorethol class (Table 2B). However, it is also possible that within S. natans var. wingei, several types of phlorotannins were co-extracted, namely phlorethols, fucols, and/or fucophlorethols, which would then give both ether and phenyl bond signals on the 2D NMR spectra. As the extracts were not analysed by mass-spectrometry, no affirmation can yet be made on the chemical structure of S. natans var. wingei and S. natans var. natans phlorotannin; nevertheless, this information is expected to be useful in the differentiation of two of the three morphotypes occurring in rafts.
The high variety and complexity of structural bonds resulting from polymerisation of phloroglucinol units and additional hydroxyl functions, in addition to the possibility of several classes of phenolic compounds occurring at the same time, render phenolic compound structures difficult to elucidate. Thus, structural identification of phenolic compounds in the Sargassum genus is yet to be widespread. The presence of fuhalols and S. ringgoldianum has been demonstrated [68,69], as well as phlorethols in S. carpophyllum and S. stenophyllum [70,71]. Sargassum muticum was reported to contain phlorethols, fuhalols, and hydroxyfuhalols [35,72,73,74,75,76], while S. spinuligerum and S. vulgare contained fuhalols, hydroxyfuhalols, phlorethols, and fucophlorethols [77,78,79,80,81,82]. Sargassum fusiforme contained fuhalols, phlorethols, fucophlorethols, and eckols [74,83].
5. Conclusions
The differentiation of the three common morphotypes of holopelagic Sargassum species present in drifting rafts in the tropical North Atlantic Ocean was achieved through morphometry and chemical markers. Two dichotomous determination keys, based respectively on morphological variables and on the ratios of three fatty acids, allow the identification of each morphotype. A differentiation of S. natans var. wingei and S. natans var. natans could also be possible through the structural elucidation of their phlorotannins; S. natans var. natans contains phlorethol-type phlorotannins, while S. natans var. wingei contains phlorethol-, fucol- and/or fucophlorethol-type phlorotannins. The terpene fingerprint did not allow us to distinguish the morphotypes of holopelagic Sargassum. Further analysis is underway to see if terpene purification could lead us to find other chemomarkers. Moreover, we are pursuing analysis with the aim of finding efficient molecular and other chemical markers in order to raise S. natans var. wingei to the rank of a new species. Indeed, Siuda et al. [16] took the decision to place S. natans VIII at the level of variety (S. natans var. wingei), and this decision was judged as conservative by the authors. Nevertheless, questions remain: how two varieties of the same species (S. natans) could live together in sympatry in rafts in having different morphologies (this study, [16,27]), different growth rates [84], different contaminants [27], and different secondary metabolites?
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/phycology4030018/s1. Figure S1: Thin Layer Chromatography of terpene extracts from different floating specimens of the three holopelagic Sargassum morphotypes across several stations within the Central Atlantic Ocean; Table S1: Pigment level of the three Sargassum morphotypes sampled during the Caribbean cruise; Table S2: Results of statistical analyses (Sheirer-Hare tests and post-hoc test when needed) on pigment contents except zeaxanthin due to low occurrence and pigment ratios.
Author Contributions
Conceptualization, N.K., M.H., V.S.-P. and S.C. (Solène Connan); Methodology, N.K., M.H., V.S.-P. and S.C. (Solène Connan); Validation, V.S.-P., S.C. (Solène Connan) and F.L.G.; Formal Analysis, N.K., M.H., A.B., C.N., V.S.-P. and S.C. (Solène Connan); Investigation, N.K., M.H., V.S.-P. and A.B.; Resources, V.S.-P., S.C. (Solène Connan), F.L.G., S.C. (Stéphane Cérantola) and G.S.; Writing—Original Draft Preparation, N.K., M.H., V.S.-P. and S.C. (Solène Connan); Writing—Review & Editing, N.K., M.H., C.N., L.B., A.B., T.C., S.C. (Solène Connan) and V.S.-P.; Visualization, N.K., M.H., V.S.-P. and S.C. (Solène Connan); Supervision, V.S.-P. and S.C. (Solène Connan); Project Administration, V.S.-P. and S.C. (Solène Connan); Funding Acquisition, V.S.-P., L.B. and T.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded through the projects SAVE-C (Fundamental and applied research, Grant No. ANR-19-SARG-0008), SARGASSUM ORIGINS (Grant No. ANR-19-SARG-0004), and BIOMAS (Grant No, ANR-22-SARG-0003) which are international partnerships funded by the ANR under the call SARGASSUM Programme 2019 and 2022.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data sets generated and/or analysed during the present study are available upon reasonable request.
Acknowledgments
This study is part of the master course work carried out by N.K. and M.H. within the Laboratory of Marine Sciences (LEMAR UMR 6539) set at IUEM (UBO). The authors sincerely thank A. Bideau for his fruitful help in the lipid extractions.
Conflicts of Interest
The authors declare that the research used to construct the manuscript was presented in the absence of any commercial or financial relationships that could be viewed as a potential conflict of interest. The funder has no role in the design of the study, in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- De Szechy, M.T.M.; Guedes, P.M.; Baeta-Neves, M.H.; Oliveira, E.N. Verification of Sargassum natans (Linnaeus) Gaillon (Heterokontophyta: Phaeophyceae) from the Sargasso Sea Off the Coast of Brazil, Western Atlantic Ocean. Checklist 2012, 8, 638–641. [Google Scholar] [CrossRef]
- Smetacek, V.; Zingone, A. Green and Golden Seaweed Tides on the Rise. Nature 2013, 504, 84–88. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. In World-Wide Electronic Publication; National University of Irlande: Galway, Ireland, 2013; Available online: http://www.algaebase.org (accessed on 18 April 2024).
- Stiger-Pouvreau, V.; Mattio, L.; De Ramon N’Yeurt, A.; Uwai, S.; Dominguez, H.; Flórez-Fernández, N.; Connan, S.; Critchley, A.T. A Concise Review of the Highly Diverse Genus Sargassum C. Agardh with Wide Industrial Potential. J. Appl. Phycol. 2023, 35, 1453–1483. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Dragone, N.B.; Schell, J.; Slikas, B.; Murphy, L.G.; Morrall, C.E.; Zettler, E.R. Comparative Mitochondrial and Chloroplast Genomics of a Genetically Distinct Form of Sargassum Contributing to Recent “Golden Tides” in the Western Atlantic. Ecol. Evol. 2017, 7, 516–525. [Google Scholar] [CrossRef]
- Dibner, S.; Martin, L.; Thibaut, T.; Aurelle, D.; Blanfuné, A.; Whittaker, K.; Cooney, L.; Schell, J.M.; Goodwin, D.S.; Siuda, A.N.S. Consistent Genetic Divergence Observed among Pelagic Sargassum Morphotypes in the Western North Atlantic. Mar. Ecol. 2022, 43, e12691. [Google Scholar] [CrossRef]
- Schell, J.M.; Goodwin, D.S.; Siuda, A.N.S. Recent Sargassum Inundation Events in the Caribbean: Shipboard Observations Revea Dominance of a Previously Rare Form. Oceanography 2015, 28, 8–10. [Google Scholar] [CrossRef]
- Sissini, M.N.; De Barros Barreto, M.B.B.; Széchy, M.T.M.; De Lucena, M.B.; Oliveira, M.C.; Gower, J.; Liu, G.; De Oliveira Bastos, E.; Milstein, D.; Gusmão, F.; et al. The Floating Sargassum (Phaeophyceae) of the South Atlantic Ocean—Likely Scenarios. Phycologia 2017, 56, 321–328. [Google Scholar] [CrossRef]
- van Tussenbroek, B.I.; Hernández Arana, H.A.; Rodríguez-Martínez, R.E.; Espinoza-Avalos, J.; Canizales-Flores, H.M.; González-Godoy, C.E.; Barba-Santos, M.G.; Vega-Zepeda, A.; Collado-Vides, L. Severe impacts of brown tides caused by Sargassum spp. on near-shore Caribbean seagrass communities. Mar. Pollut. Bull. 2017, 122, 272–281. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, R.E.; Medina-Valmaseda, A.E.; Blanchon, P.; Monroy-Velázquez, L.V.; Almazán-Becerril, A.; Delgado-Pech, B.; Vásquez-Yeomans, L.; Francisco, V.; García-Rivas, M.C. Faunal mortality associated with massive beaching and decomposition of pelagic Sargassum. Mar. Pollut. Bull. 2019, 146, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Resiere, D.; Valentino, R.; Nevière, R.; Banydeen, R.; Gueye, P.; Florentin, J.; Cabié, A.; Lebrun, T.; Mégarbane, B.; Guerrier, G.; et al. Sargassum seaweed on Caribbean islands: An international public health concern. Lancet 2018, 392, 2691. [Google Scholar] [CrossRef] [PubMed]
- Changeux, T. Sargasses transatlantiques cruise, RV Yersin. Fr. Oceanogr. Cruises 2017. [Google Scholar] [CrossRef]
- Thibaut, T. Sargasses. Fr. Oceanogr. Cruises 2017. [Google Scholar] [CrossRef]
- Winge, Ø. The Sargasso Sea, Its Boundaries and Vegetation. Report on the Danish Oceanographical Expeditions 1908–1910 to the Mediterranean and Adjacent Seas; A.F. Høst&søn.: Copenhagen, Denmark, 1923. [Google Scholar]
- Parr, A.E. Quantitative Observations on the Pelagic Sargassum Vegetation of the Western North Atlantic. Bull. Bingham Oceanogr. Collect. 1939, 6, 1–94. [Google Scholar]
- Siuda, A.N.S.; Blanfuné, A.; Dibner, S.; Verlaque, M.; Boudouresque, C.-F.; Connan, S.; Goodwin, D.S.; Stiger-Pouvreau, V.; Viard, F.; Rousseau, F.; et al. Morphological and Molecular Characters Differentiate Common Morphotypes of Atlantic Holopelagic Sargassum. Phycology 2024, 4, 256–275. [Google Scholar] [CrossRef]
- Pellegrini, M.; Valls, R.; Pellegrini, L. Chemotaxonomy and chemical markers in brown algae (in French). Lagascalia 1997, 19, 145–164. [Google Scholar]
- Le Lann, K.; Rumin, J.; Cérantola, S.; Culioli, G.; Stiger-Pouvreau, V. Spatiotemporal variations of diterpene production in the brown macroalga Bifurcaria bifurcata from the western coasts of Brittany (France). J. Appl. Phycol. 2014, 26, 1207–1214. [Google Scholar] [CrossRef]
- Jégou, C.; Culioli, G.; Stiger-Pouvreau, V. Meroditerpene from Cystoseira nodicaulis and its taxonomic significance. Biochem. Syst. Ecol. 2012, 44, 202–204. [Google Scholar] [CrossRef]
- Hernández-Bolio, G.I.; Fagundo-Mollineda, A.; Caamal-Fuentes, E.E.; Robledo, D.; Freile-Pelegrin, Y.; Hernández-Núñez, E. NMR metabolic profiling of Sargassum species under different stabilization/extraction processes. J. Phycol. 2021, 57, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.; Kervarec, N.; Cérantola, S. HRMAS NMR analysis of algae and identification of molecules of interest via conventional 1D and 2D NMR: Sample preparation and optimization of experimental conditions. Methods Mol. Biol. 2015, 1308, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Le Lann, K.; Kraffe, E.; Kervarec, N.; Cérantola, S.; Payri, C.E.; Stiger-Pouvreau, V. Isolation of turbinaric acid as a chemomarker of Turbinaria conoides (J. Agardh) Kützing from South Pacific Islands. J. Phycol. 2014, 50, 1048–1057. [Google Scholar] [CrossRef]
- Le Lann, K.; Kervarec, N.; Payri, C.E.; Deslandes, E.; Stiger-Pouvreau, V. Discrimination of allied species within the genus Turbinaria (Fucales, Phaeophyceae) using HRMAS NMR spectroscopy. Talanta 2008, 74, 1079–1083. [Google Scholar] [CrossRef]
- Jégou, C.; Culioli, G.; Kervarec, N.; Simon, G.; Stiger-Pouvreau, V. LC/ESI-MSn and 1H HR-MAS NMR analytical methods as useful taxonomical tools within the genus Cystoseira C. Agardh (Fucales; Phaeophyceae). Talanta 2010, 83, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Tanniou, A.; Vandanjon, L.; Gonçalves, O.; Kervarec, N.; Stiger-Pouvreau, V. Rapid geographical differentiation of the European spread brown macroalga Sargassum muticum using HRMAS NMR and Fourier-Transform Infrared spectroscopy. Talanta 2015, 132, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Jégou, C.; Kervarec, N.; Cérantola, S.; Bihannic, I.; Stiger-Pouvreau, V. NMR use to quantify phlorotannins. The case of Cystoseira tamariscifolia, a phloroglucinol-producing brown macroalga in Brittany (France). Talanta 2015, 135, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gobert, T.; Gautier, A.; Connan, S.; Rouget, M.L.; Thibaut, T.; Stiger-Pouvreau, V.; Waeles, M. Trace metal content from holopelagic Sargassum spp. sampled in the tropical North Atlantic Ocean: Emphasis on spatial variation of arsenic and phosphorus. Chemosphere 2022, 308, 136186. [Google Scholar] [CrossRef] [PubMed]
- Lalegerie, F.; Lajili, S.; Bedoux, G.; Taupin, L.; Stiger-Pouvreau, V.; Connan, S. Photo-protective compounds in red macroalgae from Brittany: Considerable diversity in mycosporine-like amino acids (MAAs). Mar. Environ. Res. 2019, 147, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Stengel, D.B. Intra-thallus differentiation of fatty acid and pigment profiles in some temperate Fucales and Laminariales. J. Phycol. 2015, 51, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Resuge, M.; Le Grand, F.; Brosset, P.; Lebigre, C.; Soudant, P.; Vagner, M.; Pecquerie, L.; Sardenne, F. Red muscle of small pelagic fishes’ fillets are high-quality sources of essential fatty acids. J. Food Compos. Anal. 2023, 120, 105304. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Sardenne, F.; Puccinelli, E.; Vagner, M.; Pecquerie, L.; Bideau, A.; Le Grand, F.; Soudant, P. Post-mortem storage conditions and cooking methods affect long-chain omega-3 fatty acid content in Atlantic mackerel (Scomber scombrus). Food Chem. 2021, 359, 129828. [Google Scholar] [CrossRef]
- Mathieu-Resuge, M.; Kraffe, E.; Le Grand, F.; Boens, A.; Bideau, A.; Lluch-Cota, S.E.; Racotta, I.S.; Schaal, G. Trophic ecology of suspension-feeding bivalves inhabiting a north-eastern Pacific coastal lagoon: Comparison of different biomarkers. Mar. Environ. Res. 2019, 145, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Ar Gall, E.; Lelchat, F.; Hupel, M.; Jégou, C.; Stiger-Pouvreau, V. Extraction and purification of phlorotannins from brown algae. Methods Mol. Biol. 2015, 1308, 131–143. [Google Scholar] [CrossRef]
- Stiger-Pouvreau, V.; Jégou, C.; Cérantola, S.; Guérard, F.; Le Lann, K. Chapter 13—Phlorotannins in Sargassaceae species from Brittany (France): Interesting molecules for ecophysiological and valorisation purposes. In Advances in Botanical Research; Bourgougnon, N., Ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2014; pp. 379–411. [Google Scholar]
- Zubia, M.; Fabre, M.S.; Kerjean, V.; Le Lann, K.; Stiger-Pouvreau, V.; Fauchon, M.; Deslandes, E. Antioxidant and antitumoural activities of some Phaeophyta from Brittany coasts. Food Chem. 2009, 116, 693–701. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Shen, J.; Silva, A.; Dennis, D.A.; Barrow, C.J. A simple 96-well microplate method for estimation of total polyphenol content in seaweeds. J. Appl. Phycol. 2006, 18, 445–450. [Google Scholar] [CrossRef]
- Cérantola, S.; Breton, F.; ar Gall, E.; Deslandes, E. Co-occurrence and antioxidant activities of fucol and fucophlorethol classes of polymeric phenols in Fucus spiralis. Bot. Mar. 2006, 49, 347–351. [Google Scholar] [CrossRef]
- Jégou, C.; Connan, S.; Bihannic, I.; Cérantola, S.; Guérard, F.; Stiger-Pouvreau, V. Phlorotannin and Pigment Content of Native Canopy-Forming Sargassaceae Species Living in Intertidal Rockpools in Brittany (France): Any Relationship with Their Vertical Distribution and Phenology? Mar. Drugs 2021, 19, 504–523. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R; Posit: Hong Kong, China, 2023. [Google Scholar]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster Analysis Basics and Extensions; R Package Version; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Bache, S.M.; Wickham, H. Magrittr: A Forward-Pipe Operator for R. 2020. Available online: https://cran.r-project.org/web/packages/magrittr/index.html (accessed on 18 April 2024).
- Wickham, H. Tidyr: Tidy Messy Data. 2021. Available online: https://tidyr.tidyverse.org/ (accessed on 18 April 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O'Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 18 April 2024).
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in {R}. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://rpkgs.datanovia.com/factoextra/ (accessed on 18 April 2024).
- Lê, S.; Josse, J.; Husson, F. {FactoMineR}: A package for multivariate analysis. J. Stat. Software 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Mangiafico, S. Rcompanion: Functions to Support Extension Education Program Evaluation. 2022. Available online: https://www.researchgate.net/publication/307608990_rcompanion_Functions_to_support_extension_education_program_evaluation_R_statistical_package (accessed on 18 April 2024).
- Dinno, A. Dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. 2017. Available online: https://cran.r-project.org/web/packages/dunn.test/dunn.test.pdf (accessed on 18 April 2024).
- Davis, D.; Simister, R.; Campbell, S.; Marston, M.; Bose, S.; McQueen-Mason, S.J.; Gomez, L.D.; Gallimore, W.A.; Tonon, T. Biomass composition of the golden tide pelagic seaweeds Sargassum fluitans and S. natans (morphotypes I and VIII) to inform valorisation pathways. Sci. Total Environ. 2021, 762, 143134. [Google Scholar] [CrossRef]
- Robledo, D.; Vázquez-Delfín, E.; Freile-Pelegrín, Y.; Vásquez-Elizondo, R.M.; Qui-Minet, Z.N.; Salazar-Garibay, A. Challenges and opportunities in relation to Sargassum events along the Caribbean Sea. Front. Mar. Sci. 2021, 8, 1000–1012. [Google Scholar] [CrossRef]
- Franks, J.S.; Johnson, D.R.; Ko, D.S. Pelagic Sargassum in the Tropical North Atlantic. Gulf Carib. Res. 2016, 27, 6–11. [Google Scholar] [CrossRef]
- Podlejski, W.; Berline, L.; Nerini, D.; Doglioli, A.; Lett, C. A new Sargassum drift model derived from features tracking in MODIS images. Mar. Poll. Bull. 2023, 188, 114629. [Google Scholar] [CrossRef]
- Putman, N.F.; Lumpkin, R.; Olascoaga, M.J.; Trinanes, J.; Goni, G.J. Improving transport predictions of pelagic Sargassum. J. Exp. Mar. Biol. Ecol. 2020, 529, 151398. [Google Scholar] [CrossRef]
- Stiger, V.; Horiguchi, T.; Yoshida, T.; Coleman, A.W.; Masuda, M. Phylogenetic Relationships of Sargassum (Sargassaceae, Phaeophyceae) with Reference to a Taxonomic Revision of the Section Phyllocystae Based on ITS-2 nrDNA Sequences. Phycol. Res. 2000, 48, 251–260. [Google Scholar] [CrossRef]
- Yoshida, T.; Stiger, V.; Horiguchi, T. Sargassum boreale sp. Nov. (Fucales, Phaeophyceae) from Hokkaido, Japan. Phycol. Res. 2000, 48, 125–131. [Google Scholar] [CrossRef]
- Salaün, S.; Kervarec, N.; Potin, P.; Haras, D.; Piotto, M.; La Barre, S. Whole-cell spectroscopy is a convenient tool to assist molecular identification of cultivatable marine bacteria and to investigate their adaptive metabolism. Talanta 2010, 80, 1758–1770. [Google Scholar] [CrossRef]
- Obando, C.Z.; Linossier, I.; Kervarec, N.; Zubia, M.; Turquet, J.; Faÿ, F.; Rehel, K. Rapid identification of osmolytes in tropical microalgae and cyanobacteria by 1H HR-MAS NMR spectroscopy. Talanta 2016, 153, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, G.B.; Barison, A.; Ferreira, A.G.; Honda, N.K.; Ferreira, M.M.C. Chemotaxonomic Discrimination of Lichens by 1H NMR, solution and HR-MAS, and Chemometric Analysis. Ann. Magn. Reson. 2006, 5, 1–4. [Google Scholar]
- Dutra, L.M.; da Conceicao Santos, A.D.; Lourenço, A.V.F.; Nagata, N.; Heiden, G.; Campos, F.R.; Barison, A. 1H HR-MAS NMR and chemometric methods for discrimination and classification of Baccharis (Asteraceae): A proposal for quality control of Baccharis trimera. J. Pharm. Biomed. Anal. 2020, 184, 113200. [Google Scholar] [CrossRef]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef]
- Van Ginneken, V.J.T.; Helsper, J.P.F.G.; De Visser, W.; van Keulen, H.; Brandenburg, W.A. Polyunsaturated fatty acids in various macroalgal species from north Atlantic and tropical seas. Lipids Health Dis. 2011, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Milledge, J.J.; Maneein, S.; López, E.A.; Bartlett, D. Sargassum inundations in Turks and Caicos: Methane potential and proximate, ultimate, lipid, amino acid, metal and metalloid analyses. Energies 2020, 13, 1523. [Google Scholar] [CrossRef]
- Turner, J.P.; Rooker, J.R. Fatty acid composition of flora and fauna associated with Sargassum mats in the Gulf of Mexico. Mar. Biol. 2006, 149, 1025–1036. [Google Scholar] [CrossRef]
- Lalegerie, F.; Gager, L.; Stiger-Pouvreau, V.; Connan, S. The stressful life of red and brown seaweeds on the temperate intertidal zone: Effect of abiotic and biotic parameters on the physiology of macroalgae and content variability of particular metabolites. Adv. Bot. Res. 2020, 95, 247–287. [Google Scholar] [CrossRef]
- Stiger-Pouvreau, V.; Deslandes, E.; Payri, C.E. Phenolic contents of two brown algae, Turbinaria ornata and Sargassum mangarevense on Tahiti (French Polynesia): Interspecific, ontogenic and spatio-temporal variations. Bot. Mar. 2004, 47, 402–409. [Google Scholar] [CrossRef]
- Nakai, M.; Kageyama, N.; Nakahara, K.; Miki, W. Phlorotannins as radical scavengers from the extract of Sargassum ringgoldianum. Mar. Biotechnol. 2006, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Gu, Y.; Zhang, C.; Sun, C.; Qin, L.; Yu, C.; Qi, H. Metabolomic approach for characterization of polyphenolic compounds in Laminaria japonica, Undaria pinnatifida, Sargassum fusiforme and Ascophyllum nodosum. Foods 2021, 10, 192. [Google Scholar] [CrossRef]
- Taniguchi, R.; Ito, C.; Keitoku, S.; Miyake, Y.; Itoigawa, M.; Matsui, T.; Shibata, T. Analysis on the structure of phlorethols isolated from the warm-temperate brown seaweed Sargassum carpophyllum and their antioxidant properties. Nat. Prod. Com. 2022, 17, 1934578X221109406. [Google Scholar] [CrossRef]
- Urrea-Victoria, V.; Furlan, C.M.; dos Santos, D.Y.A.C.; Chow, F. Antioxidant potential of two Brazilian seaweeds in response to temperature: Pyropia spiralis (red alga) and Sargassum stenophyllum (brown alga). J. Exp. Mar. Biol. Ecol. 2022, 549, 151706. [Google Scholar] [CrossRef]
- Agregán, R.; Munekata, P.E.S.; Franco, D.; Dominguez, R.; Carballo, J.; Lorenzo, J.M. Phenolic compounds from three brown seaweed species using LC-DAD–ESI-MS/MS. Food Res. Internat. 2017, 99, 979–985. [Google Scholar] [CrossRef]
- Glombitza, K.W.; Forster, M.; Farnham, W.F. Antibiotics from algae—Part 25: Polyhydroxyphenyl ethers from the brown alga Sargassum muticum (Yendo) Fensholt, Part II. Bot. Mar. 1982, 25, 449–454. [Google Scholar] [CrossRef]
- Montero, L.; Sánchez-Camargo, A.P.; García-Cañas, V.; Tanniou, A.; Stiger-Pouvreau, V.; Russo, M.; Rastrelli, L.; Cifuentes, A.; Herrero, M.; Ibáñez, E. Anti-proliferative activity and chemical characterization by comprehensive two-dimensional liquid chromatography coupled to mass spectrometry of phlorotannins from the brown macroalga Sargassum muticum collected on North-Atlantic coasts. J. Chrom. A 2016, 1428, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Tanniou, A.; Vandanjon, L.; Incera, M.; Serrano Leon, E.; Husa, V.; Le Grand, J.; Nicolas, J.-L.; Poupart, N.; Kervarec, N.; Stiger-Pouvreau, V.; et al. Assessment of the spatial variability of phenolic contents and associated bioactivities in the invasive alga Sargassum muticum sampled along its European range from Norway to Portugal. J. Appl. Phycol. 2014, 26, 1215–1230. [Google Scholar] [CrossRef]
- Catarino, M.D.; Pires, S.M.G.; Silva, S.; Costa, F.; Braga, S.S.; Pinto, D.C.G.A.; Silva, A.M.S.; Cardoso, S.M. Overview of Phlorotannins’ Constituents in Fucales. Mar. Drugs 2022, 20, 754. [Google Scholar] [CrossRef] [PubMed]
- Chouh, A.; Nouadri, T.; Catarino, M.D.; Silva, A.M.; Cardoso, S.M. Phlorotannins of the brown algae Sargassum vulgare from the Mediterranean Sea coast. Antioxidants 2022, 11, 1055. [Google Scholar] [CrossRef] [PubMed]
- Glombitza, K.W.; Keusgen, M.; Hauperich, S. Fucophlorethols from the brown algae Sargassum spinuligerum and Cystophora torulosa. Phytochemistry 1997, 46, 1417–1422. [Google Scholar] [CrossRef]
- Glombitza, K.W.; Keusgen, M. Fuhalols and deshydroxyfuhalols from the brown alga Sargassum spinuligerum. Phytochemistry 1995, 38, 987–995. [Google Scholar] [CrossRef]
- Keusgen, M.; Glombitza, K.W. Pseudofuhalols from the brown alga Sargassum spinuligerum. Phytochemistry 1997, 46, 1403–1415. [Google Scholar] [CrossRef]
- Keusgen, M.; Glombitza, K.W. Phlorethols, fuhalols and their derivatives from the brown alga Sargassum spinuligerum. Phytochemistry 1995, 38, 975–985. [Google Scholar] [CrossRef]
- Glombitza, K.W.; Hauperich, S.; Keusgen, M. Phlorotannins from the brown algae Cystophora torulosa and Sargassum spinuligerum. Nat. Tox. 1997, 5, 58–63. [Google Scholar] [CrossRef]
- Li, Y.; Fu, X.; Duan, D.; Liu, X.; Xu, J.; Gao, X. Extraction and identification of phlorotannins from the brown alga, Sargassum fusiforme (Harvey) Setchell. Mar. Drugs 2017, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Changeux, T.; Berline, L.; Podlejski, W.; Guillot, T.; Stiger-Pouvreau, V.; Connan, S.; Thibaut, T. Variability in growth and tissue composition (CNP, natural isotopes) of the three morphotypes of holopelagic Sargassum. Aq. Bot. 2023, 187, 103644. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).