Abstract
This manuscript summarizes information on the diverse range of RNA molecules and their role as competing endogenous RNAs (ceRNAs). Moreover, it provides an overview of ceRNA regulatory networks and their applications in ruminant biology. Knowledge of co-expression networks has increased with microarrays, RNA-seq, and scRNA-seq characterizing molecular mediators across various biological scales, using sequences from numerous blood and tissue samples. By synthesizing existing knowledge, this study summarizes interactions between coding and non-coding RNAs through microRNA response elements (MREs), elucidating large-scale regulatory networks throughout the transcriptome that influence the expression and activities of various ceRNAs. Identification of non-coding RNAs with important regulatory functions will revolutionize understanding of RNA biology, shifting from an mRNA-centric model to a complex network of RNA crosstalk. The ceRNA networks offer a more comprehensive and arguably more realistic perspective compared to protein–protein interaction (PPI) networks and weighted gene co-expression networks (WGCN). These ceRNA regulatory networks can describe potential molecular regulatory mechanisms related to functional and economically important traits in ruminants, plus contribute to disease and pathology research, by elucidating pathogenesis and potential drug effects in disease and cancer models. Furthermore, they can provide insights into farm animal biology, e.g., reproductive traits in goats and sheep, regulation of fat metabolism in beef cattle, heat stress responses, and lactation regulation in dairy cattle, fertility and muscle characteristics in buffalo, and resistance to high-salt and water-deprivation conditions in camels. In conclusion, ceRNA and associated regulatory networks should promote a new understanding of molecular mechanisms and identify candidate genes and metabolic-signaling pathways in ruminants.
Keywords:
ruminant breeding; circRNAs; competing endogenous RNA; lincRNAs; lncRNAs; miRNAs; gene expression 1. Introduction
A broad range of RNA molecules, including both coding and non-coding RNA (ncRNA), have been categorized, leading to the identification of an increasingly diverse set of subgroups and expanding our understanding of RNA’s complexity and diversity. Furthermore, these molecules have been the focus of specialized investigations and numerous applied annotation resources [1,2]. The most prominent RNA subtypes are ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and coding messenger RNAs (mRNAs). In addition, a recent study has highlighted pseudogenes, long non-coding RNAs (lncRNAs), long interfering non-coding RNAs (lincRNAs), and circular RNAs (circRNAs) [3]. Several studies demonstrated that non-coding elements represented the largest portions of transcribed molecules [4], with important roles in a wide range of biological processes in cells [5], cell-fate programming [6], cellular aging [7], and infectious diseases [8].
The lncRNAs are RNA molecules > 200 bp that regulate mRNA stability or inhibit mRNA via a competitive endogenous RNA (ceRNA) mechanism [6,9]. CircRNAs, a recently identified class of RNA molecules, have a unique closed-loop structure that imparts stability and enables them to serve as miRNA sponges. Like lncRNAs, they are also capable of functioning as competitive endogenous RNAs (ceRNAs). Recent studies revealed that pseudogenes (along with microRNAs (miRNAs)), although previously considered inactive “genomic fossils” have important roles in regulating cognate genes [10]. miRNAs are a type of small non-coding RNA, usually 20–22 nucleotides [11,12]. These molecules become part of a multi-protein complex, RNA-induced silencing complex (RISC), accountable for targeting special transcripts; miRNAs can either interfere with the translation of these transcripts or promote their destruction [13]. Recognition of a target is facilitated by a mechanism that relies on partial sequence complementarity. This mechanism involves a short sequence known as the miRNA response element (MRE), present in the transcript, and the “seed” sequence on the miRNA, a 6–8 nucleotide sequence highly conserved across species. The regulatory mechanism of this process is complex; thus, various miRNAs can affect the expression of many target transcripts, and conversely, multiple miRNAs can regulate each transcript with various MREs [14]. As minor mutations can influence this mechanism, modifying a seed sequence can affect targets regulated by a specific miRNA, whereas altering the MRE can eliminate a target from miRNA modulation [15].
Many studies have uncovered the roles of non-coding RNA molecules, including lncRNAs, circRNAs, and pseudogenes, in cellular processes. These molecules have MREs and can be targeted by miRNAs, implying a potential role as ceRNAs. The ceRNA regulatory networks represent a novel mechanism of interaction among RNAs and have crucial roles in multiple biological processes and essential cellular functions [16,17]. Moreover, they hold promise for a wide range of improving complex traits and mitigating diseases, e.g., increasing the productive and reproductive capabilities of ruminants, improving quality of ruminant products, and reducing the incidence of disease in ruminants [18]. This article consolidates information on a diverse range of RNA molecules and their role as ceRNAs in response to research questions related to understanding insights into molecular genetic factors and mechanisms associated with complex traits and diseases of ruminants. Moreover, it offers an overview of ceRNA theory, biological networks (including protein–protein interaction (PPI), weighted gene co-expression network analysis (WGCNA), and ceRNA regulatory networks), and their applications in animal science.
2. Literature Search Strategy to Identify Studies Associated with ceRNA Networks in Ruminants
Various online search databases and papers, including Web of Science and PubMed (most recently accessed on 8 March 2024), from 2010 to 2024 were used to discover ceRNA studies relevant to comprehensive literature mining with no language restrictions. Search terms consisted of both keywords and database-specific subject headings for the ceRNA regulatory network, complex traits, and diseases in ruminants: Breeds—dairy cattle, beef cattle, sheep, goats, buffalo, and camel; practical tools—RNA-seq and microarray technologies; and outcome—regulatory RNAs or ceRNA networks—complex traits and diseases. Keywords included ruminants (dairy cattle, beef cattle, sheep, goats, buffalo, and camels), complex traits, diseases, regulatory RNAs (circRNA, lincRNA, lncRNA, miRNA), mRNAs, and ceRNA networks. For this purpose, first, identifiers and synonyms for each framework element were merged by applying the boolean operator “OR”. Then, elements of the framework were merged by applying the boolean operator “AND”. In total, 65 relevant papers were identified using online search databases. All identified papers were imported into Covidence (Covidence systematic review software, Veritas Health Innovation), and duplicates were removed. Two researchers independently evaluated the studies to assess suitability; articles were further screened for relevant references and a citation check was performed. After the final screening, 26 candidate papers with the final qualified literature related to ceRNA networks were chosen (Table 1).

Table 1.
Phenotype, species, breed, and origin of data (country) for 26 articles used in the study.
3. Differential Gene Expression Analysis and Its Role in Economically Complex Traits and Diseases
Physiological processes are contingent upon the coordinated expression of numerous genes acting in concert. In recent years, considerable effort has been expended in examining mechanisms that govern gene expression and their regulation by biological and external factors, such as genetic determinants, nutritional and physiological factors, and animal management [45]. Various scientific disciplines have independently studied DNA, RNA, proteins, and biological functions, using various approaches (e.g., molecular genetics, biochemistry, metabolism, and molecular biology). Research has concentrated on understanding mechanisms that control gene expression, including the effects of both internal and external factors on gene expression. These factors include genetic determinants and nutritional elements in tissues associated with metabolism, reproduction, growth, and milk and meat production traits [46].
Differential gene expression can provide insights into biological differences between two groups of samples, e.g., control and case. One specific focus is differential gene expression analysis of different samples (e.g., from diseased and healthy animals), which involves the identification of genes that are differentially expressed in complex traits and diseases [47]. Differentially expressed genes can be valuable for identifying significant biomarkers, a monitor for tracking health status, and gene signatures for diagnostics. Reporting previous literature that associates gene expression analysis with the incidence of some economically complex traits and diseases can affect our appreciation of biological processes and mechanisms associated with complex traits and diseases in ruminants [48]. Gene expression profiling at the mRNA or protein levels provides a deeper understanding of the regulation of physiological functions in livestock species [47,49]. The abundance of transcripts is considered a heritable endophenotype and is linked to chromosomal polymorphisms, according to genetic genomics theory [50]. Based on this approach, combining information on chromosomal variants and gene expression could improve our understanding of the genetic basis underlying the onset of a disease in ruminants. Quantitative trait loci (QTLs) are polymorphisms associated with gene expression [51,52,53].
Two primary strategies have been developed to detect genes of significance. The first has focused on the expression of candidate genes for key physiological pathways at both transcript and protein levels. With the advent of genomics, a second original strategy has emerged, which addresses the same issues by examining the molecular signatures of all genes and proteins utilizing high-throughput techniques, e.g., transcriptomics, proteomics, and metabolomics [45]. Despite the decreasing costs of next-generation sequencing, transcriptomic studies in livestock are frequently conducted on limited samples due to financial constraints, reducing their ability to detect significant differentially expressed genes (DEGs). However, using meta-analysis techniques to combine and analyze data from multiple related studies enhances statistical power and the robustness of the results [54,55,56]. In the future, the information acquired from these studies could be integrated to optimize livestock production systems. This could be achieved by identifying desirable animals, propagating them, and integrating them into innovative management systems [57,58].
4. Competing Endogenous RNA (ceRNA) Theory
In 2011, Pier Paolo Pandolfi’s research group at Harvard Medical College proposed the ceRNA theory that miRNAs regulate various transcript components by identifying target sites on RNA molecules [59]. Complementary RNA molecules, including lncRNAs, circRNAs, and pseudogenes, can modify miRNA regulation by acting as ceRNAs and competitively binding miRNAs. This action prevents them from inhibiting their target genes and has an important role in various biological conditions, providing additional post-transcription gene regulation. The ceRNA theory offers a fresh perspective on the role of non-coding RNAs and expands the number of regulatory possibilities for the 3′ untranslated region (UTR) (Figure 1) of genes [60]. Furthermore, when multiple RNAs have common MREs on their 3′UTR regions, they may be targeted by the same miRNA(s), leading to indirect cross-regulation, as they compete for the same sequences [61], thereby enabling transcripts to communicate with each other and regulate their expression using a specific language where MREs represent codes (Figure 2). Some pairs of genes share common binding miRNAs, small RNA molecules that regulate gene expression. These pairs of genes are called ceRNAs, and they can interact with each other through competition for a limited pool of miRNAs. When one ceRNA’s expression level increases in a tissue, it attracts the miRNAs away from the other ceRNAs, protecting the expression of its gene partner. Conversely, if one ceRNA’s expression level decreases in a tissue, it releases the miRNAs toward the other ceRNAs, degrading the expression of its gene partner. This mechanism of ceRNA regulation provides an alternative way for genes to regulate each other without direct interactions [62]. It has a crucial role in essential biological processes and molecular functions. In this regard, almost all families of RNA molecules can participate in regulatory strategies, extending gene regulation network relationships and adding a new layer of indirect interactions [14].
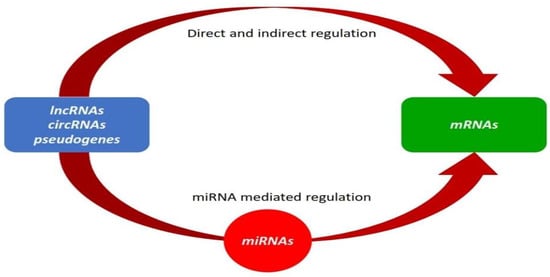
Figure 1.
mRNA transcription and post-transcription can be regulated through both direct and indirect mechanisms involving lncRNAs, circRNAs, and pseudogenes. Some modulate transcription rates via nuclear RNA–RNA complexes, whereas others affect mRNA stability in the cytoplasm. Additionally, the ceRNA mechanism offers parallel and complementary control through protein–coding and non-coding RNAs. Rather than direct interactions, ceRNA is mediated by miRNA-binding competition [18].
Bioinformatics tools and their predictions demand caution in accepting these suggestions derived from indirect biological evidence and statistical methods, but they demonstrate that molecules can participate in regulatory strategies and, at the same time, offer numerous relationships that can be tested experimentally [63]. It is common to use combined experimental strategies, including bioinformatics analyses, to identify relevant biomarkers by conducting studies on animal models due to specific metabolic characteristics, as a proxy for other species, and then use human cell lines for a better understanding of cellular processes and to support the hypothesis [64]. All types of RNA and their associated information, including protein and RNA expression and co-expression networks, miRNA target predictions, as well as other layers of gene regulation such as transcription factor activity, epigenetics, and genomic topological knowledge, should be considered to enhance understanding of molecular and cellular mechanisms in ruminant bioscience. Future studies should be integrated with previous information on ceRNAs to make the wealth of biological interconnections immediately accessible. This will benefit research on various cells, tissues, organs, stages of development, and complex traits in ruminants [20,39]. It was recently reported that lncRNA- and circRNA-mediated ceRNA networks have an essential role in numerous biological processes, with species distinctions in goats [39], sheep [34], beef [20], and dairy cattle [28].
It has become increasingly evident that regulation of gene expression through competition for miRNA binding is crucial. Understanding gene regulation at this complexity can provide a functional explanation linking known and novel genes that lack direct interactions. To enhance understanding of the role of ceRNAs and their regulatory networks in complex traits and diseases of ruminants, more needs to be known about [14,65]:
- Rates of production and turnover of miRNAs, their target RNAs, and how ceRNAs can determine how much and for how long genes are regulated. Therefore, there must be significant variations in the expression of ceRNAs to ease miRNA repression of target mRNAs.
- How the expression level of sequestered miRNAs (in very low or abundant conditions) can override competition.
- How competition among ceRNAs is affected by various factors such as the number of miRNAs they can sponge, their subcellular distribution, and their interactions with RNA-binding proteins and ribosomes. For competition to occur, ceRNAs and miRNAs must be concurrently present in the same tissue, cell type, or cell compartment.
- How the nucleotide composition of MREs on ceRNAs alters the efficiency of binding a specific miRNA.
The regulatory mechanisms of the competing endogenous RNA (ceRNA) network are highly intricate, with ceRNA activity affected by several factors (e.g., ceRNA/miRNA-binding affinity, RNA editing, and the abundance and subcellular localization of ceRNA components) [66]. Among these factors, RNA editing is an important post-transcriptional modification mechanism that alters RNA molecules by modifying their sequences by inserting, deleting, or converting nucleotides, generating genetic consequences akin to DNA mutations at the genomic level [67,68]. The predominant type of RNA editing is the conversion of adenosine (A) nucleotides to inosines (I); in this conversion, enzymes encoded by the adenosine deaminase acting on RNA (ADAR) gene family catalyze the deamination of A to I nucleotides [69]. Increased diversity of protein isoforms and their respective functions can be the result of RNA editing in pre-mRNA coding regions, which is related to changes in codons [70]. In this regard, analysis of the downstream functional consequences for RNA editing sites may result in different steps, including: (1) Elucidating the influence of RNA editing sites on amino acid sequences and alternative splicing events; (2) determining the impacts of RNA editing sites on miRNA target binding; and (3) assessing effects of RNA editing sites on corresponding gene expression levels [71]. As a result, the change in an RNA molecule following RNA editing technology can drastically change its function at the proteomics level as the type of encoded amino acid, resulting in the creation of ceRNA regulatory networks with new or different functions, which regulate gene expression for different traits [72,73]. Genome and transcriptome editing techniques have been used successfully in various types of livestock, including goats, cattle, and sheep [74,75,76]. These techniques have been used in dairy cattle to modify the genome and produce hypoallergenic milk types, e.g., milk containing less β-lactoglobulin protein [77]. Additionally, genome editing generated mastitis-resistant cattle, which has improved mammary gland health [78]. Therefore, combining genomic prediction with genome and transcriptome editing techniques could be beneficial for complex traits and diseases [79].
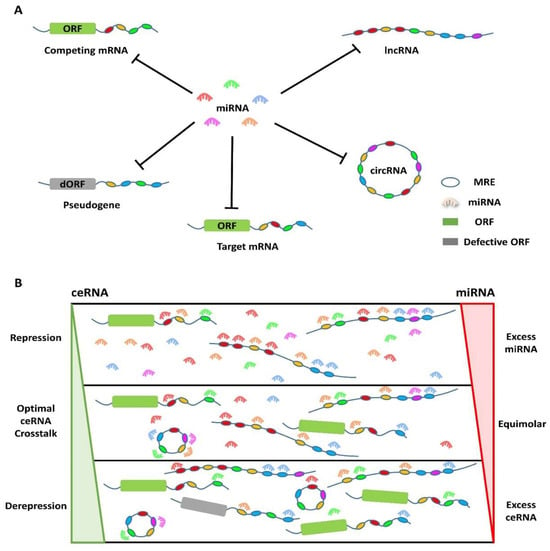
Figure 2.
ceRNA networks are composed of various kinds of RNAs, including mRNA, miRNA, lncRNA, circRNA, and pseudogene. (A) In these networks, miRNAs bind to specific MREs, which are present on coding RNAs (target mRNA and competing mRNA) as well as non-coding RNAs (miRNA, lncRNA, circRNA, and pseudogenes) to suppress gene expression. RNAs that share MREs for the same miRNAs can compete for binding to a common set of miRNAs, which reduces their availability. Transcripts containing multiple MREs can cooperatively bind miRNAs, promoting more effective competition. MREs are denoted by colored ovals that correspond to their targeting miRNAs; (B) overall conditions for optimal ceRNA crosstalk are outlined. Target repression occurs when miRNAs exceed their targets, whereas target depression results from excess ceRNAs and/or limited miRNA. For optimal ceRNA crosstalk, equimolar miRNA and target concentrations are required [80,81].
5. Biological Networks: From PPI Networks to WGCNA and ceRNA Regulatory Networks
Systems biology uses bioinformatics approaches to study cells, organs, and organisms in a systemic, integrated manner, especially cellular processes such as molecular interactions, intercellular communication, cell division, homeostasis, and environmental adaptation. Increasing availability and use of high-throughput data and a growing emphasis on systematic research have increased interest in systems biology [57,82]. Proteins have crucial roles in biological processes, regulating molecular and cellular mechanisms that affect well-being and disease susceptibility. As proteins generally impact biological processes as components of protein complexes, understanding protein–protein interactions is key to elucidating protein function within the cell. PPI networks characterize physical interactions between proteins that facilitate assembly into complexes that mediate regulatory, signaling, transport, and other events. Analyzing PPI networks can thus identify protein complexes, functional modules, and various signaling pathways. In summary, delineating PPI networks provides insights into the orchestration of proteins into functional units that control cell biology [83].
Weighted gene co-expression network analysis (WGCNA) represents a framework to apply systems biology approaches to transcriptomic data [84,85]. Rather than analyzing the expression of individual genes, WGCNA focuses on the coordinated co-expression of gene sets. This technique constructs weighted gene co-expression networks by clustering genes into modules displaying coordinated expression patterns. Module-based analysis correlates these co-expressed gene modules, rather than individual genes, with traits of interest. Consequently, WGCNA implements a shift towards systems-level analysis of gene expression by emphasizing co-expression modules rather than individual genes in relation to biological traits and outcomes. This network-based approach provides insights into higher-order organizations and relationships of transcriptomic data [82].
A comparison between weighted gene co-expression networks (WGCN) and PPI networks reveals complementary strengths and limitations. WGCN leverages transcriptomic data to construct an integrated overview of modular gene expression patterns and links these to phenotypes. This systems-level perspective benefits from the wide availability and high throughput of transcriptome profiling. PPI networks provide more detailed biochemical information about direct interactions between proteins, albeit with reduced coverage compared to other approaches. Whereas WGCN provides a macro-level understanding of coordinated gene activity, PPI networks detail molecular mechanisms underpinning biological systems; together, these two network-based approaches can supply valuable and distinct characterization of genomic regulation and protein activities [86].
Identification of abundant regulatory non-coding RNAs, such as miRNAs, lncRNAs, and circRNAs, has prompted a shift from the traditional notion of mRNAs being the exclusive biological regulators. This revelation expanded post-transcriptional control beyond modulating mRNA levels. Appreciating these overlooked non-coding RNA functions facilitated the emergence of the ceRNA theory, transforming RNA biology from an mRNA-centric model to a complex network of RNA crosstalk [60,87]. In summary, ceRNA networks can provide a more complete and realistic insight than the previous two types of networks; they encompass both protein–protein interactions (physical and chemical associations between proteins) plus synchronized expression of various coding and non-coding RNAs. This comprehensive approach provides a more holistic understanding of cellular processes. In addition, ceRNA networks model miRNA-mediated crosstalk between RNAs and can provide insights into post-transcriptional regulation and RNA crosstalk affecting many complex traits and diseases in ruminants [60].
Overall, by considering interactions and regulatory relationships between different types of RNAs, ceRNA networks offer a more comprehensive perspective compared to PPI and WGC networks, because ceRNA networks contain different types of RNAs (e.g., mRNAs, lncRNAs, and circRNAs), which can regulate each other by competing for shared miRNA binding sites. Also, this approach provides more comprehensive insights into gene regulation in transcriptomic research. In addition, these networks capture complex regulatory relationships among various regulatory RNAs, which can influence various metabolic and signaling pathways [20,39]. This analysis also reveals novel regulatory mechanisms and interactions that PPI and WGC networks may not capture clearly, as these networks focus on disparate levels of gene regulation [82]. Other applications of these networks provide insights into the functional roles of ncRNAs, implicated in various biological processes, complex traits, and diseases in ruminants. Finally, this approach should reveal potential biomarkers and therapeutic targets by identifying significant regulatory RNAs and their interactions. Table 2 represents a brief overview of the comparison these three types of networks.

Table 2.
Summary of differences among ceRNA, PPI, and WGC networks.
6. Applications of ceRNA Regulatory Network in Animal Biosciences
Competing endogenous RNA (ceRNA) network analysis is increasingly used across diverse interdisciplinary research. Many studies have demonstrated the application of the ceRNA regulatory network. For example, Zhang et al. [88] used ceRNA networks to elucidate mechanisms in disease pathology, particularly cancer research. Li et al. [89] investigated physiologic cellular and tissue function and adopted ceRNA networks to identify pathogenesis. In pharmaceutical studies, ceRNA networks have shed light on drug effects in cancer models [90,91]. Additionally, ceRNA network analysis contributed to gene function annotation and the characterization of oncology, including brain tumors [92,93].
There is growing evidence that non-coding RNAs are emerging as important regulators of a variety of economically important and complex traits in ruminants. Therefore, the role of ceRNA regulatory networks is very important to understand the regulatory effects of non-coding RNAs in animal studies. More recently, ceRNA network analyses have been used in many complex traits and diseases of ruminants (various species) to identify genes, metabolic pathways, and signaling interactions and to investigate mechanisms of RNA macromolecule interactions in biological processes [20,39].
6.1. Dairy Cattle
According to Zeng et al. [28], in tissues related to the hypothalamus-pituitary-mammary axis (HPM) of heat-stressed (HS) and control cows, a total of 1680, 1112, and 521 DE circRNAs, 120, 493, and 108 DE miRNAs, 274, 6475, and 3134 DE mRNAs were identified in hypothalamus, pituitary, and mammary gland tissues, respectively. They also identified the MAPK signaling pathway as a key pathway in heat stress response and lactation regulation. Therefore, in various species, ceRNA regulatory networks have been used to identify molecular regulatory mechanisms related to functional and complex traits. In another study on milk fat in dairy cattle [27], 290 circRNAs were significantly differentially expressed in high milk fat percentage (HMF) cows compared to low milk fat percentage (LMF) cows, of which 142 were significantly up-regulated and 148 were significantly down-regulated. Also, enrichment analysis identified four DE circRNAs (circ_0001122, circ_0007367, circ_0018269, and circ_0015179) that potentially regulate milk fat metabolism. Furthermore, the regulatory interactions of circ_0001122:miR-12043:LIPG, circ_0007367:miR-331-3p:CIDEA/PML, and circ_0018269:miR-11989:RORC/HPX were the potential interactions in the ceRNA network to explore the mechanism of milk fat regulation. In research performed by Wang et al. [26], 8003 mRNAs, 579 lncRNAs, and 205 miRNAs were differentially expressed in granulosa cells (GCs) of cows with cystic versus normal ovarian follicles. Concerning GO and KEGG pathway analysis as well as the ceRNA regulatory network analysis, it was confirmed that NONBTAT027373.1 LncRNA sponged miR-664b in GCs and prevented miR-664b from binding to the HSD17B7 3′-UTR. It was concluded that mRNAs and lncRNAs relevant to steroid hormone synthesis and energy metabolism could have important roles in the development of cystic ovarian follicles through the ceRNA mechanism and represent candidate targets for further research.
6.2. Beef Cattle
Huang et al. [24] investigated differences in muscle growth between the offspring of a cow–yak crossbred versus a yak, with 7126 mRNAs, 791 lncRNAs, and 1057 circRNAs identified as differentially expressed (DE) RNAs. Also, DE RNAs were significantly enriched in myoblast differentiation and some signaling pathways related to muscle growth (e.g., HIF-1 and PI3K-AKT signaling pathways). A study conducted by Dehghanian Reyhan et al. [20] that analyzed regulation of intramuscular fat content (IMF) and fat metabolism in five breeds of beef cattle (Aberdeen Angus, Chinese Simmental, Luxi, Nanyang, and Shandong Black) identified 34 circRNA, 57 lncRNA, 15 miRNA, and 374 mRNA that have a major role in this process. The research also identified seven key subnets that consist of 16 circRNAs, 43 lncRNAs, 7 miRNAs, and 237 mRNAs through cluster analyses. Furthermore, 48, 13, and 28 significantly enriched GO terms were related to IMF in biological process, molecular function, and cellular component categories, respectively. Moreover, the study identified several metabolic-signaling pathways associated with IMF, including metabolic, calcium, cGMP-PKG, thyroid hormone, and oxytocin signaling pathways. Finally, MCU, CYB5R1, and BAG3 genes were introduced as important candidate marker genes for fat metabolism in beef cattle.
6.3. Sheep
Sadeghi et al. [34] identified important RNAs and pathways related to sheep fertility, and by combining GO and KEGG enrichment analyses, they identified 264 mRNAs, 14 lncRNAs, and 34 miRNAs. Through ceRNA network clustering, they further obtained 44 mRNAs, 7 lncRNAs, 7 miRNAs, and 6 critical modules. The identified RNAs that were overexpressed were involved in actin cytoskeleton organization, cell adhesion, proteolysis, cell differentiation, immune system process, and lipid metabolic process (Padj < 0.01). To discover molecular mechanisms involved in the formation of muscle fibers, Cui et al. [33] compared 2 muscle tissues, longissimus dorsi and biceps femoris of Tan versus Dorper sheep breeds. Transcriptome analysis of these sheep revealed 214 DE lncRNAs, 25 DE mRNAs, 4 DE miRNAs, and 91 DE circRNAs for the longissimus dorsi muscle and 172 DE lncRNAs, 35 DE mRNAs, 12 DE miRNAs, and 95 DE circRNAs biceps femoris tissues. Also, in GO and KEGG pathway analyses, Ca2+, FoxO, and AMPK signaling pathways were enriched for the DE RNAs associated with muscle fiber formation. Finally, a total of 10 lncRNAs, 12 miRNAs, 20 circRNAs, and 19 genes formed lncRNA–circRNA–miRNA–mRNA ceRNA regulatory networks, indicating that muscle fiber formation in sheep was controlled by complex regulatory networks of coding and non-coding genes. Moreover, the ACACB, ATP6V0A1, ASAH1, EFHB, MYL3, C1QTNF7, SFSWAP, and FBXL5 genes were identified as key genes in muscle fiber formation. According to Cui et al. [33], ceRNA subnetworks may be critical in regulating muscle fiber development and are a valuable resource for future studies of muscle fiber development in sheep. According to the Bao et al. [35] study on the meat quality of longissimus thoracis (LT) muscle in Tibetan sheep at various growth stages, meat tenderness decreased (p < 0.05) with age. Furthermore, functional annotation analysis of the DE RNAs was mainly enriched in protein binding, and myofibril and organelle assembly. In addition, DE RNAs were mainly involved in the adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) and calcium signaling pathways. Also, circRNA–miRNA–mRNA ceRNA network analysis revealed that circRNAs such as circ_000631, circ_000281, and circ_003400 interacted with miR-29-3p and miR-185-5p to regulate expression of LEP, SCD, and FASN genes relevant to the transformation of muscle fiber types in the AMPK signaling pathway. With increasing age, oxidized muscle fibers transform into glycolytic muscle fibers, intramuscular fat content (IMF) decreases, muscle fiber diameter in glycolytic muscle fibers increases, and finally meat tenderness increases. Additionally, this study validated the significant role of circRNAs in the conversion of muscle fiber types in Tibetan sheep and their impacts on meat quality.
6.4. Goat
Shang et al. [40] investigated the circRNA, miRNA, and mRNA expression profiles of the skin of Inner Mongolia cashmere goat fetuses (days 45, 55, 65, and 75) to discover signaling molecules and pathways associated with hair follicle development. In total, 113 DE circRNAs related to the development of secondary hair follicles were identified. Following principles governing ceRNA regulatory networks, a ceRNA regulatory network was constructed, composed of 13 circRNAs, 21 miRNAs, and 110 mRNAs involved in the development of secondary hair follicles. Afterward, circRNA2034, circRNA5712, circRNA888, and circRNA9127 were identified by qRT-PCR and Sanger sequencing. In the next step, the circRNA5712-miR-27b-3p-Dll4 pathway was verified by a dual-luciferase reporting assay. This research established a basis for future analyses of how circRNAs control the morphogenesis and development of cashmere goat secondary hair follicles through the ceRNA mechanism. In the longissimus dorsi tissues of 1-month-old (n = 3) and 9-month-old (n = 3) Wu’an black goats, a total of 36 DE lncRNAs were identified, of which 30 DE lncRNAs were located in the neighborhood of 71 mRNAs; several of these genes were enriched in pathways related to muscle development, including APC, IFRD1, NKX2-5, and others. Finally, an lncRNA–miRNA–mRNA ceRNA network composed of 4 lncRNAs, 3 miRNAs, and 8 mRNAs was constructed; it was concluded that XR_001296113.2 may regulate the PDLIM7 gene expression by sponging to the chi-miR-1296 and affect skeletal muscle development. This study characterized expression patterns of lncRNAs in goats, provided a resource for future research studies, and improved our understanding of molecular mechanisms governing skeletal muscle development in goats [94]. Ghafouri et al. [39] examined the transcriptome of goats with high versus low fertility, involving lincRNAs in the ceRNA regulatory network, adopted a new approach to identify genes and regulatory factors involved in goat fertility, and added a new layer of non-coding RNA molecules involved in physiological functions to the large ceRNA network. There were 18 genes implicated in goat fertility (SMAD1, SMAD2, SMAD3, SMAD4, TIMP1, ERBB2, BMP15, TGFB1, MAPK3, CTNNB1, BMPR2, AMHR2, TGFBR2, BMP4, ESR1, BMPR1B, AR, and TGFB2). In addition, cytokine-cytokine receptor interactions, regulating pluripotency of stem cells, ovarian steroidogenesis, progesterone-mediated oocyte maturation, oocyte meiosis, cortisol synthesis and secretion, parathyroid and growth hormone synthesis, as well as prolactin, Hippo, TGF-beta, PI3K-Akt, FoxO, and MAPK signaling pathways were detected in KEGG and biological pathway enrichment analyses.
6.5. Buffalo
Low fertility or infertility problems can lead to additional inseminations, an increased need for veterinary attention, and hormonal treatments that can affect current and future lactations in ruminants, especially dairy cattle and buffalos [95]. Moreover, fertility issues can result in additional expenses due to culling and replacing animals. Consequently, enhancing livestock fertility is crucial to reducing management costs, increasing farm profits, and sustaining milk production [96]. In this regard, granulosa cells (GCs) are the primary supportive cells in follicles that have a crucial role in regulating oocyte maturation and follicular atresia. Pan et al. [42] defined healthy and atretic follicles (HFs and AFs) based on both the apoptosis rate of granulosa cells and hormone concentrations in follicular fluid. Granulosa cells were collected from ovarian follicles (HFs and AFs) from 5 to 8 mm (n = 15) for whole-transcriptome analysis using second-generation high-throughput sequencing. There were a total of 1861 and 1075 mRNAs, 159 and 24 miRNAs, and 123 and 100 lncRNAs that were up-regulated and down-regulated between HFs and AFs, respectively. They reported enrichment analysis of differentially expressed genes (DEGs) and found that many of the DEmRNAs and DEmiRNA targets were associated with ECM-receptor interaction and focal adhesion, as well as several signaling pathways such as Rap1, PI3K-AKT, TGF-beta, mTOR, and estrogen. Then, based on the ceRNA theory, a competing endogenous RNA (CeRNA) network was constructed to reveal the regulatory roles of these DERNAs in the GCs of buffalo follicles. These results demonstrate that lncRNAs interact with target genes in a ceRNA network, implying their crucial roles in follicular development and atresia. According to Li et al. [43], the regulatory mechanisms of long non-coding RNA (lncRNA) and their impact on differences in meat quality are not well understood. The chemical-physical parameters of buffalo and cattle muscles can be similar, but there were significant differences in shearing force and muscle fiber structure. In a comparison between buffalo and cattle muscle tissue, 16,236 lncRNA candidates were detected, including 865 up-regulated lncRNAs and 1296 down-regulated lncRNAs. Based on reconstructed co-expression and ceRNA networks, MSTRG.30030.4, MSTRG.203788.46, and MSTRG.48330.7 lncRNAs could potentially bind with miR-1/206 and miR-133a miRNAs as competitive endogenous RNA (ceRNA). Tissue expression analysis indicated that these identified lncRNAs are highly and specifically expressed in muscle tissue.
6.6. Camel
Zhang et al. [44] utilized high-throughput sequencing to identify genes involved in resistance to water deprivation and salt absorption in the ileum and liver of the Bactrian camel (Camelus bactrianus) under stress conditions by investigating stress-induced alternative splicing events. In this regard, genes driven by alternative splicing were enriched to molecular functions potentially fixed by organ and stress types. Using qRT-PCR detection, they identified functionally important genes including AQP5, MUC6, LOC105076960, CDH11, PKP4, SDS, TENM1, LOC105061856, UPP2, and PLIN2, along with miR-484, miR-29b, miR-128, miR-362-5p, miR-195, miR-96, and miR-148a. These genes contributed to resistance to cellular stress. Furthermore, the underlying competing endogenous RNAs were in the ileum (Let-7e and LOC105076960 mRNAs) and liver (LNC001770, LNC001438, LNC003417, miR-199c, and TENM1 mRNA).
7. Conclusions
In conclusion, we considered interactions between coding and non-coding RNAs via MREs that formed large-scale regulatory networks throughout the transcriptome. These ceRNA interactions can provide answers to evolutionary questions, as they may partially explain associations between genome size and the complexity of regulatory mechanisms. Furthermore, ceRNA networks can explain molecular interactions involved in biological functions within the cell and, thus, can be used to improve complex traits and reduce the incidence of diseases in ruminants. Although comprehension of ceRNA networks remains nascent, current experimental techniques can elucidate the regulatory mechanisms of ceRNA by comprehensive identification of miRNA binding sites, facilitated by high-throughput sequencing technologies such as RNA-seq and databases compiling ncRNAs–miRNA interactions. Therefore, attention to ceRNA regulatory networks and their application to identify molecular regulatory mechanisms related to functional and economic traits in ruminants can be useful. Further understanding of the regulatory roles of non-coding RNAs, as well as their prominent role in network structure and metabolic and signaling pathways involved in complex traits, along with other information that can be integrated into these networks, are among future challenges.
8. Future Directions
One of the best potential areas for future research is a clear understanding of the role of ceRNAs in disease and the complex economic traits of ruminants. With the emergence of ceRNA regulatory mechanisms and functions, there is a need for comprehensive analyses of genes, miRNAs, and other types of regulatory RNA expression in tissue or organs with pathology; this will provide practical approaches for the identification of novel metabolic and signaling pathways that are deregulated in complex diseases (e.g., mastitis and Johne’s disease) and the discovery of novel interactions between metabolic and signaling pathways associated with economically important diseases in ruminants. Sequencing technologies, bioinformatics tools, integrated omics technology and system biology approaches have contributed largely to comprehensive analyses. Additionally, with the discovery and definition of ceRNA mechanisms, sequencing and analysis pipelines will provide insights beyond coding regions and non-coding regions (including UTRs, introns, etc.) because these can cause changes in MREs that can affect complex regulatory circuits. In this regard, genomic loss and amplification can have important consequences for ceRNAs [60].
In addition, point mutations can result in genetic disorders that cause inborn errors in metabolic processes. Such mutations can abolish protein function if mutant transcripts retain complete ceRNA function. Moreover, these mutations in the MREs can eliminate potential ceRNA functions, thereby disrupting the regulatory network function. Gene regulation mediated by ceRNAs could also have implications for targeted therapy and therapeutic responses. Additionally, it can strengthen the immune system, increase milk production, and improve productive and reproductive performance in ruminants. The complex relationship among RNAs (circRNAs, lncRNAs, miRNAs, and mRNAs) makes it increasingly difficult to comprehend the regulation of genes. Hence, it is critical to gain a better understanding of spatial and temporal relationships between the associated ceRNAs, miRNAs, and mRNAs based on the ceRNA regulatory networks such as circRNA–lncRNA–miRNA–mRNA ceRNA regulatory network. In the future, the development of these networks could identify complex mechanisms of gene regulation involved in various economic traits associated with ruminants.
Author Contributions
Conceptualization, F.G., V.D.R., M.S. (Mostafa Sadeghi) and M.S. (Masoud Shirali); methodology, F.G., V.D.R. and M.S. (Mostafa Sadeghi); investigation, F.G., V.D.R. and M.S. (Masoud Shirali); writing—original draft preparation, F.G., V.D.R., M.S. (Mostafa Sadeghi) and M.S. (Masoud Shirali); writing—review and editing, S.R.M.-A., J.P.K. and H.W.B.; visualization, S.R.M.-A.; supervision, M.S. (Mostafa Sadeghi) and M.S. (Masoud Shirali); project administration, M.S. (Masoud Shirali). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Brosius, J.; Raabe, C.A. What is an RNA? A top layer for RNA classification. RNA Biol. 2016, 13, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Rao, M.R.S. Long noncoding RNAs in pluripotency of stem cells and cell fate specification. Adv. Exp. Med. Biol. 2017, 1008, 223–252. [Google Scholar] [PubMed]
- Flynn, R.A.; Chang, H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef]
- Degirmenci, U.; Lei, S. Role of lncRNAs in cellular aging. Front. Endocrinol. 2016, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Thakkar, N.; Chhatai, J.; Pal Bhadra, M.; Bhadra, U. Long non-coding RNA: Functional agent for disease traits. RNA Biol. 2017, 14, 522–535. [Google Scholar] [CrossRef]
- Ghafouri, F.; Sedeghi, M.; Bahrami, A. Long non-coding RNAs (LncRNAs): Roles, functions, and mechanisms. Genet. Eng. Biosaf. J. 2018, 7, 245–266. [Google Scholar]
- Tutar, Y. Pseudogenes. Int. J. Genom. 2012, 2012, 424526. [Google Scholar] [CrossRef]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri, F.; Bahrami, A.; Sadeghi, M.; Miraei-Ashtiani, S.R.; Bakherad, M.; Barkema, H.W.; Larose, S. Omics multi-layers networks provide novel mechanistic and functional insights into fat storage and lipid metabolism in poultry. Front. Genet. 2021, 12, 646297. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Ala, U.; Karreth, F.A.; Bosia, C.; Pagnani, A.; Taulli, R.; Léopold, V.; Tay, Y.; Provero, P.; Zecchina, R.; Pandolfi, P.P. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl. Acad. Sci. USA 2013, 110, 7154–7159. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, X.; Cao, Y.H.; Pokharel, K.; Hu, X.J.; Chen, Z.H.; Xu, S.S.; Peippo, J.; Honkatukia, M.; Kantanen, J.; et al. Comparative mRNA and miRNA expression in European mouflon (Ovis musimon) and sheep (Ovis aries) provides novel insights into the genetic mechanisms for female reproductive success. Heredity 2019, 122, 172–186. [Google Scholar] [CrossRef]
- Gao, L.; Zhao, Y.; Ma, X.; Zhang, L. Integrated analysis of lncRNA–miRNA–mRNA ceRNA network and the potential prognosis indicators in sarcomas. BMC Med. Genom. 2021, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Ala, U. Competing endogenous RNAs, non-coding RNAs and diseases: An intertwined story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, X.; Bai, X.; Xiao, C.; Dong, Y. Different expression of lipid metabolism-related genes in Shandong black cattle and Luxi cattle based on transcriptome analysis. Sci. Rep. 2020, 10, 21915. [Google Scholar] [CrossRef]
- Dehghanian Reyhan, V.; Ghafouri, F.; Sadeghi, M.; Miraei-Ashtiani, S.R.; Kastelic, J.P.; Barkema, H.W.; Shirali, M. Integrated Comparative Transcriptome and circRNA-lncRNA-miRNA-mRNA ceRNA Regulatory Network Analyses Identify Molecular Mechanisms Associated with Intramuscular Fat Content in Beef Cattle. Animals 2023, 13, 2598. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, J.; Yang, Y.; Wang, Q.; Yu, Q. Metagenomic and transcriptomic analyses reveal the differences and associations between the gut microbiome and muscular genes in Angus and Chinese Simmental cattle. Front. Microbiol. 2022, 13, 815915. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, Y.; Zhao, X.; Zhao, Y.; Jing, Y.; Liu, G.; Wang, S.; Li, H.; Ma, Y. Transcriptome profiling of mRNAs in muscle tissue of Pinan cattle and Nanyang cattle. Gene 2022, 825, 146435. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Bai, Y.; Liu, J.; Cai, W.; Liu, L.; He, Y.; Song, J. Metabolic regulations by lncRNA, miRNA, and ceRNA under grass-fed and grain-fed regimens in Angus beef cattle. Front. Genet. 2021, 12, 579393. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Ma, X.; Dai, R.; Dingkao, R.; Zhaxi, Z.; Burenchao, G.; Bao, P.; Wu, X.; Guo, X.; et al. Comprehensive analysis of mRNA, lncRNA, circRNA, and miRNA expression profiles and Their ceRNA networks in the longissimus dorsi muscle of Cattle-Yak and Yak. Front. Genet. 2021, 12, 772557. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Liu, M.; Li, M.; Chen, H. Characterization of lncRNA–miRNA–mRNA network to reveal potential functional ceRNAs in bovine skeletal muscle. Front. Genet. 2019, 10, 430773. [Google Scholar] [CrossRef]
- Wang, K.; Cheng, Y.; Guo, T.; Guo, X.; Zhang, H.; Ma, X.; Pan, Y.; Kebreab, E.; Wang, D.; Lyu, L. Analyzing the interactions of mRNAs, miRNAs and lncRNAs to predict ceRNA networks in bovine cystic follicular granulosa cells. Front. Vet. Sci. 2022, 9, 1028867. [Google Scholar] [CrossRef]
- Feng, X.; Cai, Z.; Mu, T.; Yu, B.; Wang, Y.; Ma, R.; Liu, J.; Wang, C.; Zhang, J.; Gu, Y. CircRNA screening and ceRNA network construction for milk fat metabolism in dairy cows. Front. Vet. Sci. 2022, 9, 995629. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xia, H.; Wang, X.; Wang, Y.; Fang, J.; Li, S.; Zhai, Y.; Han, Z. Comprehensive profiling of ceRNA (circRNA-miRNA-mRNA) networks in hypothalamic-pituitary-mammary gland axis of dairy cows under heat stress. Int. J. Mol. Sci. 2023, 24, 888. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.R.; Salazar, N.A.; Ayoola, A.O.; Memili, E.; Thomas, B.N.; Morenikeji, O.B. Regulatory network of miRNA, lncRNA, transcription factor and target immune response genes in bovine mastitis. Sci. Rep. 2021, 11, 21899. [Google Scholar] [CrossRef]
- Liang, R.; Han, B.; Li, Q.; Yuan, Y.; Li, J.; Sun, D. Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 2017, 7, 6396. [Google Scholar] [CrossRef]
- Mu, T.; Hu, H.; Feng, X.; Ma, Y.; Wang, Y.; Zhang, J.; Gu, Y. Screening and conjoint analysis of key lncRNAs for milk fat metabolism in dairy cows. Front. Genet. 2022, 13, 772115. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, S.; Malik, W.A.; Feng, H.; Liu, T.; Xie, L.; Miao, X. Genome wide identification and characterization of fertility associated novel CircRNAs as ceRNA reveal their regulatory roles in sheep fecundity. J. Ovarian Res. 2023, 16, 115. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Kang, X.; Liu, Y.; Liu, X.; Chan, S.; Wang, Y.; Li, Z.; Ling, Y.; Feng, D.; Li, M.; et al. Integrated analysis of the whole transcriptome of skeletal muscle reveals the ceRNA regulatory network related to the formation of muscle fibers in Tan sheep. Front. Genet. 2022, 13, 991606. [Google Scholar] [CrossRef]
- Sadeghi, M.; Bahrami, A.; Hasankhani, A.; Kioumarsi, H.; Nouralizadeh, R.; Abdulkareem, S.A.; Ghafouri, F.; Barkema, H.W. lncRNA–miRNA–mRNA ceRNA Network Involved in Sheep Prolificacy: An Integrated Approach. Genes 2022, 13, 1295. [Google Scholar] [CrossRef]
- Bao, G.; Zhao, F.; Wang, J.; Liu, X.; Hu, J.; Shi, B.; Wen, Y.; Zhao, L.; Luo, Y.; Li, S. Characterization of the circRNA–miRNA–mRNA network to reveal the potential functional ceRNAs associated with dynamic changes in the meat quality of the longissimus thoracis muscle in Tibetan sheep at different growth stages. Front. Vet. Sci. 2022, 9, 803758. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wang, Y.; He, J. Transcriptomic analysis reveals the involvement of lncRNA–miRNA–mRNA networks in hair follicle induction in Aohan fine wool sheep skin. Front. Genet. 2020, 11, 533225. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Liu, Y.; He, X.; Fang, M.; Chu, M. Uterus proliferative period ceRNA network of Yunshang black goat reveals candidate genes on different kidding number trait. Front. Endocrinol. 2023, 14, 1165409. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Wang, J.; Mao, M.; Zhao, X.; Li, Q.; Xuan, R.; Li, F.; Chao, T. Analyses of lncRNAs, circRNAs, and the Interactions between ncRNAs and mRNAs in Goat Submandibular Glands Reveal Their Potential Function in Immune Regulation. Genes 2023, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri, F.; Sadeghi, M.; Bahrami, A.; Naserkheil, M.; Reyhan, V.D.; Javanmard, A.; Miraei-Ashtiani, S.R.; Ghahremani, S.; Barkema, H.W.; Abdollahi-Arpanahi, R.; et al. Construction of a circRNA–lincRNA–lncRNA–miRNA–mRNA ceRNA regulatory network identifies genes and pathways linked to goat fertility. Front. Genet. 2023, 14, 1195480. [Google Scholar] [CrossRef]
- Shang, F.; Ma, R.; Rong, Y.; Pan, J.; Wang, M.; Niu, S.; Qi, Y.; Li, Y.; Wang, Z.; Lv, Q.; et al. Construction and functional analysis of ceRNA regulatory network related to the development of secondary hair follicles in Inner Mongolia cashmere goats. Front. Vet. Sci. 2022, 9, 959952. [Google Scholar] [CrossRef]
- Jin, M.; Fan, W.; Lyu, S.; Cong, L.; Xue, T. Screening and bioinformatics analysis of a potential ceRNA network in melatonin-induced cashmere growth in Liaoning cashmere goats. Arch. Anim. Breed. 2024, 67, 97–109. [Google Scholar] [CrossRef]
- Pan, Y.; Yang, S.; Cheng, J.; Lv, Q.; Xing, Q.; Zhang, R.; Liang, J.; Shi, D.; Deng, Y. Whole-Transcriptome Analysis of LncRNAs Mediated ceRNA Regulation in Granulosa Cells Isolated from Healthy and Atresia Follicles of Chinese Buffalo. Front. Vet. Sci. 2021, 8, 680182. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Feng, T.; Qian, Q.; Liu, Q. Comparison of long non-coding RNA expression profiles of cattle and buffalo differing in muscle characteristics. Front. Genet. 2020, 11, 492141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Pan, J.; Zhou, H.; Cao, Y. Evidence from ileum and liver transcriptomes of resistance to high-salt and water-deprivation conditions in camel. Zool. Lett. 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Cassar-Malek, I.; Picard, B.; Bernard, C.; Hocquette, J.F. Application of gene expression studies in livestock production systems: A European perspective. Aust. J. Exp. Agric. 2008, 48, 701–710. [Google Scholar] [CrossRef]
- Cesar, A.S.; Regitano, L.C.; Reecy, J.M.; Poleti, M.D.; Oliveira, P.S.; de Oliveira, G.B.; Moreira, G.C.; Mudadu, M.A.; Tizioto, P.C.; Koltes, J.E.; et al. Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits. BMC Genom. 2018, 19, 1–20. [Google Scholar] [CrossRef]
- Loging, W.; Harland, L.; Williams-Jones, B. High-throughput electronic biology: Mining information for drug discovery. Nat. Rev. Drug Discov. 2007, 6, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.J.; Gaulton, A.; Marshall, J.; Bichko, D.; Martin, S.; Brouwer, C.; Harland, L. Visualizing the drug target landscape. Drug Disco. Today 2010, 15, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Reyhan, V.D.; Sadeghi, M.; Miraei-Ashtiani, S.R.; Ghafouri, F.; Kastelic, J.P.; Barkema, H.W. Integrated transcriptome and regulatory network analyses identify candidate genes and pathways modulating ewe fertility. Gene Rep. 2022, 28, 101659. [Google Scholar] [CrossRef]
- Jansen, R.C.; Nap, J.P. Genetical genomics: The added value from segregation. Trends Genet. 2001, 17, 388–391. [Google Scholar] [CrossRef]
- Fairfax, B.P.; Knight, J.C. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 2014, 30, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cloney, R. Integrating gene variation and expression to understand complex traits. Nat. Rev. Genet. 2016, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Essa, B.; Al-Sharif, M.; Abdo, M.; Fericean, L.; Ateya, A. New insights on nucleotide sequence variants and mRNA levels of candidate genes assessing resistance/susceptibility to mastitis in Holstein and montbéliarde dairy cows. Vet. Sci. 2023, 10, 35. [Google Scholar] [CrossRef]
- Rau, A.; Marot, G.; Jaffrézic, F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinform. 2014, 15, 91. [Google Scholar] [CrossRef] [PubMed]
- Keel, B.N.; Lindholm-Perry, A.K. Recent developments and future directions in meta-analysis of differential gene expression in livestock RNA-Seq. Front. Genet. 2022, 13, 983043. [Google Scholar] [CrossRef] [PubMed]
- Lindholm-Perry, A.K.; Meyer, A.M.; Kern-Lunbery, R.J.; Cunningham-Hollinger, H.C.; Funk, T.H.; Keel, B.N. Genes involved in feed efficiency identified in a meta-analysis of rumen tissue from two populations of beef steers. Animals 2022, 12, 1514. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri, F.; Mehrabani Yeganeh, H.; Mohamadian Jeshvaghani, S. Big data and the role of high-throughput technologies in livestock and poultry breeding. Prof. J. Domest. 2020, 20, 34–40. [Google Scholar]
- Ghafouri, F.; Alipour, S.; Mohamadian Jeshvaghani, S. Application of machine learning approach and its subset algorithms in estimating genomic breeding values. Prof. J. Domest. 2020, 20, 19–29. [Google Scholar]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Sumazin, P.; Yang, X.; Chiu, H.S.; Chung, W.J.; Iyer, A.; Llobet-Navas, D.; Rajbhandari, P.; Bansal, M.; Guarnieri, P.; Silva, J.; et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 2011, 147, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.S.; Martínez, M.R.; Bansal, M.; Subramanian, A.; Golub, T.R.; Yang, X.; Sumazin, P.; Califano, A. High-throughput validation of ceRNA regulatory networks. BMC Genom. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cao, F.; Gu, X.; Chen, J.; Xu, R.; Huang, Y.; Ying, L. LncRNA XIST, as a ceRNA of miR-204, aggravates lipopolysaccharide-induced acute respiratory distress syndrome in mice by upregulating IRF2. Int. J. Clin. Exp. Pathol. 2019, 12, 2425. [Google Scholar] [PubMed]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Wahlstedt, H.; Daniel, C.; Ensterö, M.; Öhman, M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009, 19, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Zipeto, M.A.; Jiang, Q.; Melese, E.; Jamieson, C.H. RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 2015, 21, 549–559. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, X.; Jin, Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci. China Life Sci. 2013, 56, 944–952. [Google Scholar] [CrossRef]
- Hwang, T.; Park, C.K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Lagergren, J.; Öhman, M. RNA editing of non-coding RNA and its role in gene regulation. Biochimie 2015, 117, 22–27. [Google Scholar] [CrossRef]
- Shevchenko, G.; Morris, K.V. All I’s on the RADAR: Role of ADAR in gene regulation. FEBS Lett. 2018, 592, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Cui, C.; Liu, J.; Luo, Y.; Quan, F.; Jin, Y.; Zhang, Y. The growth and reproduction performance of TALEN-mediated β-lactoglobulin-knockout bucks. Transgenic Res. 2016, 25, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, Y.; Zhang, Y.; Yang, M.; Lv, J.; Liu, J.; Zhang, Y. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc. Natl. Acad. Sci. USA 2015, 112, 1530–1539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, L.; Ren, G.; Li, Z.; Ren, C.; Zhang, T.; Xu, K.; Zhang, Z. Targeted disruption of the sheep MSTN gene by engineered zinc-finger nucleases. Mol. Biol. Rep. 2014, 41, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Song, Y.; Liu, J.; Ge, H.; Li, Q.; Huang, H.; Hu, L.; Zhu, H.; Jin, Y.; Zhang, Y. Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci. Rep. 2015, 5, 10482. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Guo, W.; Chang, B.; Liu, J.; Guo, Z.; Quan, F.; Zhang, Y. Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat. Commun. 2013, 4, 2565. [Google Scholar] [CrossRef] [PubMed]
- Jenko, J.; Gorjanc, G.; Cleveland, M.A.; Varshney, R.K.; Whitelaw, C.B.A.; Woolliams, J.A.; Hickey, J.M. Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs. Genet. Sel. Evol. 2015, 47, 1–14. [Google Scholar]
- Wang, Y.; Hou, J.; He, D.; Sun, M.; Zhang, P.; Yu, Y.; Chen, Y. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016, 32, 211–224. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA regulatory networks in cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Dehghanian Reyhan, V.; Sadeghi, M.; Ghafouri, F. Method of weighted gene co-expression network analysis and its application in animal and poultry breeding and genetics. Prof. J. Domest. 2022, 22, 5–13. [Google Scholar]
- Walhout, M.; Vidal, M.; Dekker, J. (Eds) Handbook of Systems Biology: Concepts and Insights; Academic Press: New York, NY, USA, 2012. [Google Scholar]
- Zhao, X.; Wang, C.; Wang, Y.; Zhou, L.; Hu, H.; Bai, L.; Wang, J. Weighted gene co-expression network analysis reveals potential candidate genes affecting drip loss in pork. Anim. Genet. 2020, 51, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, Q.; Wu, Y.; Zhang, Y.; Zhang, B.; Zhang, H. Identification of candidate genes that specifically regulate subcutaneous and intramuscular fat deposition using transcriptomic and proteomic profiles in Dingyuan pigs. Sci. Rep. 2022, 12, 2844. [Google Scholar] [CrossRef]
- Ovens, K.; Eames, B.F.; McQuillan, I. Comparative analyses of gene co-expression networks: Implementations and applications in the study of evolution. Front. Genet. 2021, 12, 695399. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, Y.; Xin, S.; Yang, L.; Jiang, M.; Xin, Y.; Wang, Y.; Yang, J.; Lu, J. Insight into LncRNA-and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review. Cancers 2022, 14, 4564. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qiu, X.; He, Q.; Fu, X.; Ji, F.; Tian, X. CCND1-associated ceRNA network reveal the critical pathway of TPRG1-AS1-hsa-miR-363-3p-MYO1B as a prognostic marker for head and neck squamous cell carcinoma. Sci. Rep. 2023, 13, 11831. [Google Scholar] [CrossRef]
- Ma, S.C.; Li, Q.; Peng, J.Y.; Zhouwen, J.L.; Zhang, D.N.; Zhang, C.B.; Jiang, W.G.; Jia, W. CLDN5 affects lncRNAs acting as ceRNA dynamics contributing to regulating blood-brain barrier permeability in tumor brain metastasis. Oncol. Rep. 2018, 39, 1441–1453. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, B.; Huang, J.; Li, W.; Yi, P.; Yi, M.; Peng, W. Identification of the protective effect of Polygonatum sibiricum polysaccharide on d-galactose-induced brain ageing in mice by the systematic characterization of a circular RNA-associated ceRNA network. Pharm. Biol. 2021, 59, 345–364. [Google Scholar] [CrossRef]
- Zhu, X.; Jiang, L.; Yang, H.; Chen, T.; Wu, X.; Lv, K. Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell Int. 2020, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.R.; Zhang, Y.B.; Zheng, S.F.; Xu, Y.W.; Lin, P.; Shang-Guan, H.C.; Lin, Y.X.; Kang, D.Z.; Yao, P.S. Decreased SPTBN2 expression regulated by the ceRNA network is associated with poor prognosis and immune infiltration in low-grade glioma. Exp. Ther. Med. 2023, 25, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, X.; Li, W.; Liu, J.; Fan, Y.; Zhang, H.; Yang, J.; Gao, Y.; Liu, Y. Identification and characterization of lncRNAs expression profile related to goat skeletal muscle at different development stages. Animals 2022, 12, 2683. [Google Scholar] [CrossRef] [PubMed]
- Boichard, D. Estimation of the economic value of conception rate in dairy cattle. Livest. Prod. Sci. 1990, 24, 187–204. [Google Scholar] [CrossRef]
- Dekkers, J.C.M. Estimation of economic values for dairy cattle breeding goals: Bias due to sub-optimal management policies. Livest. Prod. Sci. 1991, 29, 131–149. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).