Preclinical Development of Costimulatory Switch Protein (CSP)-Armored NY-ESO-1/LAGE-1a-Specific TCR-T Cells for Therapy of Hard-to-Treat PD-L1-Positive Solid Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of NY-ESO-1/LAGE1a-Specific TCR-T Cells (MDG1015)
2.2. Generation of Transduced Tumor Cell Lines and 3D Tumor Cell Spheroids
2.3. Quantitative Real-Time PCR (qPCR)
2.4. IFN-γ/IL-2 Release Assay
2.5. Bulk Poly-Cytokine Analysis
2.6. Single-Cell Poly-Cytokine Secretion Analysis
2.7. Live-Cell Imaging Cytotoxicity Assay
2.8. Cell Surface Staining and Flow Cytometry
2.9. Proliferation and TCR-T Cell Expansion Analysis
3. Results
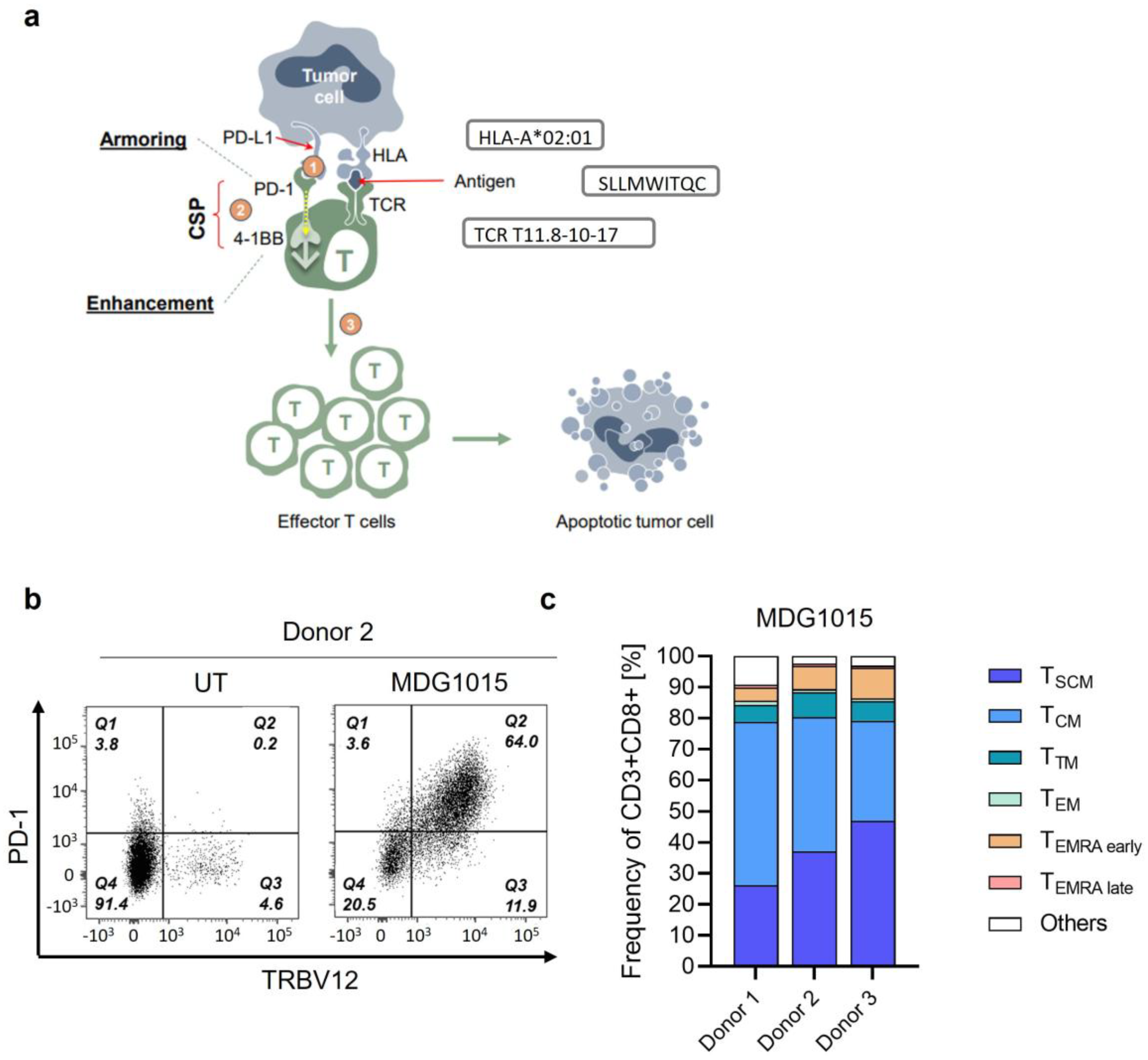
3.1. Phenotype Characteristics of MDG1015
3.2. Functionality of MDG1015
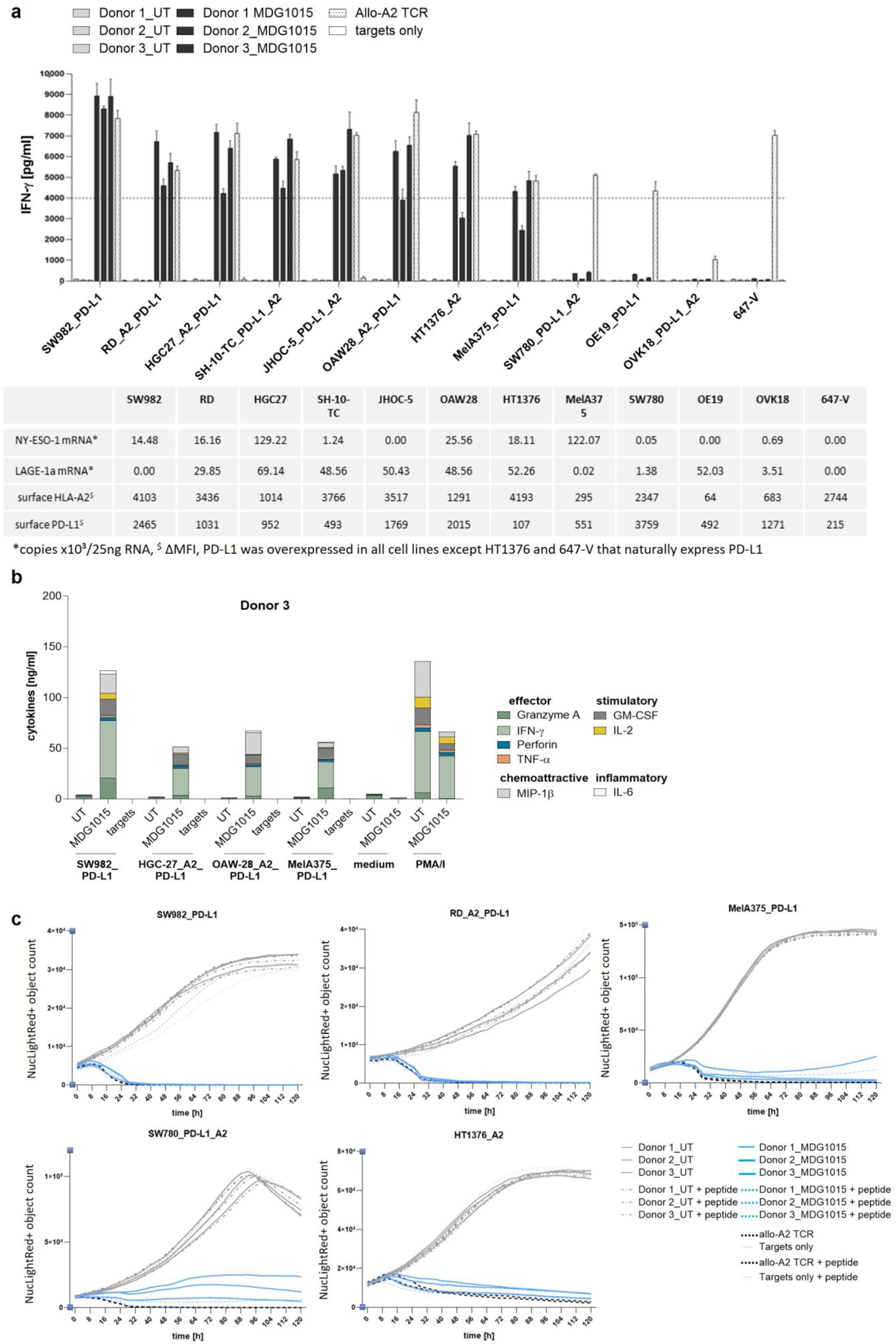
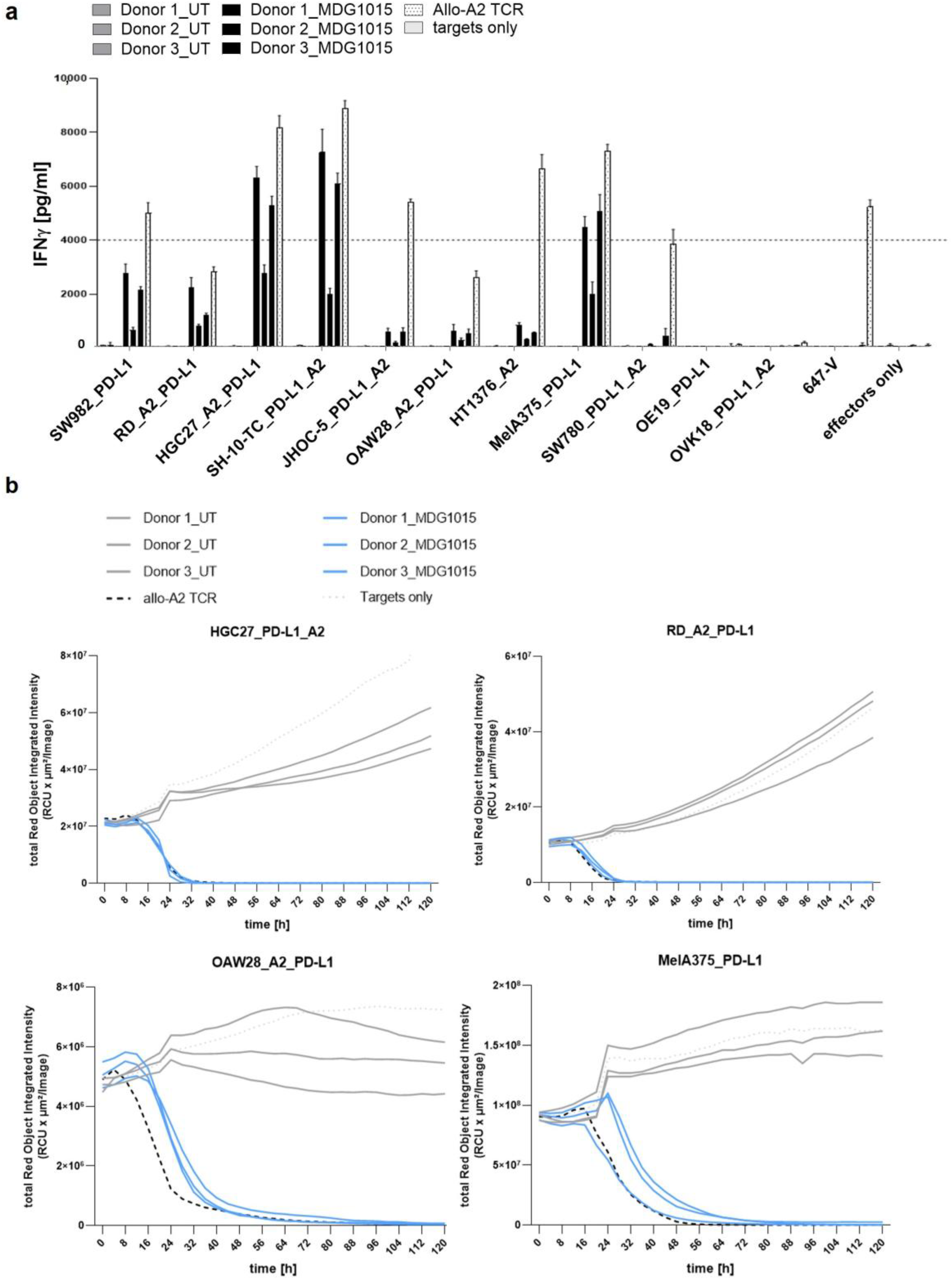
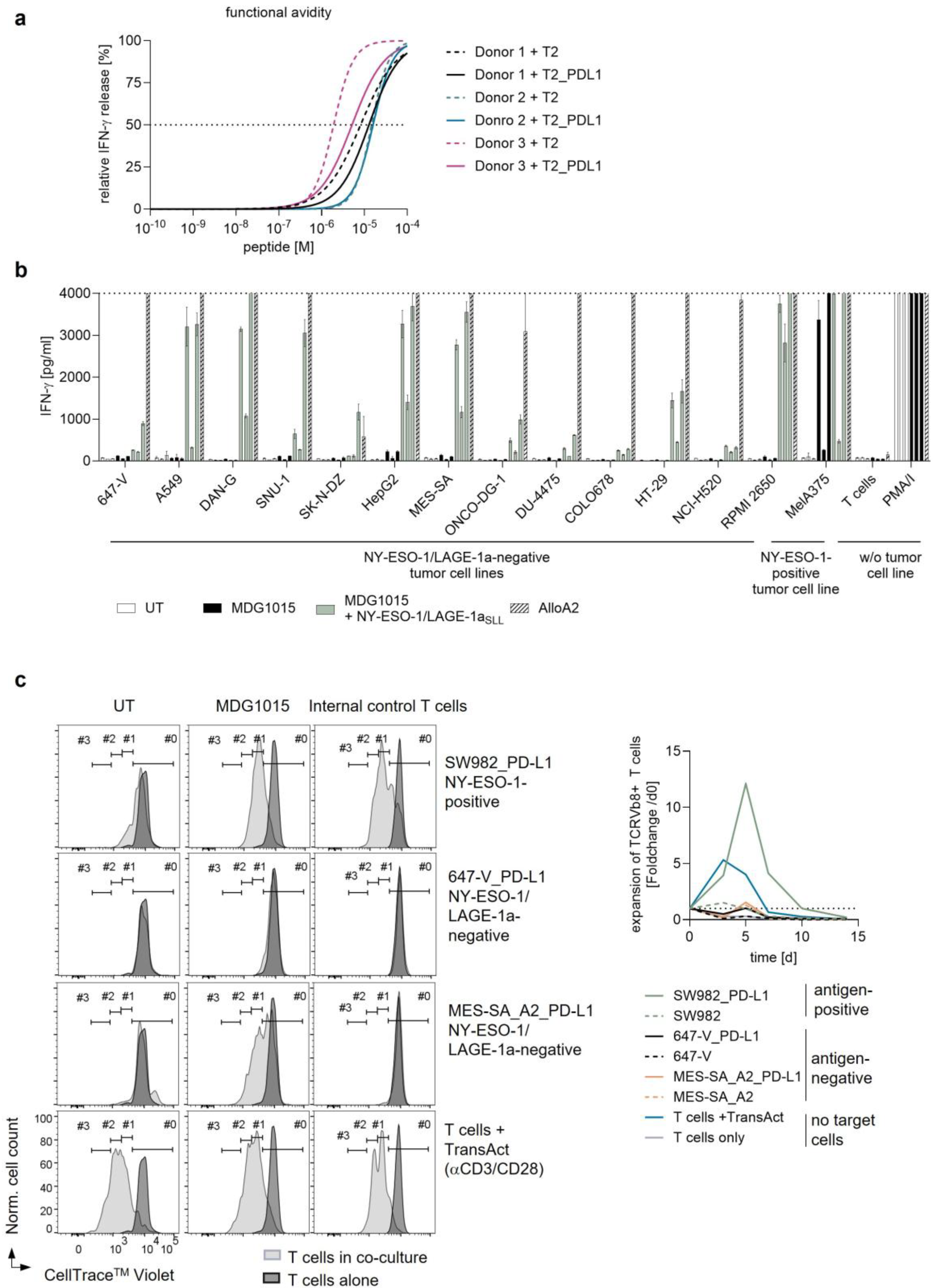
Functionality of MDG1015 in Response to PD-L1-Expressing Tumor Cell Lines
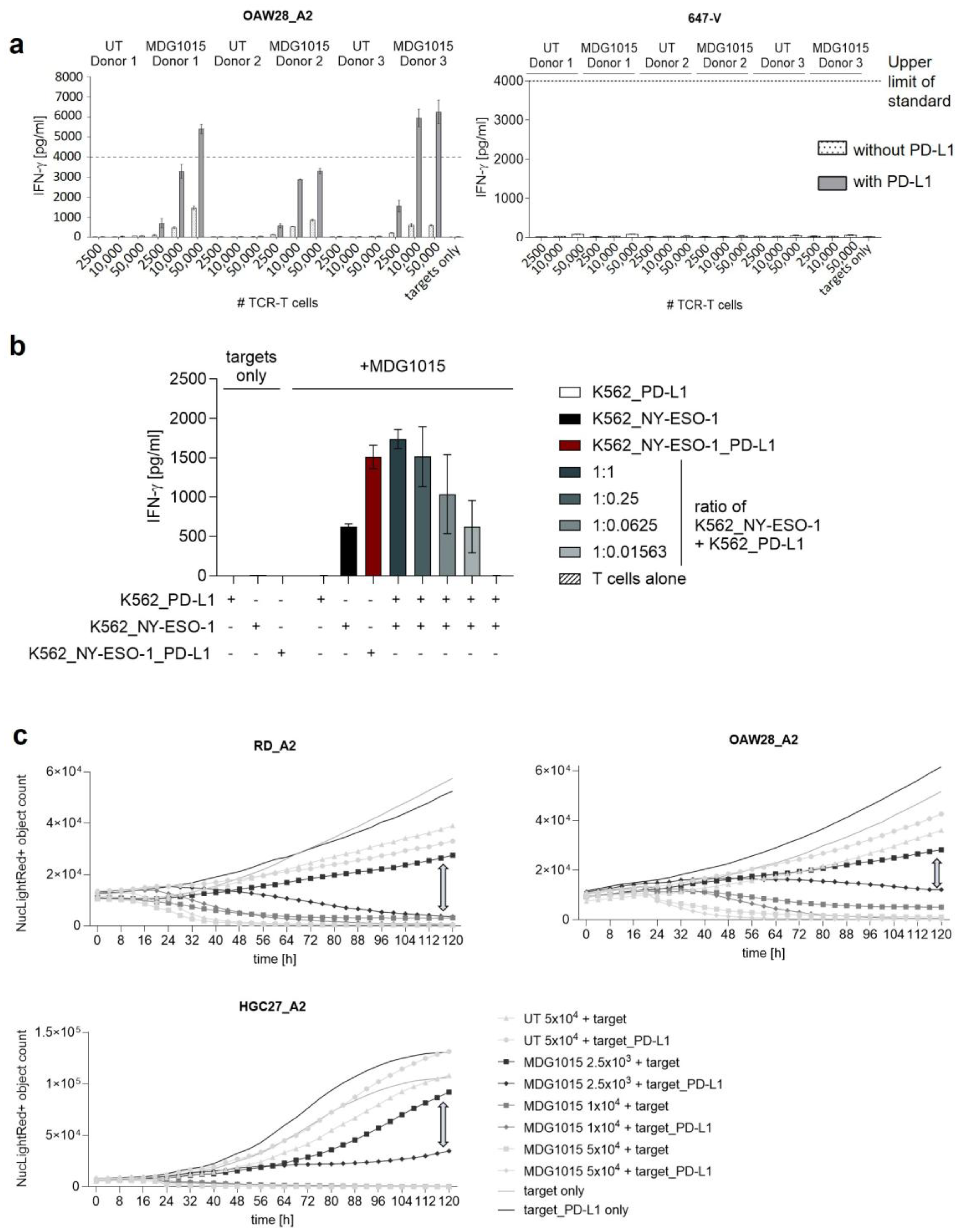
3.3. Enhanced Functionality of MDG1015 upon PD-L1:CSP Interaction
3.3.1. Increased IFN-γ Secretion and Tumor Cell Killing
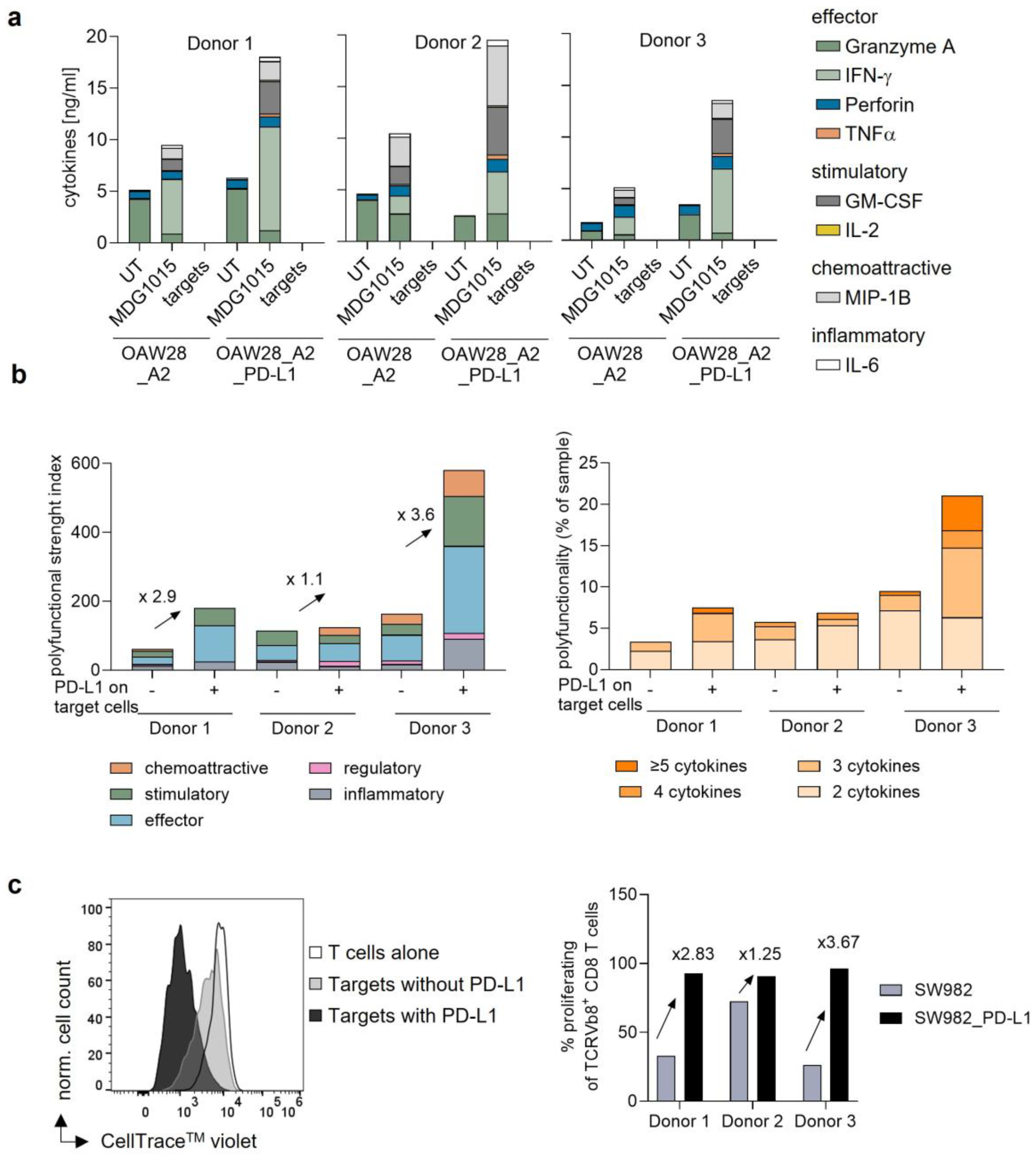
3.3.2. Increased Polyfunctionality and Proliferation
3.4. Safety of MDG1015
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Tumor Cell Line | Culture Medium |
|---|---|
| MelA375_NucLightRed_PD-L1 | RPMI 1640 + 2 mM L-glutamine + 1 mM sodium pyruvate + 1% MEM NEAA (all Thermo Scientific™, Waltham, MA, USA) + 10% FCS (Sigma-Aldrich, St. Louis, MO, USA) + 0.5 µg/mL Puromycin (invivogen, San Diego, CA, USA) |
| SW982_PD-L1_NucLightRed | DMEM 1 g/l glucose + 2 mM L-glutamine + 1% MEM NEAA (all Thermo Scientific™, Waltham, MA, USA)+ 10% FCS + 0.5 µg/mL Puromycin |
| RD_HLA-A*02:01_NucLightRed_PD-L1 | DMEM 1 g/l glucose + 10% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| HGC27_HLA-A*02:01_NucLightRed_PD-L1 | EMEM (ATCC, Manassas, VA, USA)+ 10% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| SW780_ PD-L1_HLA-A*02:01_NucLightRed | DMEM 1 g/l glucose + 10% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| OE19_PD-L1_NucLightRed | RPMI 1640 + 10% FCS + 2 mM L-glutamine + 1 mM sodium pyruvate + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| HT1376_HLA-A*02:01_NucLightRed | DMEM 4.5 g/l glucose (Thermo Scientific™, Waltham, MA, USA)+ 15% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| OVK18_PD-L1_HLA-A*02:01_NucLightRed | EMEM + 10% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| JHOC-5_PD-L1_HLA-A*02:01_NucLightRed | DMEM/Ham’s F12 + 10% FCS + 1% MEM NEAA + 0.5 µg/mL Puromycin |
| SH-10-TC_PD-L1_HLA-A*02:01_NucLightRed | RPMI 1640 + 10% FCS + 2 mM L-glutamine + 1 mM sodium pyruvate + 1% MEM NEAA + 0.25 µg/mL Puromycin |
| OAW28_HLA-A*02:01_NucLightRed_PD-L1 | DMEM 1 g/l glucose + 2 mM L-Glutamine + 1 mM pyruvate + 10% FCS + 20 IU/mL insulin + 0.5 µg/mL Puromycin |
| 647-V_NucLightRed | DMEM 4.5 g/l glucose + 15% FCS + 2 mM L-glutamine + 1% MEM NEAA + 0.5 µg/mL Puromycin |
References
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef]
- Cappell, K.M.; Kochenderfer, J.N. Long-term outcomes following CAR T cell therapy: What we know so far. Nat. Rev. Clin. Oncol. 2023, 20, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jäger, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef]
- Abramson, J.S.; Palomba, M.L.; Gordon, L.I.; Lunning, M.A.; Wang, M.; Arnason, J.; Mehta, A.; Purev, E.; Maloney, D.G.; Andreadis, C.; et al. Lisocabtagene maraleucel for patients with relapsed or refractory large B-cell lymphomas (TRANSCEND NHL 001): A multicentre seamless design study. Lancet 2020, 396, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Berdeja, J.G.; Madduri, D.; Usmani, S.Z.; Jakubowiak, A.; Agha, M.; Cohen, A.D.; Stewart, A.K.; Hari, P.; Htut, M.; Lesokhin, A.; et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): A phase 1b/2 open-label study. Lancet 2021, 398, 314–324. [Google Scholar] [CrossRef]
- Park, K.; Veena, M.S.; Shin, D.S. Key Players of the Immunosuppressive Tumor Microenvironment and Emerging Therapeutic Strategies. Front. Cell Dev. Biol. 2022, 10, 830208. [Google Scholar] [CrossRef] [PubMed]
- Callahan, M.K.; Postow, M.A.; Wolchok, J.D. CTLA-4 and PD-1 Pathway Blockade: Combinations in the Clinic. Front. Oncol. 2015, 4, 385. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune Checkpoint Targeting in Cancer Therapy: Toward Combination Strategies with Curative Potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef]
- Berman, D.; Korman, A.; Peck, R.; Feltquate, D.; Lonberg, N.; Canetta, R. The development of immunomodulatory monoclonal antibodies as a new therapeutic modality for cancer: The Bristol-Myers Squibb experience. Pharmacol. Ther. 2015, 148, 132–153. [Google Scholar] [CrossRef]
- Su, C.; Wang, H.; Liu, Y.; Guo, Q.; Zhang, L.; Li, J.; Zhou, W.; Yan, Y.; Zhou, X.; Zhang, J. Adverse Effects of Anti-PD-1/PD-L1 Therapy in Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 554313. [Google Scholar] [CrossRef]
- Davari, K.; Holland, T.; Prassmayer, L.; Longinotti, G.; Ganley, K.P.; Pechilis, L.J.; Diaconu, I.; Nambiar, P.R.; Magee, M.S.; Schendel, D.J.; et al. Development of a CD8 co-receptor independent T-cell receptor specific for tumor-associated antigen MAGE-A4 for next generation T-cell-based immunotherapy. J. Immunother. Cancer 2021, 9, e002035. [Google Scholar] [CrossRef]
- Wilde, S.; Sommermeyer, D.; Frankenberger, B.; Schiemann, M.; Milosevic, S.; Spranger, S.; Pohla, H.; Uckert, W.; Busch, D.H.; Schendel, D.J. Dendritic cells pulsed with RNA encoding allogeneic MHC and antigen induce T cells with superior anti-tumor activity and higher TCR functional avidity. Blood 2009, 114, 2131–2139. [Google Scholar] [CrossRef]
- Sailer, N.; Fetzer, I.; Salvermoser, M.; Braun, M.; Brechtefeld, D.; Krendl, C.; Geiger, C.; Mutze, K.; Noessner, E.; Schendel, D.; et al. T-Cells Expressing a Highly Potent PRAME-Specific T-Cell Receptor in Combination with a Chimeric PD1-41BB Co-Stimulatory Receptor Show a Favorable Preclinical Safety Profile and Strong Anti-Tumor Reactivity. Cancers 2022, 14, 1998. [Google Scholar] [CrossRef]
- Lai, J.-P.; Robbins, P.F.; Raffeld, M.; Aung, P.P.; Tsokos, M.; Rosenberg, S.A.; Miettinen, M.M.; Lee, C.-C.R. NY-ESO-1 expression in synovial sarcoma and other mesenchymal tumors: Significance for NY-ESO-1-based targeted therapy and differential diagnosis. Mod. Pathol. 2012, 25, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.-P.; Rosenberg, A.Z.; Miettinen, M.M.; Lee, C.-C.R. NY-ESO-1 expression in sarcomas A diagnostic marker and immunotherpy target. Oncoimmunology 2012, 1, 1409–1410. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K.; Jungbluth, A.A.; Stockert, E.; Qian, F.; Gnjatic, S.; Tammela, J.; Intengan, M.; Beck, A.; Keitz, B.; Santiago, D.; et al. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res. 2003, 63, 6076–6083. [Google Scholar] [PubMed]
- Resnick, M.B.; Sabo, E.; Kondratev, S.; Kerner, H.; Spagnoli, G.C.; Yakirevich, E. Cancer-testis antigen expression in uterine malignancies with an emphasis on carcinosarcomas and papillary serous carcinomas. Int. J. Cancer 2002, 101, 190–195. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Araujo, D.M.; Abdul Razak, A.R.; Agulnik, M.; Attia, S.; Blay, J.-Y.; Carrasco Garcia, I.; Charlson, J.A.; Choy, E.; Demetri, G.D.; et al. Afamitresgene autoleucel for advanced synovial sarcoma and myxoid round cell liposarcoma (SPEARHEAD-1): An international, open-label, phase 2 trial. Lancet 2024, 403, 1460–1471. [Google Scholar] [CrossRef]
- D’angelo, S.P.; Melchiori, L.; Merchant, M.S.; Bernstein, D.; Glod, J.; Kaplan, R.; Grupp, S.; Tap, W.D.; Chagin, K.; Binder, G.K.; et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1c259T cells in synovial sarcoma. Cancer Discov. 2018, 8, 944–957. [Google Scholar] [CrossRef]
- Ishihara, M.; Kitano, S.; Kageyama, S.; Miyahara, Y.; Yamamoto, N.; Kato, H.; Mishima, H.; Hattori, H.; Funakoshi, T.; Kojima, T.; et al. NY-ESO-1-specific redirected T cells with endogenous TCR knockdown mediate tumor response and cytokine release syndrome. J. Immunother. Cancer 2022, 10, e003811. [Google Scholar] [CrossRef]
- Mahnke, Y.D.; Brodie, T.M.; Sallusto, F.; Roederer, M.; Lugli, E. The who s who of T-cell differentiation Human memory T-cell subsets. Eur. J. Immunol. 2013, 43, 2797–2809. [Google Scholar] [CrossRef] [PubMed]
- Jaravine, V.; Mösch, A.; Raffegerst, S.; Schendel, D.J.; Frishman, D. Expitope 2.0: A tool to assess immunotherapeutic antigens for their potential cross-reactivity against naturally expressed proteins in human tissues. BMC Cancer 2017, 17, 892. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Sanomachi, T.; Katsuya, Y.; Nakatsura, T.; Koyama, T. Next-Generation CAR-T and TCR-T Cell Therapies for Solid Tumors: Innovations, Challenges, and Global Development Trends. Cancers 2025, 17, 1945. [Google Scholar] [CrossRef]
- Beenen, A.C.; Sauerer, T.; Schaft, N.; Dörrie, J. Beyond Cancer: Regulation and Function of PD-L1 in Health and Immune-Related Diseases. Int. J. Mol. Sci. 2022, 23, 8599. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Simon, S.; Labarriere, N. PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? Oncoimmunology 2018, 7, e1364828. [Google Scholar] [CrossRef]
- Baumeister, S.H.; Freeman, G.J.; Dranoff, G.; Sharpe, A.H. Coinhibitory Pathways in Immunotherapy for Cancer. Annu. Rev. Immunol. 2016, 34, 539–573. [Google Scholar] [CrossRef]
- Boussiotis, V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway Immune Checkpoint Blockade as Cancer Therapy. N. Engl. J. Med. 2017, 375, 1767–1778. [Google Scholar] [CrossRef]
- LaFleur, M.W.; Muroyama, Y.; Drake, C.G.; Sharpe, A.H. Inhibitors of the PD-1 Pathway in Tumor Therapy. J. Immunol. 2018, 200, 375–383. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Yin, Q.; Wu, L.; Han, L.; Zheng, X.; Tong, R.; Li, L.; Bai, L.; Bian, Y. Immune-related adverse events of immune checkpoint inhibitors: A review. Front. Immunol. 2023, 14, 1167975. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef]
- Mardiana, S.; Solomon, B.J.; Darcy, P.K.; Beavis, P.A. Supercharging adoptive T cell therapy to overcome solid tumor–induced immunosuppression. Sci. Transl. Med. 2019, 11, eaaw2293. [Google Scholar] [CrossRef]
- Rasmussen, J.H.; Lelkaitis, G.; Håkansson, K.; Vogelius, I.R.; Johannesen, H.H.; Fischer, B.M.; Bentzen, S.M.; Specht, L.; Kristensen, C.A.; von Buchwald, C.; et al. Intratumor heterogeneity of PD-L1 expression in head and neck squamous cell carcinoma. Br. J. Cancer 2019, 120, 1003–1006. [Google Scholar] [CrossRef]
- Nicoś, M.; Krawczyk, P.; Crosetto, N.; Milanowski, J. The Role of Intratumor Heterogeneity in the Response of Metastatic Non-Small Cell Lung Cancer to Immune Checkpoint Inhibitors. Front. Oncol. 2020, 10, 569202. [Google Scholar] [CrossRef]
- Palicelli, A.; Croci, S.; Bisagni, A.; Zanetti, E.; De Biase, D.; Melli, B.; Sanguedolce, F.; Ragazzi, M.; Zanelli, M.; Chaux, A.; et al. What Do We Have to Know About PD-L1 Expression in Prostate Cancer? A Systematic Literature Review (Part 6): Correlation of PD-L1 Expression with the Status of Mismatch Repair System, BRCA, PTEN, and Other Genes. Biomedicines 2022, 10, 236. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J. Tumor microenvironment in PD1/PD-1-mediated tumor immune escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Filin, I.Y.; Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Recent Advances in Experimental Dendritic Cell Vaccines for Cancer. Front. Oncol. 2021, 11, 730824. [Google Scholar] [CrossRef]

| Assessment | Test System | Method | Results |
|---|---|---|---|
| On-target/off-tumor toxicity | NY-ESO-1/LAGE-1a mRNA-positive healthy cells, with or without NY-ESO-1/LAGE-1a SLL peptide | Co-cultures of MDG1015 and healthy test cells: IFN-γ secretion at 24 h; live-cell imaging assay 0–120 h | Recognition of one batch of NY-ESO-1/LAGE-1a mRNA-positive mDC |
| Off-target/off-tumor toxicity | Antigen-negative tumor cell lines (9 PD-L1-positive) derived from various tissues; unloaded or loaded with NY-ESO-1/LAGE-1a SLL peptide | Co-cultures of MDG1015 and tumor cells: IFN-γ secretion at 24 h; proliferation over 15 d | No cross-recognition of antigen-negative, PD-L1-positive tumor cell lines; no cytokine-independent expansion of MDG1015 in the absence of NY-ESO-1/LAGE-1a-positive target cells |
| Human LCL panel with common HLA allotypes; unloaded or loaded with NY-ESO-1/LAGE-1a SLL peptide | Co-cultures of MDG1015 and LCL panel cells: IFN-γ secretion at 24 h | Cross-recognition of 3 unloaded LCL; relevant cross-recognized HLA allotypes: HLA-A*02:02 and HLA-A*02:03; NY-ESO-1/LAGE-1a SLL-loaded LCL were recognized that expressed HLA-A*02:04 and HLA-A*02:09 | |
| Mismatched peptide test panel identified using Expitope® 2.0 database: 318 peptides loaded on T2_PD-L1 cells; 5 peptides were expressed as ivtRNA in tumor cell lines | Co-cultures of MDG1015 and mismatched peptide-pulsed T2_PD-L1 cells or ivtRNA-transfected tumor cell lines: IFN-γ secretion at 24 h | 27 mismatched peptides cross-recognized on peptide- loaded T2_PD-L1 cells; recognition of 1 mismatched peptide-encoding ivtRNA in tumor cell lines; and no recognition of full-length gene-encoding ivtRNA in tumor cell lines | |
| NY-ESO-1/LAGE-1a mRNA-negative panel of primary human healthy cell types; unloaded or loaded with SLL peptide | Co-cultures of MDG1015 and healthy test cells: IFN-γ secretion at 24 h; live-cell imaging assay 0–120 h | No cross-recognition of NY-ESO-1/LAGE-1a mRNA-negative test cells of the healthy cell panel |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bürdek, M.; Prinz, P.U.; Mutze, K.; Bosch, M.; Tippmer, S.; Coluccio, A.; Geiger, C.; Majumder, S.; Longinotti, G.; Schendel, D.J. Preclinical Development of Costimulatory Switch Protein (CSP)-Armored NY-ESO-1/LAGE-1a-Specific TCR-T Cells for Therapy of Hard-to-Treat PD-L1-Positive Solid Tumors. Int. J. Transl. Med. 2025, 5, 45. https://doi.org/10.3390/ijtm5040045
Bürdek M, Prinz PU, Mutze K, Bosch M, Tippmer S, Coluccio A, Geiger C, Majumder S, Longinotti G, Schendel DJ. Preclinical Development of Costimulatory Switch Protein (CSP)-Armored NY-ESO-1/LAGE-1a-Specific TCR-T Cells for Therapy of Hard-to-Treat PD-L1-Positive Solid Tumors. International Journal of Translational Medicine. 2025; 5(4):45. https://doi.org/10.3390/ijtm5040045
Chicago/Turabian StyleBürdek, Maja, Petra U. Prinz, Kathrin Mutze, Miriam Bosch, Stefanie Tippmer, Andrea Coluccio, Christiane Geiger, Snigdha Majumder, Giulia Longinotti, and Dolores J. Schendel. 2025. "Preclinical Development of Costimulatory Switch Protein (CSP)-Armored NY-ESO-1/LAGE-1a-Specific TCR-T Cells for Therapy of Hard-to-Treat PD-L1-Positive Solid Tumors" International Journal of Translational Medicine 5, no. 4: 45. https://doi.org/10.3390/ijtm5040045
APA StyleBürdek, M., Prinz, P. U., Mutze, K., Bosch, M., Tippmer, S., Coluccio, A., Geiger, C., Majumder, S., Longinotti, G., & Schendel, D. J. (2025). Preclinical Development of Costimulatory Switch Protein (CSP)-Armored NY-ESO-1/LAGE-1a-Specific TCR-T Cells for Therapy of Hard-to-Treat PD-L1-Positive Solid Tumors. International Journal of Translational Medicine, 5(4), 45. https://doi.org/10.3390/ijtm5040045






