Abstract
After an initial positive response to chemotherapy, cancer patients often acquire chemoresistance and tumor relapse, which makes cancer one of the most lethal diseases worldwide. Exosomes are essential mediators of cell-to-cell communication by delivering their cargo, such as proteins, RNAs and DNA, from cell to cell. They participate in cancer progression, metastasis, immune response and therapy resistance. Their ability to shuttle between cells makes them efficient drug delivery systems. As drug transporters, they provide novel strategies for cancer therapy by advancing targeted drug therapy and improving the therapeutic effects of anti-cancer medications. In this review, a comprehensive overview of the potential of exosomes as therapeutic agents and targeted molecules in the treatment of cancer patients is given. The current challenges of preparation of exosomes loaded with drugs and delivering them to the recipient tumor cells as well as a consequent exosome-mediated cancer therapy are also discussed.
1. Introduction
Cancer progression is a multi-step process and eventually leads to the development of metastases and patient death. In 1863, Virchow postulated that cancer originates at sites of chronic inflammation, because these regions were observed to cause enhanced cell proliferation [1]. Nowadays, it is recognized that proliferation of cells alone does not cause cancer. Continued cell proliferation occurs in an environment rich with growth/survival factors, activated stroma and DNA-damage-promoting agents, as well as inflammatory cells that may cause neoplastic risk [2]. As soon as a cluster of neoplastic cells is established, angiogenesis, the development of blood vessels, occurs to provide the tumor with oxygen, nutrients and growth factors, as well as to allow tumor cells to disseminate to distant organs [3]. The invasion and metastasis of solid tumors is accompanied by the epithelial–mesenchymal transition (EMT) [4], a process in which epithelial cells acquire mesenchymal features for their movement to distant organs. Metastatic dissemination can occur early in the malignant progression [5]. In the last decade, the 5-year survival of tumor patients has increased since solid tumors are earlier detected, locally confined and treated by improved adjuvant therapies. However, clinicians are confronted with an increase in late relapse rates, prolonged disease courses and chemoresistance. Therefore, new targeted therapies have to be developed. Such therapies could be performed by therapeutic exosomes loaded with tumor-specific drugs or genetic material, which in turn are specifically directed to the tumor [6]. Exosomes are a part of the tumor microenvironment and participate in this process regulating multiple tumor stages, for example angiogenesis, immune response, chemoresistance epithelial–mesenchymal transition (EMT) and metastasis [7]. To date, tumor cells release higher levels of exosomes than normal healthy cells [8].
In 1983, exosomes were first detected in maturing mammalian reticulocytes by Harding et al. [9] and Johnstone et al. [10]. Later, in 1987, the name of ‘exosomes’ was given by Johnstone et al. [10]. During the maturation process of reticulocytes into erythrocytes, exosomes were observed to selectively remove plasma membrane proteins. In this respect, their lipid composition contains high sphingomyelin content and is similar to the origin cell [11].
Exosomes are a subgroup of extracellular vesicles (EVs) [12]. They are small, at a size of 30–200 nm and of endosomal origin. They mediate intercellular communication by transferring their cargo containing proteins, RNA, DNA and lipids from cell to cell [8,13]. By delivering these molecules to neighboring and distant cells, they may influence the phenotype of recipient cells. Since their content presents the features of the origin cell, they may transmit the characteristics of the origin cell. Thus, the movement of exosomes from cancer cells may propagate cancerous attributes to healthy cells and can help cancer cells to spread genetic information, leading to the development and maintenance of metastases [14]. Exosomes can be found in different body fluids, including blood, bronchoalveolar lavage, breast milk, malignant effusions, urine and nasal lavage and are formed, among others, by lymph cells, blood platelets, mast cells, dendritic cells, nerve cells, astrocytes and tumor cells, indicating that they can shuttle in different body regions [15].
So far, a variety of methods has been used for the extraction and purification of exosomes by different studies. Techniques for the preparation of exosomes include differential density and gradient centrifugation, size exclusion chromatography, filtration, polymer-based precipitation and isolation by filtering and chips, which have been described in detail by Shtam et al. [16]. In addition, immunological separation using a wide range of antibodies that are directed against target molecules, such as the exosomal surface markers tetraspanins (CD9, CD63, CD81, CD82) and heat-shock proteins (Hsp60, Hsp70, and Hsp90), and carrying out experiments with both magnetic beads and nanowires are also reliable methods [17]. Numerous laboratories use commercial kits based on polymers, which can be carried out in a few steps. The main advantage of these kits is a quick purification and the high yields of the exosomes, but their main disadvantage is the co-precipitation of proteins of non-exosomal origin. However, the gold standard method seems to be the ultracentrifugation method of isolation, but this technique is labor-intensive and time-consuming, and leads to lower exosome amounts. Quality, concentration and biological activity of the extracted exosomes can be verified by Western blot, nanoparticle tracking analysis (NTA), and confocal microscopy [18,19,20].
Considering the involvement of exosomes in cancer development and progression, the translation of exosome shuttle into the cancer therapy may be of clinical relevance. Since exosomes can deliver their cargo to specific cells, investigation of these vehicles for targeted drug or signal delivery may be a promising approach. Thus, exosomes as carriers of anti-tumor compounds may be promising in the treatment of tumors. In addition, an alternative therapy approach is to target cancer-derived exosomes to prevent cancer progression [21,22].
In this review, a comprehensive overview of the pivotal roles of exosomes in the potential clinical application as novel therapeutic agents and targets is presented. Improved treatment strategies to enhance drug effects mediated by improved drug delivery of this vehicles to the region of the disease are also described.
2. Exosome Biogenesis
Exosomes are created by the endosomal pathway (Figure 1) [23]. Beginning with the (receptor-mediated) endocytosis of extracellular materials by the invagination of the plasma membrane, vesicles are formed and converged into early endosomes. These early endosomes contain extracellular components and plasma membrane proteins. Their maturation into late endosomes occurs by ATPase-mediated acidification. Late endosomes can even obtain cargo-loaded vesicles from the Trans-Golgi network. Multivesicular bodies (MVBs) mature from late endosomes through membrane in-folding processes to form small intra-vesicular vesicles (IVLs). To date, diverse mechanisms of generation of IVLs have been described. For example, MVB formation is coordinated by ESCRT (endosomal sorting complex required for transport) which encompasses four soluble multi-protein complexes, namely ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III [24]. It started with the binding of ubiquitinated proteins with the ESCRT-0 subunit in the endosomal membrane The association with the cytosolic side of the endosomal membrane allows that particular proteins are sorted into ILVs. Alternatively, another course of EV formation occurs in the absence of ESCRT and is based on ceramide- or tetraspanin-enriched microdomains within the endosomal membrane [25]. Upon fusion of the MVB with the plasma membrane, the ILVs, which are contained within the MVBs, are released into the extracellular space as exosomes. The subsequent exosomal DNA and RNA uptake in the recipient cell occurs by either membrane fusion followed by release of these molecules in the cytoplasm or by macro-pinocytosis or receptor/raft-mediated endocytosis. This uptake of exosomes by neighboring and distant cells occurs by docking of the exosomes with specific proteins, sugars, and lipids or by micro-pinocytosis [26]. In turn, the internalized exosomes are targeted to the endosomes, which release their content in the recipient cell. The cargo containing genetic material and proteins is then active in the guest cell.
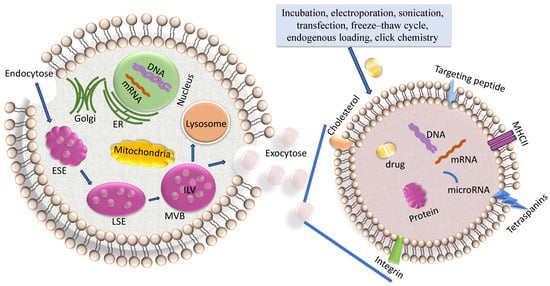
Figure 1.
Biogenesis of exosomes. On the left side, the biogenesis of exosomes, and on the right side, an enhancement of an exosome loaded with a drug are shown. The biogenesis of exosomes is described in the text and comprises endocytosis, MVB formation and exosome secretion into the extracellular microenvironment. Lading of exosomes, as described in the text, is carried out by diverse methods. ER, endoplasmatic reticulum; ESE, early sorting endosome; LSE, late sorting endosome; MVB, multivesicular body; ILV, intraluminal vesicles.
Another pathway is that MVBs can also fuse with lysosomes, leading to the degradation of their contents [8] (Figure 1).
3. Characteristics of Exosomes and Their Cargo
The exosome membrane resembles that of the origin cells from which the exosomes are released. The phospholipid belayer consists of diverse proteins, such as anexin II, heat shock proteins, major histocompatibility complex (MHC) class II complexes, integrins and tetraspanins [27]. The cell-specific proteins are dependent on the origin cell—for example, exosomes derived from B-lymphocytes are also enriched in MHC-II peptides [28], while exosomes derived from glioma cells carry epidermal growth factor receptor (EGFR) [29].
Exosomes are found in different body fluids, such as blood, lymph and interstitial fluid. They use the circulation as a transport system to reach distal cellular sites where they can be uptaken by other cells. Although cancer-derived exosomes only form a small fraction of all exosomes in body fluids, cancer cells can release more than 10,000 of these vesicles during a day [30]. While normal blood is assumed to contain as many as 2000 trillion exosomes, the blood of cancer patients is expected to even contain the double yield, possibly due to altered cellular physiology [31]. This high yield of exosomes in the blood fluid demonstrates that exosomes stably circulate in the body fluids. This is because of their ability to circumvent the mononuclear phagocytic system by owing their relatively small size but also, in particular, their negatively charged phospholipid membrane and presentation of the surface marker CD47 [32]. CD47, a surface protein, seems to reduce phagocytic uptake by binding to its receptor signal-regulatory protein (SIRP) α, which is expressed on cells of the myeloid lineage. Exosomes without CD47 showed a lower half-life than their CD47-expressing counterparts [33]. In addition, exosomes are relatively large molecules, which means they avoid being rapidly cleared by the kidney. Amazingly, exosomes even have the aptitude to cross the brain barrier that excludes most molecules, namely a size bigger than 400 Da. However, exosomes modified to specifically target the brain by expressing a brain-targeting peptide overcome this barrier [34,35].
Exosomes contain different components, such as RNAs, including, among others, mRNAs, microRNAs (miRNAs) and long noncoding RNAs (lncRNA), snRNAs (small nuclear RNAs), proteins, ceramides and cholesterol, lipids, and DNA [22]. Considering the movement of exosomes from tumor cells to healthy cells to propagate genetic information leading to tumor progression, their content of exosomal RNAs and proteins are of particular interest [14]. They may alter the characteristics of the recipient cells to which they are transported [36]. In addition, exosomes are also delivered back from the surrounding cells into the tumor cells.
Exosomes contain variable amounts of RNAs, comprising over a dozen different RNA forms, including miRNAs forming the predominant amounts. They belong to the non-coding RNAs (ncRNAs) and are not translated into proteins, but function in both RNA silencing and post-transcriptional regulation of gene expression [37]. In this respect, they inhibit the expression of mRNA. MiRNAs and mRNAs are incorporated into an argonaute-containing protein complex termed RISC (RNA-induced silencing complex), in which mRNA silencing takes place. The RISC was even found in exosomes [38]. MiRNAs can also interact with another ncRNA, namely lncRNAs, and so abrogate the regulatory effects mediated by lncRNAs [39,40].
Some examples of exosomal miRNAs that play a role in cancer include, among others, miR-155 which is a highly conserved and an early detected miRNA. It exhibits unique expression profiles and multifunctionality. This miRNA can bind to a repertoire of over 241 genes and so influence diverse signaling pathways. MiR-155 plays a critical role in various physiological and pathological processes, notably in cancer [41]. Another miRNA, miR-23a-3p was found to have high expression levels in breast cancer-derived exosomes, which can distinguish between adriamycin-resistant and sensitive patients [42]. In addition, the exosomal levels of miR-200 are associated with the tumor marker CA125, cell proliferation, apoptosis and overall survival of epithelial ovarian cancer [43]. The exosomal levels of miR-146a can differentiate hepatocellular carcinoma from liver cirrhosis patients [44].
Besides, studies have shown that specific cell-derived exosomes transfer their biological cargo from cell to cell to regulate a variety of processes, and so exosomes are involved in diverse processes, e.g., in tumor progression, angiogenesis, tissue repair and immune functions [45]. These specific characteristics make exosomes a promising source of a cellular therapy for various conditions, which have the advantage of further engineering these exosomes for the delivery of therapeutics. Hereby, it is important to mention that the origin cell that produces exosomes can be modulated by therapeutics, as well as by targeting ligands to be loaded.
4. Loading of Exosomes with Drugs or Nucleic Acids
Studies have shown that the contents of exosomes can lead to tumor progression, invasion and metastasis [14,46]. By specifically manipulating their cargo, exosomes can be excellent carriers of therapeutics because they are body-intrinsic molecules and so may have potential biocompatibitly with the different body regions. In addition, they possess further advantages over traditional synthetic delivery vectors, including lower immunogenicity and high stability in the blood circulation. Moreover, loading genetic material into exosomes stimulates the immune system [21]. Finally, they can directly deliver drugs to cells because their lipid bilayer membrane can be modified to enhance their targeting specificity [47].
There are different strategies for exosome drug loading, including incubation, electroporation, transfection, sonication, freeze–thaw cycle, endogenous loading and click chemistry (Figure 1) [48].
For example, the incubation and electroporation method are carried out by the co-incubation of exosomes with small polar molecules. The loading efficiency depends on the polarity of these molecules. Following isolation and purification of exosomes, purified exosomes are mixed with a specific hydrophobic drug or nucleic acids and then incubated or electroporated. The applied electric charge disrupts the exosome membrane and forms transient, temporary micro-pores in the membrane. These micro-pores enhance the membrane permeability and develop an electrical potential, which facilitates efficient loading of the drugs. However, the major disadvantages of electroporation are the apoptotic cells caused by the high voltage applied. To confirm the encapsulation efficiency, the absorption intensity of free drugs, exosomes and drug-loaded exosomes are analyzed by a UV-spectrophotometer. In addition, the fluorescence spectra can also be carried out to confirm the successful loading of drugs. Afterwards, the co-localization of drugs and fluorescence-labeled exosomes can be observed in cell cultures by fluorescence microscopy. Liang et al. loaded exosomes with 5-Fluorouracil (5-FU) and the miRNA inhibitor for miR-21 by electroporation. When administered to mice with 5-FU resistant colon tumors, the exosomes enhanced cell cytotoxicity and antitumor activity [49].
The transfection method involves the transfection of molecules, especially small proteins, siRNAs or miRNAs, into the parent cells, and the cell packaging into exosomes. Kooijmans et al. transfected neuroblastoma cells with human glycosylphosphatidylinositol (GPI)-anchored protein decay-accelerating factor, which was fused to targeting ligands for the EGFR, and demonstrated that the cells generated exosomes harboring EGFR-targeting ligands on their surface [50].
Another method for exosome loading is sonication, which uses high-frequency ultrasound energy to partly disrupt the exosome membrane, helping the diffusion of therapeutic agents. Kim et al. sonicated a mixture of exosomes and paclitaxel with a 20% amplitude, with six cycles of 30 s on/off for three minutes followed by a two-minute cooling period between each cycle. They showed that a high amount of paclitaxel could be loaded into exosomes, which was measured by a high-performance liquid chromatography. In a model of murine Lewis lung carcinoma pulmonary metastases, the exosomes successfully integrated paclitaxel by sonication, and showed an almost complete co-localization of delivered exosomes with the cancer cells and a potent anticancer effect [51].
Freeze-thaw cycles involve the incubation of exosomes with drugs in several alternate cycles at room and below freezing temperatures (e.g., −80 °C). They permeabilize the membrane and allow the drugs to enter the exosome membrane. Sato et al. [52] applied the freeze-thaw method to fuse exosomes and liposomes containing connexin. Fusion of the exosomes with liposomes to form exosome–liposome hybrids could significantly increase their loading capacity and the half-life of exosomes in plasma.
The endogenous loading method uses small drug molecules that are co-incubated together with donor cells or treated with other loading strategies so that they can be captured by the cell lipid bilayer and encapsulated in exosomes. The subsequent exosomes contain, then, the desired small-molecule drugs [48].
Using the click chemistry, small molecules and macromolecules can be attached to the exosome surface by covalent chemical bonds. An example of such a bioconjugation is the copper-catalyzed azide-alkyne cycloaddition, which is a relatively rapid technique specific for the action between an alkyne and an azide to form a triazole linkage [53].
5. Delivery of Drug-Loaded Exosomes to Cancer Cells
As soon as the exosomes are loaded with the particular therapeutic, they have to be directed to the target cell. However, the targeted delivery of drugs to recipient cancer cells is one of the essential challenges in cancer therapy research. Typically, nanoparticles as drug carriers are used. They possess prolonged bioavailability, an enhanced permeation and retention effects on tumors, as well as few side effects. Siemer et al. [54] identified the ion channel LRRC8A as a critical component for cisplatin resistance of head and neck cancer patients. To avoid LRRC8A-mediated cisplatin resistance, they constructed cisplatin-loaded, polysarcosine-based core cross-linked polymeric nanoparticles with low immunogenicity, low toxicity and prolonged in vivo circulation. By circumventing the LRRC8A-transport pathway via the endocytic delivery pathway, the directed delivery of cisplatin by these nanoparticles was able to overcome cisplatin resistance and successfully eliminate cancer cells in a cell spheroid model of head and neck cancer. Furthermore, Lu et al. [55] observed that copper (II) bis (diethyldithiocarbamate) nanoparticles effectively induced copper-dependent programmed cell death in non-small lung cancer cells and potent anti-tumor effects in a cisplatin-resistant tumor model in vivo.
As demonstrated by Ye et al. [56], exosomes combined with low-density lipoprotein (LDL) could improve their uptake by a human primary glioma cell line and permeation into three-dimensional glioma spheroids, compared with unmodulated exosomes. In vivo imaging experiments exposed that LDL could clearly promote exosome extravasation across the blood brain barrier and distribution at the glioma site. The conjugation of apolipoprotein A-1 peptides with lipids allowed for the targeted delivery of methotrexate-loaded exosomes to primary glioma cells.
Triple-negative breast cancer (TNBC) is the most metastatic and recurrent subtype of breast cancer. Owing to the lack of estrogen and progesterone receptors as well as human epidermal grow factor receptor 2 (HER2) and, consequently, to the lack of therapeutic targets, chemotherapy and surgical intervention are the only treatments for TNBC. Li et al. [57] constructed a macrophage-derived exosome-coated poly (lactic-co-glycolic acid) nanoplatform that successfully targeted TNBC cells that overexpressed the mesenchymal–epithelial transition factor (c-Met) in vitro and in vivo. The constructed exosome-coated nanoparticles significantly upgraded the cellular uptake efficiency and the antitumor efficacy of doxorubicin.
Since the uptake of exosomes by recipient cells occurs, among others, by receptor-mediated binding, their surface can be modulated for cell-specific targeting via ligand-receptor binding. For pancreatic cancer therapy, Faruque et al. [58] engineered human pancreatic cancer cell-derived exosomes by conjugating the functional ligand Arg-Gly-Asp (RGD) and magnetic nanoparticles on their surface. The increased therapeutic effect was contributed to the modulation of the exosome surface using RGD, which has an affinity for the highly expressed αvβ3 integrin in pancreatic cancer cells. The RGD-modified autologous exosomes effectively penetrated and internalized tumor cells, and eventually regressed the tumors by stimulating the spontaneous removal of α-smooth muscle actin and collagen type 1 in the extracellular matrix of mouse xenografts.
For the specific drug delivery to lung tumors, Pham et al. [59] coupled exosomes with EGFR-targeting peptides and nanoantibodies via protein ligases that facilitated the specific uptake by EGFR-positive lung cancer cells. Systemic delivery of paclitaxel by EGFR-targeting exosomes at a low dose significantly increased the efficacy of drugs in a xenografted mouse model of EGFR-positive lung cancer.
Zhou et al. [60] used exosomes-based biomimetic nanoparticles and designed hybrid exosomes loaded with the protein kinase inhibitor dasatinib by fusing human pancreatic cancer cell-derived exosomes with dasatinib-loaded liposomes. Dasatinib-loaded hybrid exosomes displayed significantly higher uptake rates and cytotoxicity in parent pancreatic ductal adenocarcinoma cells than free drugs or liposomal formulations.
A further option for exosome delivery is the use of antibody-like affibodies that are based on the immunoglobulin-binding domain of protein A. Exosomes that express HER2-affibodies from genetically engineered human embryonic kidney donor cells on their surface successfully delivered paclitaxel and miR-21 to HER2-expressing colorectal cancer cells and showed anti-tumor effects in mice [49]. Genetically engineered exosomes that express a fragment of interleukin-3 (IL-3) on their surface effectively bound to IL3-receptor-overexpressing chronic myeloid leukemia cells and inhibited cell growth by delivering the tyrosine kinase inhibitor imatinib to the cells [61]. For immunotherapy of hepatocellular cancer patients, exosome vaccines can be generated by anchoring hepatocellular cancer-targeting peptides, antigens or immune adjuvants on the surface of exosomes that induce effective tumor-specific immune responses [62].
In summary, diverse technical platforms have been developed to modulate the exosome membrane by specific molecules for the production of specific exosome delivery systems in different cancer types.
6. Drug-Loaded Exosomes vs. Free Drugs in Cancer Treatment
Due to their high safety and biocompatibility, exosomes are specific drug delivery systems and enhance the uptake of the drug by the target cell, reducing the toxicity and maintaining therapeutic effects. The delivery of drugs by exosomes has a variety of advantages, compared with the administration of free drugs, without a carrier system.
Exosomes possess unique characteristics owing to their ability to travel from one cell to another cell, to across their cargo through the cell membrane and to release their contents in a biologically active form into the recipient cell. Within the target cell, the fate of drug-loaded exosomes is sealed by degradation in endosomes or lysosomes. The target-orientated release of the drugs in the cancer cells lowers the toxicity and immunogenicity. The increase in the local concentration of drugs in cancer cells maximizes the therapeutic efficacy, and thereby, the doses of drugs can be decreased. In this respect, the target-orientated delivery provides strategies for more specific therapies. In contrast, free drugs also affect non-target organs, enhancing potential side effects to non-diseased organs. Thus, the non-restricted toxicity of chemotherapeutics reduces the complete use of the therapeutic potential of the drugs. Concerning this matter, the local drug delivery is particularly important in cancer treatment, since the tumor is localized as distinct metastases in diverse organs [47,53].
7. Exosomes as Therapeutic Agents
Since exosomes have exclusive characteristics, including stability, low immunogenicity and high biocompatibility, they are ideal candidates in the treatment of cancer patients. In particular, their eligibility in regenerative outcomes of injury and disease treatment have been shown in mesemchymal stem cell-derived exosomes that play an essential role in wound repair, tissue regeneration and immune response by activating several signaling pathways [21,63]. Table 1 gives an overview of a selection of modified exosomes that may serve as therapeutic agents (Table 1).
Over the past decades, a variety of synthetic drug delivery systems have been developed and introduced to the market. However, the application of such systems is restricted since they are often inefficient, cytotoxic and/or immunogenic. Since exosomes are naturally occurring vesicles, their unique features can be used as therapeutic agents to induce immunogenic cell death. The probability of using exosomes derived from dendritic cells as a cancer therapeutic vaccine has been tested in two Phase I clinical studies in melanoma and lung cancer patients. These studies demonstrated that exosomes derived from cancer patient dendritic cells, important in the induction of antitumor immunity, can stimulate both T cells and natural killer cells, leading to adaptive, innate cellular immune responses, respectively. The relevance of these exosomes is to transfer antigen-loaded MHC I and II molecules, and other associated molecules, to naive dendritic cells, to amplify a cellular immune response [64]. They can prime antigen-specific CD4 and CD8 T-cells through MHC-I and II expression and antigen presentation [65]. In breast cancer, synthetic multivalent antibody retargeted exosomes, and activated and redirected T-cells to cancer cells that express EGFR or HER-2 on their surface by presenting the respective antibodies [66,67].
In this respect, Zhou et al. constructed a delivery system from bone marrow mesenchymal stem cell exosomes. They loaded galectin-9 siRNA into exosomes by electroporation and superficially modified them with oxaliplatin prodrug as an immunogenic cell death trigger. The approach achieved therapeutic efficacy in pancreatic cancer treatment by eliciting anti-tumor immunity through tumor-suppressive macrophage polarization and cytotoxic T lymphocyte recruitment, while also downregulating regulatory T cells (Tregs), a T-cell population that suppresses the immune response and maintains immune homeostasis [68].
CD40 signaling is critical in the activation of dendritic cells. Wang et al. [69] identified exosomes from CD40 ligand gene-modified lung tumor cells to be more immunogenic than unmodified exosomes. These modified exosomes induced a more mature phenotype of dendritic cells and stimulated them to secrete increased levels of IL-12. In a mouse model, they induced robust tumor antigen-specific CD4+ T cell proliferation and enhanced the anti-tumor activity of T-cells.
For individualized immunotherapies, Li et al. [70] developed a nanovaccine platform containing dendritic cell-derived exosomes loaded with patient-specific neoantigens. The nanovaccine elicited a potent antigen-specific, broad spectrum of T-cell and B-cell-mediated immune responses. In particular, the delivery of neoantigen-exosome nanovaccine specifically inhibited tumor growth, extended survival, delayed tumor occurrences with long-term memory and eradicated lung metastasis. Due to the presence of exosomal proteins, this exosome-based nanovaccine elicited a synergistic antitumor response superior to liposomal formulation.
The therapeutic usefulness of umbilical cord blood-derived M1 macrophage exosomes loaded with cisplatin in ovarian cancer with platinum resistance was investigated by Zhang et al. [71]. In addition, M1 macrophage-derived exosomes carried lnRNA H19, implicated in upregulation of PTEN protein and downregulation of miR-130a and Pgp gene. These engineered exosomes were able to reverse cisplatin drug resistance. Thus, cisplatin-loaded M1 macrophage exosomes derived from umbilical cord blood target tumor sites of ovarian cancer, and can be used to increase cisplatin sensitivity and cytotoxicity.
In their study, Rehman et al. [72] investigated exosomes isolated from allogeneic bone marrow mesenchymal stem cells treated with heme oxygenase-1 specific short peptide and siRNA nanocarrier for glioblastoma resistant against the cytostatic drug temozolomide. This laboratory showed the specific tumor cell targeting capability, based on the overexpression of heme oxygenase-1 in glioblastoma, by modifying bone marrow mesenchymal stem cell-derived exosomes using a specific short peptide for heme oxygenase-1 and by loading temozolomide or siRNA into these exosomes.
5-FU is one of the most widely used effective drugs for the treatment of colorectal cancer. To reduce the systemic side effects of 5-FU and chemoresistance, Pang et al. [73] established colorectal cancer cells that overexpressed miR-323a-3p, a tumor suppressor that targets both EGFR and thymidylate synthase. MiR-323a-3p-loaded exosomes could effectively induce apoptosis in colorectal cancer cells by targeting EGFR and thymidylate synthase, and enhanced the therapeutic effects of 5-FU, demonstrating the potency of miRNA-loaded exosomes for advanced colorectal cancer biotherapy.

Table 1.
A selection of modified exosomes as therapeutic agents.
Table 1.
A selection of modified exosomes as therapeutic agents.
| Cancer | Exosome-Derived Cell | Loading | Exosome Extraction | Refs. |
|---|---|---|---|---|
| Breast | tumor | CD3, EGFR | differential centrifugation | [66,67] |
| antibody cloning | ||||
| Pancreatic | mesenchymal stem | galectin-9 siRNA | electroporation | [68] |
| electroporation | ||||
| Lewis lung | tumor | CD40 | ultracentrifugation | [69] |
| cell transfection | ||||
| Melanoma | dendritic | neoantigens | ultracentrifugation | [70] |
| cell transfection | ||||
| Ovarian | M1 macrophage | cisplatin | magnetic beads | [71] |
| cell treatment | flow cytometry | |||
| Glioblastoma | mesenchymal | heme oxygenase-1 | ultracentrifugation | [72] |
| cell transfrection | ||||
| Colorectal | cancer | miR-323a-3p | differential centrifugation | [73] |
| cell transfection |
8. Exosomes as Therapeutic Targets
Numerous studies have shown the oncogenic potential of cancer-derived exosomes to promote tumor invasion [74], deliver oncogenic genetic material to normal cells [75], increase drug resistance of cancer cells [76,77], and prime distant organs for metastasis [78,79,80]. In addition, exosomes released from cancer-associated fibroblasts play a role in supporting chemoresistance, e.g., in colorectal and breast cancer cells [76,77]. Since such exosomes play a pivotal role in cancer progression, they are also attractive targets in the treatment of cancer patients. Table 2 gives an overview of a selection of exosomes that may serve as therapeutic targets (Table 2).

Table 2.
A selection of exosomes as therapeutic targets.
GW4869 is a noncompetitive neutral sphingomyelinase inhibitor that hydrolyzes sphingomyelins to produce ceramides. In addition, GW4869-induced sphingomyelinase inhibition has been reported to inhibit exosome biogenesis and release. Richards et al. [81] showed that the GW4869-caused inhibition of exosome release from cancer-associated fibroblasts could decrease the chemoresistance and, thus, the survival of pancreatic cancer cells. Furthermore, in a mouse model, the combination treatment of gemcitabine and GW4869 resulted in diminished tumor growth. Moreover, Wang et al. [82] developed an assembly of GW4869 and ferroptosis inducer via amphiphilic hyaluronic acid. Treatment with a CD44-targeting nanounit composed of this assembly induced an anti-tumor immune response to melanoma cells in mice, stimulated cytotoxic T lymphocytes and immunological memory and increased the response to programmed cell death-ligand 1 (PD-L1) checkpoint blockade.
The cytostatic drug gemcitabine is a major drug for the treatment of pancreatic ductal adenocarcinoma. However, changes in the levels of specific exosomal miRNAs play an important role in chemoresistance development. RAB27A is a member of the RAS oncogene family, belongs to the small GTPase superfamily and is involved in protein transport. Targeting of RAB27A with siRNA inhibits exosome secretion and reduces tumor growth and metastasis in mouse models [83,84]. To overcome miR-155-induced gemcitabine resistance of pancreatic ductal adenocarcinoma, Mikamori et al. [85] transfected pancreatic cancer cells with RAB27B siRNA. The reduced exosome release and, thus, decrease in miR-155 levels led to a significant decrease in chemoresistance.
Myeloid-derived suppressor cells are a population of immature myeloid cells that are involved in the process of T cell inactivation. Chalmin et al. [86] prevented the delivery of exosomes containing heat shock protein HSP72 to myeloid-derived suppressor cells by the blood pressure-lowering drug dimethyl amiloride, which reduces endocytic recycling and, in turn, exosome release. The administration with amiloride could reduce tumor growth in mice and increase cyclophosphamide-based chemotherapy efficacy.
It is not only the delivery of exosomes that can be inhibited, the uptake of exosomes can also be reduced. Heparan sulfate proteoglycans (HSPGs) serve as receptors of cancer cell-derived exosomes. Christianson et al. [87] revealed that the enzymatic depletion of cell surface HSPG effectively attenuated exosome uptake. In addition, annexin V can also inhibit exosome uptake by binding to and blocking surface phosphatidylserine, which is important for membrane adhesion. Lima et al. [88] indicated that a highly metastatic melanoma cell line released large yields of exosomes containing phosphatidylserine. These tumor-derived exosomes increased TGF-β production by cultured macrophages in vitro and enhanced the metastatic potential of melanoma cells in mice. Both effects could be reversed by annexin V. In addition, the treatment could also reduce the tumor growth rate and metastatic potential of human glioma xenografts in mice. However, the analyses with these agents have to be continued as, because of the disruption of other cellular processes, such signaling pathways, cannot be excluded.
Likewise, Zhang et al. [89] found that mesenchymal stromal cell-derived exosomes that carry miR-101 could inhibit osteosarcoma cell invasion and metastasis by downregulating B-cell lymphoma 6 expression.
9. Discussion and Conclusions
In the present review article, a short overview of the potential of both exosomes in clinical applications to function as a provider of antitumor agents and the development of exosome-derived therapeutic strategies to overcome chemoresistance and tumor progression was presented.
For clinical application, several challenges have to be overcome. First, the cell type qualified to be used to for exosome extraction has to be identified, and then they have to be loaded with drugs. For example, the exosome source could be immune cells because exosomes released by antigen-presenting cells can provide therapeutic benefits though attenuating or stimulating the immune response. Exosomes derived from dendritic cells can activate T and B cells and carry MHC and so modulate antigen-specific T cell responses [90,91]. A further challenge is to carry out a large-scale production of exosomes [92]. For example, Lamparski et al. [93] developed a quick method for the production, purification and characterization of exosomes derived from antigen-presenting cells by ultrafiltration and ultracentrifugation. However, this technique still requires further testing with different types of cells. Next, effective loading approaches have to be standardized [94]. Finally, studies regarding the potency and toxicology of exosomes are essential for bringing them into the clinic. Thus, a comprehensive evaluation of the optimal dose and drug distribution of exosomes in cancer treatment is urgently mandatory. To date, most studies have focused on exosomes in cell experiments, both in vitro and less often in mice. Hence, their efficacy and delivery to the recipient cells should be examined on long-term monitoring platforms and in vivo systems. Therefore, large multicenter and longer-term studies are required to achieve their clinical application.
However, a major drawback of using exosomes as treatment strategies is that they contain thousands of unknown molecules (proteins, RNAs) with oncogenic and tumor suppressive characteristics. They are involved in different signaling pathways and may therefore have different effects on the tumor growth—in doing so, certain modes of actions may be predominant or less relevant. This is also the reason that it is difficult to predict their precise molecular mechanism and function in cancer. Their complex nature and, thus, the diversity of their natural compounds, may, therefore, complicate the therapeutic process of the disease. It is also not predictable how robustly and durably they influence the different signal pathways. Notably, exosomes can be engineered and used as anti-tumor delivery agents. Thus, engineering exosomes with known contents that counteract tumor-promoting functions may support the progress in cancer therapy.
Higher levels of exosomes are particularly secreted by the tumor cells in their environment than by normal cells [95]. The cancer-derived exosomes may in turn be uptaken by normal cells that may then adopt cancerous characteristics. Therefore, a further challenge is to inhibit the tumor-derived exosome secretion and uptake by recipient cells, to restore tumor immunity and impair tumor progression. These investigations should also be carried out by large multicenter and longer-term studies to establish the efficacity of this approach.
Indeed, the exact efficiency of targeting of tumor-derived exosomes is also not predicable since they do not only contain tumor-stimulatory components but also tumor-suppressive components. The inhibition of these tumor-suppressive components may possibly have unforeseeable effects on the cancer therapy by inhibiting other signal pathways that are important for the fight against cancer, e.g., the immune system.
Nevertheless, the concept of using exosomes as delivery vehicles or tumor-derived exosomes as targets may have promising potential in the treatment management of cancer patients.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Balkwill, F.; Mantovani, A. Inflammation and Cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, L.M.; Parris, E.E.; Folkman, J. Tumor Angiogenesis: Therapeutic Implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Organs 2022, 211, 157–182. [Google Scholar] [CrossRef] [PubMed]
- Géraud, C.; Koch, P.S.; Damm, F.; Schledzewski, K.; Goerdt, S. The Metastatic Cycle: Metastatic Niches and Cancer Cell Dissemination. JDDG-J. Ger. Soc. Dermatol. 2014, 12, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. Cancer Progression and the Invisible Phase of Metastatic Colonization. Nat. Rev. Cancer 2020, 20, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Paskeh, M.D.A.; Entezari, M.; Mirzaei, S.; Zabolian, A.; Saleki, H.; Naghdi, M.J.; Sabet, S.; Khoshbakht, M.A.; Hashemi, M.; Hushmandi, K.; et al. Emerging Role of Exosomes in Cancer Progression and Tumor Microenvironment Remodeling. J. Hematol. Oncol. 2022, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Stahl, P. Transferrin Recycling in Reticulocytes: PH and Iron Are Important Determinants of Ligand Binding and Processing. Biochem. Biophys. Res. Commun. 1983, 113, 650–658. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammonds, J.R.; Turbide, C. Vesicle Formation during Reticulocyte Maturation. J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A Common Pathway for a Specialized Function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular Vesicles as Biomarkers and Therapeutic Targets for Cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking Back Three Decades and into the Future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Shtam, T.; Evtushenko, V.; Samsonov, R.; Zabrodskaya, Y.; Kamyshinsky, R.; Zabegina, L.; Verlov, N.; Burdakov, V.; Garaeva, L.; Slyusarenko, M.; et al. Evaluation of Immune and Chemical Precipitation Methods for Plasma Exosome Isolation. PLoS ONE 2020, 15, e0242732. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Choi, M.; Lee, H.; Kim, Y.H.; Han, J.Y.; Lee, E.S.; Cho, Y. Direct Isolation and Characterization of Circulating Exosomes from Biological Samples Using Magnetic Nanowires. J. Nanobiotechnol. 2019, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H. Methods for Quantification and Characterization of MicroRNAs in Cell-Free Plasma/Serum, Normal Exosomes and Tumor-Derived Exosomes. Transl. Cancer Res. 2018, 7, S253–S263. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Lu, Y.; Luo, X.; Huang, Y.; Xie, Y.; Pilarsky, C.; Dang, Y.; Zhang, J. Microfluidic Technology for the Isolation and Analysis of Exosomes. Micromachines 2022, 13, 1571. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of Exosomes from Whole Blood by Integrating Acoustics and Microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Gahan, P.B. Exosomes in Immune Regulation. Noncoding RNA 2021, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Gahan, P.B. Predictive Value of Exosomes and Their Cargo in Drug Response/Resistance of Breast Cancer Patients. Cancer Drug Resist. 2020, 3, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.F.; Li, W.J.; Hu, K.S.; Gao, J.; Zhai, W.L.; Yang, J.H.; Zhang, S.J. Exosome Biogenesis: Machinery, Regulation, and Therapeutic Implications in Cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Zhang, H.; Lyden, D. An Exosome Pathway without an ESCRT. Cell Res. 2021, 31, 105–106. [Google Scholar] [CrossRef] [PubMed]
- Krylova, S.V.; Feng, D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int. J. Mol. Sci. 2023, 24, 1337. [Google Scholar] [CrossRef] [PubMed]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.L.; Vader, P. Extracellular Vesicles as Drug Delivery Systems: Why and How? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Leijendekker, R.; Harding, C.V.; Melief, C.J.M.; Geuze, H.J. B Lymphocytes Secrete Antigen-Presenting Vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular Transfer of the Oncogenic Receptor EGFRvIII by Microvesicles Derived from Tumour Cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- De Jong, O.G.; Kooijmans, S.A.A.; Murphy, D.E.; Jiang, L.; Evers, M.J.W.; Sluijter, J.P.G.; Vader, P.; Schiffelers, R.M. Drug Delivery with Extracellular Vesicles: From Imagination to Innovation. Acc. Chem. Res. 2019, 52, 1761–1770. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; Lebleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of SiRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Cooper, J.M.; Wiklander, P.B.O.; Nordin, J.Z.; Al-Shawi, R.; Wood, M.J.; Vithlani, M.; Schapira, A.H.V.; Simons, J.P.; El-Andaloussi, S.; Alvarez-Erviti, L. Systemic Exosomal SiRNA Delivery Reduced Alpha-Synuclein Aggregates in Brains of Transgenic Mice. Mov. Disord. 2014, 29, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Gahan, P.B. Interplay between LncRNAs and MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 8095. [Google Scholar] [CrossRef]
- Müller, V.; Oliveira-Ferrer, L.; Steinbach, B.; Pantel, K.; Schwarzenbach, H. Interplay of LncRNA H19/MiR-675 and LncRNA NEAT1/MiR-204 in Breast Cancer. Mol. Oncol. 2019, 13, 1137–1149. [Google Scholar] [CrossRef]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.M.; Callen, D.F. The Oncogenic Role of MiR-155 in Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Xu, L.Y.; Qian, Q.; He, X.; Peng, W.T.; Zhu, Y.L.; Cheng, L. Analysis of MiRNA Signature Differentially Expressed in Exosomes from Adriamycin-Resistant and Parental Human Breast Cancer Cells. Biosci. Rep. 2018, 38, BSR20181090. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Stevic, I.; Müller, V.; Ni, Q.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Exosomal MicroRNAs as Tumor Markers in Epithelial Ovarian Cancer. Mol. Oncol. 2018, 12, 1935–1948. [Google Scholar] [CrossRef]
- Fründt, T.; Krause, L.; Hussey, E.; Steinbach, B.; Köhler, D.; von Felden, J.; Schulze, K.; Lohse, A.W.; Wege, H.; Schwarzenbach, H. Diagnostic and Prognostic Value of Mir-16, Mir-146a, Mir-192 and Mir-221 in Exosomes of Hepatocellular Carcinoma and Liver Cirrhosis Patients. Cancers 2021, 13, 2484. [Google Scholar] [CrossRef] [PubMed]
- Turturici, G.; Tinnirello, R.; Sconzo, G.; Geraci, F. Extracellular Membrane Vesicles as a Mechanism of Cell-to-Cell Communication: Advantages and Disadvantages. Am. J. Physiol. Cell Physiol. 2014, 306, C621–C633. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes–Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.S.; Chen, C.A.; Zhou, Q.A. Exosomes Nature’s Lipid Nanoparticles, a Rising Star in Drug Delivery and Diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Yi, K.; Qi, H.; Li, S.; Li, X.; Wang, Q.; Wang, Y.; Liu, C.; Qiu, M.; Yuan, X.; et al. Engineering Blood Exosomes for Tumor-Targeting Efficient Gene/Chemo Combination Therapy. Theranostics 2020, 10, 7889–7905. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered Exosomes for Targeted Co-Delivery of MiR-21 Inhibitor and Chemotherapeutics to Reverse Drug Resistance in Colon Cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Schiffelers, R.M.; Zarovni, N.; Vago, R. Modulation of Tissue Tropism and Biological Activity of Exosomes and Other Extracellular Vesicles: New Nanotools for Cancer Treatment. Pharmacol. Res. 2016, 111, 487–500. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of Exosome-Encapsulated Paclitaxel to Overcome MDR in Cancer Cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.A.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering Hybrid Exosomes by Membrane Fusion with Liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering Exosomes as Refined Biological Nanoplatforms for Drug Delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Siemer, S.; Bauer, T.A.; Scholz, P.; Breder, C.; Fenaroli, F.; Harms, G.; Dietrich, D.; Dietrich, J.; Rosenauer, C.; Barz, M.; et al. Targeting Cancer Chemotherapy Resistance by Precision Medicine-Driven Nanoparticle-Formulated Cisplatin. ACS Nano 2021, 15, 18541–18556. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Pan, Q.; Gao, W.; Pu, Y.; He, B. Reversal of Cisplatin Chemotherapy Resistance by Glutathione-Resistant Copper-Based Nanomedicine via Cuproptosis. J. Mater. Chem. B 2022, 10, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhang, T.; He, W.; Jin, H.; Liu, C.; Yang, Z.; Ren, J. Methotrexate-Loaded Extracellular Vesicles Functionalized with Therapeutic and Targeted Peptides for the Treatment of Glioblastoma Multiforme. ACS Appl. Mater. Interfaces 2018, 10, 12341–12350. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, Y.; Ding, F.; Yang, J.; Li, J.; Gao, X.; Zhang, C.; Feng, J. Engineering Macrophage-Derived Exosomes for Targeted Chemotherapy of Triple-Negative Breast Cancer. Nanoscale 2020, 12, 10854–10862. [Google Scholar] [CrossRef] [PubMed]
- Al Faruque, H.; Choi, E.S.; Kim, J.H.; Kim, E. Enhanced Effect of Autologous EVs Delivering Paclitaxel in Pancreatic Cancer. J. Control. Release 2022, 347, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.C.; Jayasinghe, M.K.; Pham, T.T.; Yang, Y.; Wei, L.; Usman, W.M.; Chen, H.; Pirisinu, M.; Gong, J.; Kim, S.; et al. Covalent Conjugation of Extracellular Vesicles with Peptides and Nanobodies for Targeted Therapeutic Delivery. J. Extracell. Vesicles 2021, 10, e12057. [Google Scholar] [CrossRef]
- Zhou, X.; Zhuang, Y.; Liu, X.; Gu, Y.; Wang, J.; Shi, Y.; Zhang, L.; Li, R.; Chen, H.; Li, J.; et al. Study on Tumour Cell-Derived Hybrid Exosomes as Dasatinib Nanocarriers for Pancreatic Cancer Therapy. Artif. Cells Nanomed. Biotechnol. 2023, 51, 532–546. [Google Scholar] [CrossRef]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3-Receptor Targeted Exosomes Inhibit in Vitro and in Vivo Chronic Myelogenous Leukemia Cell Growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Zuo, B.; Zhang, Y.; Zhao, K.; Wu, L.; Qi, H.; Yang, R.; Gao, X.; Geng, M.; Wu, Y.; Jing, R.; et al. Universal Immunotherapeutic Strategy for Hepatocellular Carcinoma with Exosome Vaccines That Engage Adaptive and Innate Immune Responses. J. Hematol. Oncol. 2022, 15, 15–46. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Liu, W.; Wei, W.; Zhao, Y.; Zhuang, P.; Wang, X.; Wang, Y.; Hu, Y.; Dai, H. Photothermal Hydrogel Encapsulating Intelligently Bacteria-Capturing Bio-MOF for Infectious Wound Healing. ACS Nano 2022, 16, 19491–19508. [Google Scholar] [CrossRef] [PubMed]
- Delcayre, A.; Shu, H.; Le Pecq, J.B. Dendritic Cell-Derived Exosomes in Cancer Immunotherapy: Exploiting Nature’s Antigen Delivery Pathway. Expert. Rev. Anticancer Ther. 2005, 5, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Fu, C.; Zhou, L.; Mi, Q.S.; Jiang, A. Dc-Derived Exosomes for Cancer Immunotherapy. Cancers 2021, 13, 3667. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Shi, X.; Han, M.; Smbatyan, G.; Lenz, H.J.; Zhang, Y. Reprogramming Exosomes as Nanoscale Controllers of Cellular Immunity. J. Am. Chem. Soc. 2018, 140, 16413–16417. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Cheng, Q.; Hou, T.; Han, M.; Smbatyan, G.; Lang, J.E.; Epstein, A.L.; Lenz, H.J.; Zhang, Y. Genetically Engineered Cell-Derived Nanoparticles for Targeted Breast Cancer Immunotherapy. Mol. Ther. 2020, 28, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhou, Y.; Chen, X.; Ning, T.; Chen, H.; Guo, Q.; Zhang, Y.; Liu, P.; Zhang, Y.; Li, C.; et al. Pancreatic Cancer-Targeting Exosomes for Enhancing Immunotherapy and Reprogramming Tumor Microenvironment. Biomaterials 2021, 268, 120546. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, L.; Lin, Z.; Tao, L.; Chen, M. More Efficient Induction of Antitumor T Cell Immunity by Exosomes from CD40L Gene-Modified Lung Tumor Cells. Mol. Med. Rep. 2014, 9, 125–131. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Peng, Y.; Du, Y.; Yang, Z.; Qi, X. Dendritic Cell Derived Exosomes Loaded Neoantigens for Personalized Cancer Immunotherapies. J. Control. Release 2023, 353, 423–433. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Liu, N.; Wu, W.; Li, H.; Lu, W.; Guo, X. Umbilical Cord Blood-Derived M1 Macrophage Exosomes Loaded with Cisplatin Target Ovarian Cancer In Vivo and Reverse Cisplatin Resistance. Mol. Pharm. 2023, 20, 5440–5453. [Google Scholar] [CrossRef] [PubMed]
- Rehman, F.U.; Liu, Y.; Yang, Q.; Yang, H.; Liu, R.; Zhang, D.; Muhammad, P.; Liu, Y.; Hanif, S.; Ismail, M.; et al. Heme Oxygenase-1 Targeting Exosomes for Temozolomide Resistant Glioblastoma Synergistic Therapy. J. Control. Release 2022, 345, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Chen, X.; Xu, B.; Zhang, Y.; Liang, S.; Hu, J.; Liu, R.; Luo, X.; Wang, Y. Engineered Multitargeting Exosomes Carrying MiR-323a-3p for CRC Therapy. Int. J. Biol. Macromol. 2023, 247, 125794. [Google Scholar] [CrossRef] [PubMed]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Chennakrishnaiah, S.; Audemard, E.; Montermini, L.; Meehan, B.; Rak, J. Oncogenic Ras-Driven Cancer Cell Vesiculation Leads to Emission of Double-Stranded DNA Capable of Interacting with Target Cells. Biochem. Biophys. Res. Commun. 2014, 451, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef] [PubMed]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Twyman-Saint Victor, C.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-Induced PTEN Loss by Exosomal MicroRNA Primes Brain Metastasis Outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Ray, K. Pancreatic Cancer: Pancreatic Cancer Exosomes Prime the Liver for Metastasis. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 371. [Google Scholar] [CrossRef]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-Associated Fibroblast Exosomes Regulate Survival and Proliferation of Pancreatic Cancer Cells. Oncogene 2017, 36, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xie, L.; Li, B.; Sang, W.; Yan, J.; Li, J.; Tian, H.; Li, W.; Zhang, Z.; Tian, Y.; et al. A Nanounit Strategy Reverses Immune Suppression of Exosomal PD-L1 and Is Associated with Enhanced Ferroptosis. Nat. Commun. 2021, 12, 5733. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a Supports Exosome-Dependent and -Independent Mechanisms That Modify the Tumor Microenvironment and Can Promote Tumor Progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells toward a Pro-Metastatic Phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef] [PubMed]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-Associated Hsp72 from Tumor-Derived Exosomes Mediates STAT3-Dependent Immunosuppressive Function of Mouse and Human Myeloid-Derived Suppressor Cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Christianson, H.C.; Svensson, K.J.; Van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer Cell Exosomes Depend on Cell-Surface Heparan Sulfate Proteoglycans for Their Internalization and Functional Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.G.; Chammas, R.; Monteiro, R.Q.; Moreira, M.E.C.; Barcinski, M.A. Tumor-Derived Microvesicles Modulate the Establishment of Metastatic Melanoma in a Phosphatidylserine-Dependent Manner. Cancer Lett. 2009, 283, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular Vesicle-Mediated Delivery of MiR-101 Inhibits Lung Metastasis in Osteosarcoma. Theranostics 2020, 10, 411–425. [Google Scholar] [CrossRef]
- Shenoda, B.B.; Ajit, S.K. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin. Med. Insights Pathol. 2016, 2016. [Google Scholar] [CrossRef]
- Quah, B.J.C.; O’Neill, H.C. The Immunogenicity of Dendritic Cell-Derived Exosomes. Blood Cells Mol. Dis. 2005, 35, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Van Der Meel, R.; Fens, M.H.A.M.; Vader, P.; Van Solinge, W.W.; Eniola-Adefeso, O.; Schiffelers, R.M. Extracellular Vesicles as Drug Delivery Systems: Lessons from the Liposome Field. J. Control. Release 2014, 195, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and Characterization of Clinical Grade Exosomes Derived from Dendritic Cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.-M.; Chen, M.; Xia, S.J.; Lu, R. Drug Loading Techniques for Exosome-Based Drug Delivery Systems. Pharmazie 2021, 76, 61–67. [Google Scholar]
- Schwarzenbach, H. Clinical Relevance of Circulating, Cell-Free and Exosomal MicroRNAs in Plasma and Serum of Breast Cancer Patients. Oncol. Res. Treat. 2017, 40, 423–429. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).