Insights into the Function of Regulatory RNAs in Bacteria and Archaea
Abstract
1. Introduction
2. Genes and mRNAs Structures in Prokaryotes
3. miRNA-Size Molecules and Small RNAs in Prokaryotes
3.1. miRNA-Size Molecules
3.2. Small RNAs
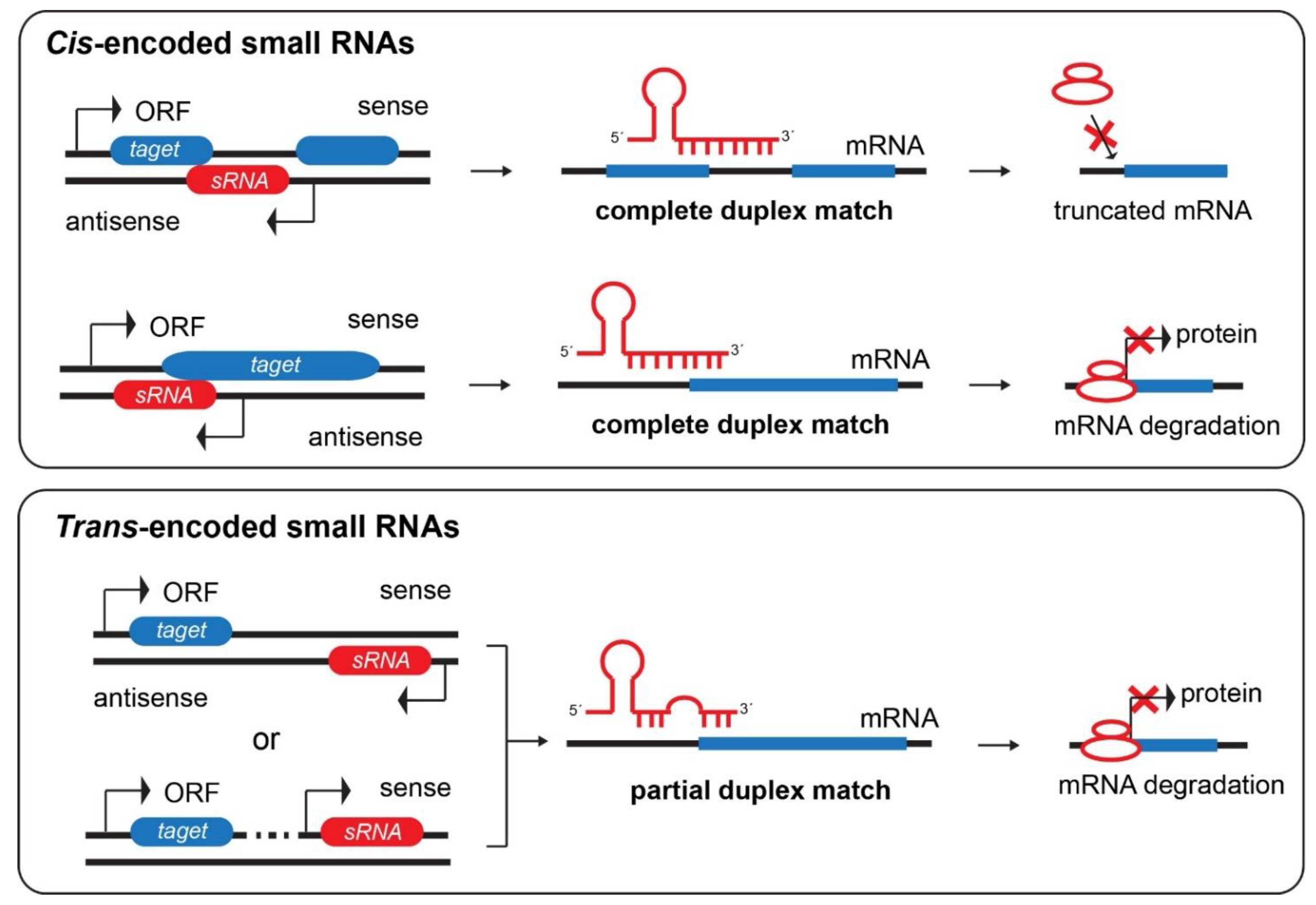
3.2.1. Cis-Encoded Small RNAs
3.2.2. Trans-Encoded Small RNAs
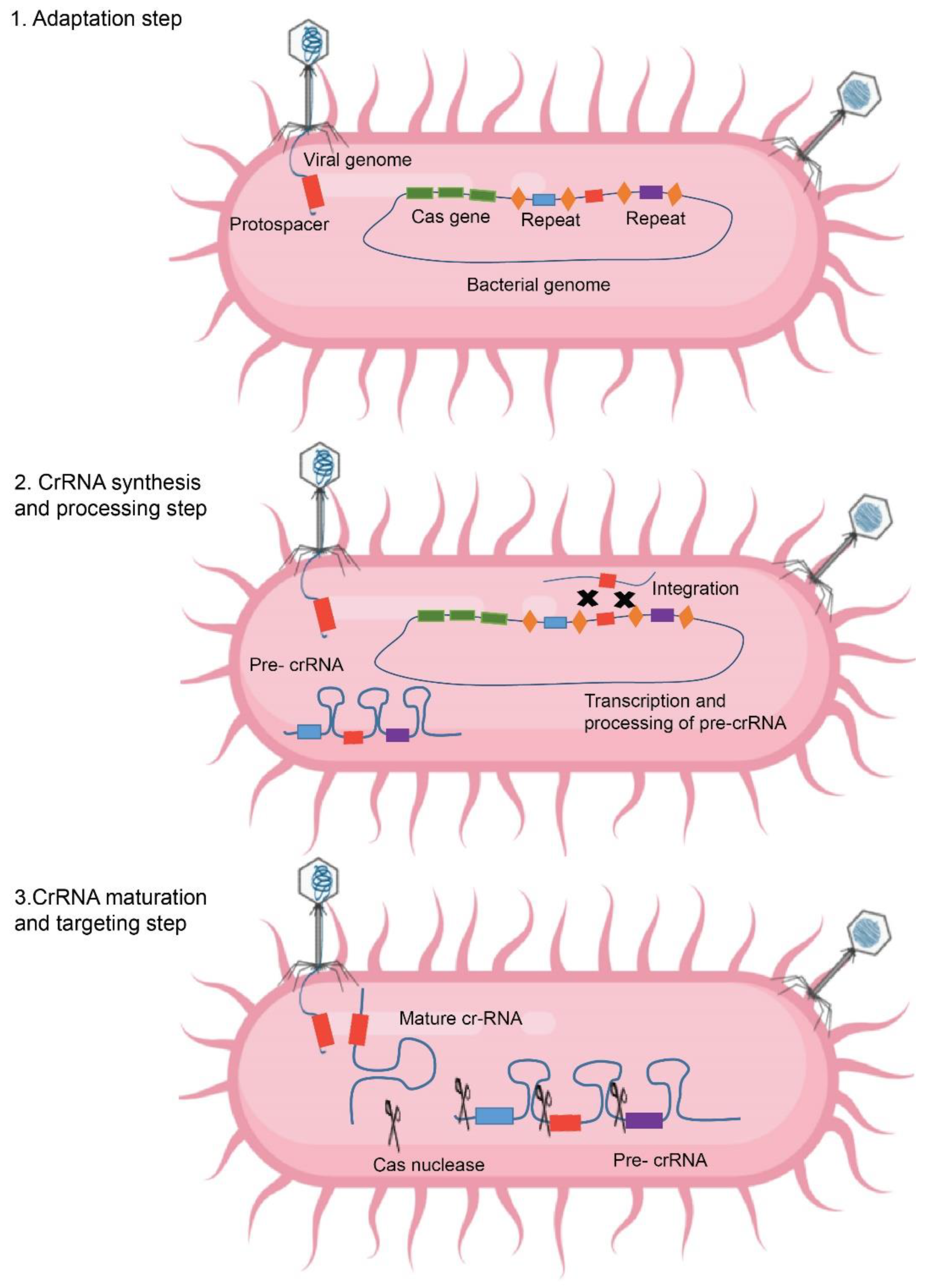
4. CRISPR
CRISPR-Cas Classification, Structure, and Mechanism of Action
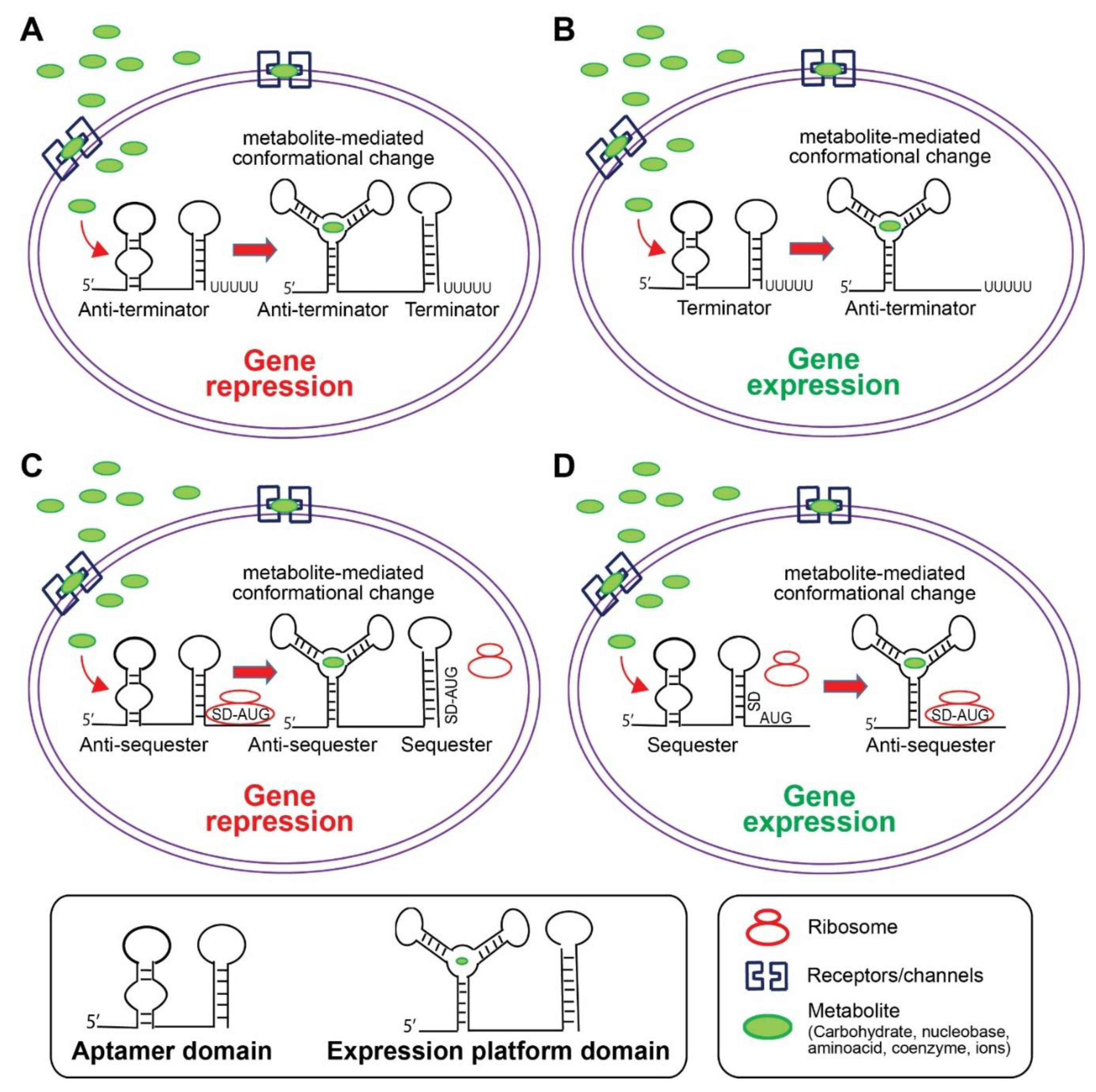
5. Riboswitch
5.1. Riboswitch Structural Classification
5.2. Riboswitches Ligands and Regulatory Mechanisms
6. Novel Bacterial-Based Therapeutic Strategies against Genetic Diseases
6.1. CRISPR System as a Potential Target of Genetic Diseases: How Do We Do It “Right”?
6.2. Riboswitches as Novel Targets in Antibacterial Therapies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-coding RNA regulatory networks. Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Narberhaus, F.; Vogel, J. Regulatory RNAs in prokaryotes: Here, there and everywhere. Mol. Microbiol. 2009, 74, 261–269. [Google Scholar] [CrossRef]
- Evguenieva-Hackenberg, E.; Klug, G. New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 2011, 14, 587–592. [Google Scholar] [CrossRef]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and Mechanisms. Cold Spring Harb. Perspect. Biol. 2010, 3, a003533. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Prévost, K.; Salvail, H.; Desnoyers, G.; Jacques, J.-F.; Phaneuf, É.; Massé, E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007, 64, 1260–1273. [Google Scholar] [CrossRef]
- Srijyothi, L.; Ponne, S.; Prathama, T.; Ashok, C.; Baluchamy, S. Roles of Non-Coding RNAs in Transcriptional Regulation. In Transcriptional and Post-Transcriptional Regulation; IntechOpen: London, UK, 2018; p. 55. [Google Scholar]
- Wang, Y.; Huang, Y.; Xiang, P.; Tian, W. LncRNA expression and implication in osteosarcoma: A systematic review and meta-analysis. OncoTargets Ther. 2017, 10, 5355–5361. [Google Scholar] [CrossRef][Green Version]
- Hallberg, Z.F.; Su, Y.; Kitto, R.Z.; Hammond, M.C. Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem. 2017, 86, 515–539. [Google Scholar] [CrossRef] [PubMed]
- Brocken, D.J.; Tark-Dame, M.; Dame, R.T. The organization of bacterial genomes: Towards understanding the interplay between structure and function. Curr. Opin. Syst. Biol. 2018, 8, 137–143. [Google Scholar] [CrossRef]
- Kang, Y.; Gu, C.; Yuan, L.; Wang, Y.; Zhu, Y.; Li, X.; Luo, Q.; Xiao, J.; Jiang, D.; Qian, M.; et al. Flexibility and Symmetry of Prokaryotic Genome Rearrangement Reveal Lineage-Associated Core-Gene-Defined Genome Organizational Frameworks. mBio 2014, 5, e01867-14. [Google Scholar] [CrossRef]
- Jackson, D.; Wang, X.; Rudner, D.Z. Spatio-Temporal Organization of Replication in Bacteria and Eukaryotes (Nucleoids and Nuclei). Cold Spring Harb. Perspect. Biol. 2012, 4, a010389. [Google Scholar] [CrossRef][Green Version]
- Harrison, P.W.; Lower, R.P.; Kim, N.K.; Young, J.P.W. Introducing the bacterial ‘chromid’: Not a chromosome, not a plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Ishii, Y.; Matsuura, Y.; Kakizawa, S.; Nikoh, N.; Fukatsu, T. Diversity of Bacterial Endosymbionts Associated with Macrosteles Leafhoppers Vectoring Phytopathogenic Phytoplasmas. Appl. Environ. Microbiol. 2013, 79, 5013–5022. [Google Scholar] [CrossRef]
- Tatusova, T.; Ciufo, S.; Federhen, S.; Fedorov, B.; McVeigh, R.; O’Neill, K.; Tolstoy, I.; Zaslavsky, L. Update on RefSeq microbial genomes resources. Nucleic Acids Res. 2015, 43, D599–D605. [Google Scholar] [CrossRef] [PubMed]
- Mira, A.; Ochman, H.; Moran, N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001, 17, 589–596. [Google Scholar] [CrossRef]
- Toro, E.; Shapiro, L. Bacterial Chromosome Organization and Segregation. Cold Spring Harb. Perspect. Biol. 2010, 2, a000349. [Google Scholar] [CrossRef] [PubMed]
- Robinow, C.; Kellenberger, E. The bacterial nucleoid revisited. Microbiol. Rev. 1994, 58, 211–232. [Google Scholar] [CrossRef]
- Thanbichler, M.; Wang, S.C.; Shapiro, L. The bacterial nucleoid: A highly organized and dynamic structure. J. Cell. Biochem. 2005, 96, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Rocha, E.P. Coevolution of the Organization and Structure of Prokaryotic Genomes. Cold Spring Harb. Perspect. Biol. 2016, 8, a018168. [Google Scholar] [CrossRef]
- Gregory, T.R.; Desalle, R. Comparative Genomics in Prokaryotes. Evol. Genome 2005, 585–675. [Google Scholar]
- Harshey, R.M.; Ramakrishnan, T. Rate of ribonucleic acid chain growth in Mycobacterium tuberculosis H37Rv. J. Bacteriol. 1977, 129, 616–622. [Google Scholar] [CrossRef]
- Dicenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Warf, M.B.; Berglund, J.A. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010, 35, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Ephrussi, A. mRNA Localization: Gene Expression in the Spatial Dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef]
- Ray, P.S.; Jia, J.; Yao, P.; Majumder, M.; Hatzoglou, M.; Fox, P.L. A stress-responsive RNA switch regulates VEGFA expression. Nat. Cell Biol. 2009, 457, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.-D.; Novoa, E.M.; Vejnar, C.E.; Yartseva, V.; Takacs, C.; Kellis, M. mRNA structure dynamics identifies the embryonic RNA regulome. bioRxiv 2018, 274290. [Google Scholar]
- Rauhut, R.; Klug, G. mRNA degradation in bacteria. FEMS Microbiol. Rev. 1999, 23, 353–370. [Google Scholar] [CrossRef]
- Belasco, J.G. All things must pass: Contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat. Rev. Mol. Cell Biol. 2010, 11, 467–478. [Google Scholar] [CrossRef]
- Mirkin, E.V.; Mirkin, S. Replication Fork Stalling at Natural Impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Gottipati, P.; Helleday, T. Transcription-associated recombination in eukaryotes: Link between transcription, replication and recombination. Mutagenesis 2009, 24, 203–210. [Google Scholar] [CrossRef]
- Shafee, T.; Lowe, R. Eukaryotic and prokaryotic gene structure. WikiJournal Med. 2017, 4, 1–5. [Google Scholar] [CrossRef]
- Sarkar, N. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 1997, 66, 173–197. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.M. The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip. Rev. RNA 2016, 8, e1370. [Google Scholar] [CrossRef]
- Carrier, M.-C.; Lalaouna, D.; Massé, E. Broadening the Definition of Bacterial Small RNAs: Characteristics and Mechanisms of Action. Annu. Rev. Microbiol. 2018, 72, 141–161. [Google Scholar] [CrossRef]
- Ying, S.-Y.; Chang, D.C.; Miller, J.D.; Lin, S.-L. The MicroRNA: Overview of the RNA Gene That Modulates Gene Functions; Humana Press: Totowa, NJ, USA, 2006; Volume 342, pp. 1–18. [Google Scholar]
- Vafadar, A.; Shabaninejad, Z.; Movahedpour, A.; Mohammadi, S.; Fathullahzadeh, S.; Mirzaei, H.R.; Namdar, A.; Savardashtaki, A.; Mirzaei, H. Long Non-Coding RNAs As Epigenetic Regulators in Cancer. Curr. Pharm. Des. 2019, 25, 3563–3577. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. In Non-Coding RNAs in Colorectal Cancer; Springer: Berlin/Heidelberg, Germany, 2016; pp. 3–17. [Google Scholar]
- Shabaninejad, Z.; Yousefi, F.; Movahedpour, A.; Ghasemi, Y.; Dokanehiifard, S.; Rezaei, S.; Aryan, R.; Savardashtaki, A.; Mirzaei, H. Electrochemical-based biosensors for microRNA detection: Nanotechnology comes into view. Anal. Biochem. 2019, 581, 113349. [Google Scholar] [CrossRef]
- Naeli, P.; Pourhanifeh, M.H.; Karimzadeh, M.R.; Shabaninejad, Z.; Movahedpour, A.; Tarrahimofrad, H.; Mirzaei, H.R.; Bafrani, H.H.; Savardashtaki, A.; Mirzaei, H.; et al. Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. 2020, 145, 102854. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef]
- Shabaninejad, Z.; Vafadar, A.; Movahedpour, A.; Ghasemi, Y.; Namdar, A.; Fathizadeh, H.; Pourhanifeh, M.H. Circular RNAs in cancer: New insights into functions and implications in ovarian cancer. J. Ovarian Res. 2019, 12, 1–12. [Google Scholar] [CrossRef]
- Savardashtaki, A.; Shabaninejad, Z.; Movahedpour, A.; Sahebnasagh, R.; Mirzaei, H.; Hamblin, M.R. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics 2019, 11, 1627–1645. [Google Scholar] [CrossRef]
- Movahedpour, A.; Ahmadi, N.; Ghasemi, Y.; Savardashtaki, A.; Shabaninejad, Z. Circulating microRNAs as potential diagnostic biomarkers and therapeutic targets in prostate cancer: Current status and future perspectives. J. Cell. Biochem. 2019, 120, 16316–16329. [Google Scholar] [CrossRef] [PubMed]
- Jamali, Z.; Taheri-Anganeh, M.; Shabaninejad, Z.; Keshavarzi, A.; Taghizadeh, H.; Razavi, Z.S. Autophagy regulation by microRNAs: Novel insights into osteosarcoma therapy. IUBMB Life 2020, 72, 1306–1321. [Google Scholar] [CrossRef]
- Alamdari-Palangi, V.; Vahedi, F.; Shabaninejad, Z.; Dokeneheifard, S.; Movehedpour, A.; Taheri-Anganeh, M.; Savardashtaki, A. microRNA in inflammatory bowel disease at a glance. Eur. J. Gastroenterol. Hepatol. 2021, 32, 140–148. [Google Scholar] [CrossRef]
- Furuse, Y.; Finethy, R.; Saka, H.A.; Xet-Mull, A.M.; Sisk, D.M.; Smith, K.L.J.; Lee, S.; Coers, J.; Valdivia, R.H.; Tobin, D.; et al. Search for MicroRNAs Expressed by Intracellular Bacterial Pathogens in Infected Mammalian Cells. PLoS ONE 2014, 9, e106434. [Google Scholar]
- Nejman-Faleńczyk, B.; Bloch, S.; Licznerska, K.; Dydecka, A.; Felczykowska, A.; Topka, G. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24 Β. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Hong, S.-H. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 2011, 326, 131–136. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Olina, A.; Kulbachinskiy, A.V.; Aravin, A.A.; Esyunina, D.M. Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes. Biochemistry 2018, 83, 483–497. [Google Scholar] [CrossRef]
- Orban, T.I.; Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005, 11, 459–469. [Google Scholar] [CrossRef]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Förstemann, K.; Tomari, Y.; Du, T.; Vagin, V.V.; Denli, A.M.; Bratu, D.P. Normal microRNA Maturation and Germ-Line Stem Cell Maintenance Requires Loquacious, a Double-Stranded RNA-Binding Domain Protein. PLoS Biol. 2005, 3, e236. [Google Scholar] [CrossRef]
- Li, C.; Feng, Y.; Coukos, G.; Zhang, L. Therapeutic MicroRNA Strategies in Human Cancer. AAPS J. 2009, 11, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Politz, J.C.R.; Hogan, E.M.; Pederson, T. MicroRNAs with a nucleolar location. RNA 2009, 15, 1705–1715. [Google Scholar] [CrossRef]
- Politz, J.C.R.; Zhang, F.; Pederson, T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA 2006, 103, 18957–18962. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Caswell, C.C.; Oglesby, M.; Murphy, E.R. Sibling rivalry: Related bacterial small RNAs and their redundant and non-redundant roles. Front. Cell. Infect. Microbiol. 2014, 4, 151. [Google Scholar] [CrossRef]
- Kawano, M.; Reynolds, A.A.; Miranda-Rios, J.; Storz, G. Detection of 5′-and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005, 33, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Axmann, I.M.; Kensche, P.; Vogel, J.; Kohl, S.; Herzel, H.; Hess, W.R. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005, 6, R73. [Google Scholar] [CrossRef]
- Pichon, C.; Felden, B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA 2005, 102, 14249–14254. [Google Scholar] [CrossRef]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Genes for Small, Noncoding RNAs under Sporulation Control in Bacillus subtilis. J. Bacteriol. 2006, 188, 532–541. [Google Scholar] [CrossRef]
- Schroeder, C.L.C.; Narra, H.P.; Rojas, M.; Sahni, A.; Patel, J.M.; Khanipov, K.; Wood, T.G.; Fofanov, Y.; Sahni, S.K. Bacterial small RNAs in the Genus Rickettsia. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Lybecker, M.C.; Samuels, D.S. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 2007, 64, 1075–1089. [Google Scholar] [CrossRef]
- Samuels, D.S. Gene Regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 2011, 65, 479–499. [Google Scholar] [CrossRef]
- Bloch, S.; Węgrzyn, A.; Węgrzyn, G.; Nejman-Faleńczyk, B. Small and Smaller—sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins 2017, 9, 181. [Google Scholar] [CrossRef]
- Gottesman, S.; McCullen, C.; Guillier, M.; Vanderpool, C.; Majdalani, N.; Benhammou, J. Small RNA Regulators and the Bacterial Response to Stress. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 1–11. [Google Scholar] [CrossRef]
- Babitzke, P.; Romeo, T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007, 10, 156–163. [Google Scholar] [CrossRef]
- Lenz, D.H.; Miller, M.B.; Zhu, J.; Kulkarni, R.V.; Bassler, B.L. CsrA and three redundant small RNAs regulate quorum sensing inVibrio cholerae. Mol. Microbiol. 2005, 58, 1186–1202. [Google Scholar] [CrossRef]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef]
- Grieshaber, N.A.; Grieshaber, S.S.; Fischer, E.R.; Hackstadt, T. A small RNA inhibits translation of the histone-like protein Hc1 inChlamydia trachomatis. Mol. Microbiol. 2006, 59, 541–550. [Google Scholar] [CrossRef]
- Brantl, S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007, 10, 102–109. [Google Scholar] [CrossRef]
- Tramonti, A.; De Canio, M.; De Biase, D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY–gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008, 70, 965–982. [Google Scholar]
- Opdyke, J.A.; Kang, J.-G.; Storz, G. GadY, a Small-RNA Regulator of Acid Response Genes in Escherichia coli. J. Bacteriol. 2004, 186, 6698–6705. [Google Scholar] [CrossRef]
- Dühring, U.; Axmann, I.M.; Hess, W.R.; Wilde, A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA 2006, 103, 7054–7058. [Google Scholar] [CrossRef]
- Brantl, S.; Jahn, N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015, 39, 413–427. [Google Scholar] [CrossRef]
- Svensson, S.L.; Sharma, C.M. Small RNAs in Bacterial Virulence and Communication. Virulence Mech. Bact. Pathog. 2016, 2016, 169–212. [Google Scholar]
- Jahn, N.; Brantl, S. One antitoxin—Two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013, 41, 9870–9880. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Unoson, C.; Wagner, E.G.H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008, 70, 258–270. [Google Scholar] [CrossRef]
- Santiago-Frangos, A.; Woodson, S.A. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 2018, 9, e1475. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Förstner, K.U.; Holmqvist, E.; Otto, A.; Günster, R.; Becher, D.; Reinhardt, R.; Vogel, J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. USA 2016, 113, 11591–11596. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Gimpel, M.; Wildenhain, T.; Brantl, S. A new role for CsrA: Promotion of complex formation between an sRNA and its mRNA target in Bacillus subtilis. RNA Biol. 2019, 16, 972–987. [Google Scholar] [CrossRef]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Repoila, F.; Cossart, P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007, 10, 182–188. [Google Scholar] [CrossRef]
- Massé, E.; Escorcia, F.E.; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003, 17, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, K.; Papenfort, K.; Fekete, A.; Vogel, J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013, 32, 2963–2979. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Sun, Y.; Miyakoshi, M.; Vanderpool, C.K.; Vogel, J. Small RNA-Mediated Activation of Sugar Phosphatase mRNA Regulates Glucose Homeostasis. Cell 2013, 153, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Soper, T.; Mandin, P.; Majdalani, N.; Gottesman, S.; Woodson, S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. USA 2010, 107, 9602–9607. [Google Scholar] [CrossRef]
- Aiba, H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007, 10, 134–139. [Google Scholar] [CrossRef]
- Massé, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved Small Non-coding RNAs that belong to the σE Regulon: Role in Down-regulation of Outer Membrane Proteins. J. Mol. Biol. 2006, 364, 1–8. [Google Scholar] [CrossRef]
- Thompson, K.M.; Rhodius, V.A.; Gottesman, S. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007, 189, 4243–4256. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Adams, P.P.; Zhang, A.; Zhang, H.; Storz, G. RNA-RNA Interactomes of ProQ and Hfq Reveal Overlapping and Competing Roles. Mol. Cell 2020, 77, 411–425.e7. [Google Scholar] [CrossRef]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Genet. 2018, 16, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, M.; Wassarman, K.M.; Biham, O.; Margalit, H. Global Regulation of Transcription by a Small RNA: A Quantitative View. Biophys. J. 2014, 106, 1205–1214. [Google Scholar] [CrossRef]
- Loureiro, A.; da Silva, G.J. Crispr-cas: Converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- McGinn, J.; Marraffini, L.A. CRISPR-Cas Systems Optimize Their Immune Response by Specifying the Site of Spacer Integration. Mol. Cell 2016, 64, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.; Yakunin, A.; et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Genet. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef]
- Burmistrz, M.; Pyrć, K. CRISPR-Cas Systems in Prokaryotes. Pol. J. Microbiol. 2015, 64, 193–202. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E.V. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Rahimpour, A.; Ahani, R.; Najaei, A.; Adeli, A.; Barkhordari, F.; Mahboudi, F. Development of Genetically Modified Chinese Hamster Ovary Host Cells for the Enhancement of Recombinant Tissue Plasminogen Activator Expression. Malays. J. Med. Sci. 2016, 23, 6–13. [Google Scholar] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Garrett, R.A.; Shah, S.A.; Erdmann, S.; Liu, G.; Mousaei, M.; León-Sobrino, C.; Peng, W.; Gudbergsdottir, S.; Deng, L.; Vestergaard, G.; et al. CRISPR-Cas Adaptive Immune Systems of the Sulfolobales: Unravelling Their Complexity and Diversity. Life 2015, 5, 783–817. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nat. Cell Biol. 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Zhang, F.; Wen, Y.; Guo, X. CRISPR/Cas9 for genome editing: Progress, implications and challenges. Hum. Mol. Genet. 2014, 23, R40–R46. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Taylor, D.W.; Chen, J.S.; Kornfeld, J.E.; Zhou, K.; Thompson, A.J.; Nogales, E.; Doudna, J.A. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. The rise and fall of the RNA world. New Boil. 1991, 3, 399–407. [Google Scholar]
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic Control by a Metabolite Binding mRNA. Chem. Biol. 2002, 9, 1043–1049. [Google Scholar] [CrossRef]
- Winkler, W.C.; Breaker, R.R. Genetic Control by Metabolite-Binding Riboswitches. ChemBioChem 2003, 4, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Breaker, R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004, 5, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, E.M.; Hejazi, M.S.; Barzegar, A. Riboswitches: From living biosensors to novel targets of antibiotics. Gene 2016, 592, 244–259. [Google Scholar] [CrossRef]
- Li, S.; Breaker, R.R. Fluoride enhances the activity of fungicides that destabilize cell membranes. Bioorganic Med. Chem. Lett. 2012, 22, 3317–3322. [Google Scholar] [CrossRef]
- Liberman, J.A.; Salim, M.; Krucinska, J.; Wedekind, J.E. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat. Chem. Biol. 2013, 9, 353–355. [Google Scholar] [CrossRef]
- Serganov, A.; Polonskaia, A.; Phan, A.T.; Breaker, R.; Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nat. Cell Biol. 2006, 441, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Bocobza, S.; Adato, A.; Mandel, T.; Shapira, M.; Nudler, E.; Aharoni, A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007, 21, 2874–2879. [Google Scholar] [CrossRef]
- Cheah, M.T.; Wachter, A.; Sudarsan, N.; Breaker, R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nat. Cell Biol. 2007, 447, 497–500. [Google Scholar] [CrossRef]
- McRose, D.; Guo, J.; Monier, A.; Sudek, S.; Wilken, S.; Yan, S.; Mock, T.; Archibald, J.M.; Begley, T.P.; Reyes-Prieto, A.; et al. Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J. 2014, 8, 2517–2529. [Google Scholar] [CrossRef]
- Machtel, P.; Bąkowska-Żywicka, K.; Żywicki, M. Emerging applications of riboswitches–from antibacterial targets to molecular tools. J. Appl. Genet. 2016, 57, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Cromie, M.J.; Shi, Y.; Latifi, T.; Groisman, E.A. An RNA Sensor for Intracellular Mg2+. Cell 2006, 125, 71–84. [Google Scholar] [CrossRef]
- Dann, C.E., III; Wakeman, C.A.; Sieling, C.L.; Baker, S.C.; Irnov, I.; Winkler, W.C. Structure and mechanism of a metal-sensing regulatory RNA. Cell 2007, 130, 878–892. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Breaker, R.R. The Power of Riboswitches. Sci. Am. 2007, 296, 50–57. [Google Scholar] [CrossRef]
- Yarnell, W.S.; Roberts, J.W. Mechanism of Intrinsic Transcription Termination and Antitermination. Science 1999, 284, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Mironov, A.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing Small Molecules by Nascent RNA: A Mechanism to Control Transcription in Bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef]
- Sudarsan, N.; Barrick, J.E.; Breaker, R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 2003, 9, 644–647. [Google Scholar] [CrossRef]
- Hollands, K.; Proshkin, S.; Sklyarova, S.; Epshtein, V.; Mironov, A.; Nudler, E.; Groisman, E.A. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. USA 2012, 109, 5376–5381. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4, a003566. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Wang, J.; Beyer, A.I.; Teque, F.; Cradick, T.; Qi, Z.; Chang, J.C.; Bao, G.; Muench, M.; Yu, J.; et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA 2014, 111, 9591–9596. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child.-Educ. Pract. 2016, 101, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Li, M.; Zhao, L.; Page-McCaw, P.S.; Chen, W. Zebrafish Genome Engineering Using the CRISPR–Cas9 System. Trends Genet. 2016, 32, 815–827. [Google Scholar] [CrossRef]
- Pineda, M.; Moghadam, F.; Ebrahimkhani, M.R.; Kiani, S. Engineered CRISPR Systems for Next Generation Gene Therapies. ACS Synth. Biol. 2017, 6, 1614–1626. [Google Scholar] [CrossRef]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chenhui, D.; Li, Y.; et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef]
- Walsh, C. Molecular mechanisms that confer antibacterial drug resistance. Nature 2000, 406, 775–781. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Toney, J.H. Nature’s clarion call of antibacterial resistance: Are we listening? Curr. Opin. Investig. Drugs 2006, 7, 158–166. [Google Scholar]
- Wolfson, W. Holding Back the Tide of Antibiotic Resistance. Chem. Biol. 2006, 13, 1–3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bermingham, A.; Derrick, J.-P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 2002, 24, 637–648. [Google Scholar] [CrossRef]
- Koch, A.L. Bacterial wall as target for attack: Past, present, and future research. Clin. Microbiol. Rev. 2003, 16, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, A. DNA gyrase as a drug target. Trends Microbiol. 1997, 5, 102–109. [Google Scholar] [CrossRef]
- Poehlsgaard, J.; Douthwaite, S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Genet. 2005, 3, 870–881. [Google Scholar] [CrossRef]
- Majdalani, N.; Vanderpool, C.K.; Gottesman, S. Bacterial Small RNA Regulators. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 93–113. [Google Scholar] [CrossRef]
- Deigan, K.E.; FerrÉ-D’AmarÉ, A.R. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res. 2011, 44, 1329–1338. [Google Scholar] [CrossRef]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef]
- Blount, K.F.; Breaker, R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006, 24, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T. Aminoglycoside antibiotics: Old drugs and new therapeutic approaches. Cell. Mol. Life Sci. 2007, 64, 1841–1852. [Google Scholar] [CrossRef]
- Lee, E.R.; Blount, K.F.; Breaker, R.R. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009, 6, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Montange, R.K.; Batey, R.T. Riboswitches: Emerging Themes in RNA Structure and Function. Annu. Rev. Biophys. 2008, 37, 117–133. [Google Scholar] [CrossRef] [PubMed]
| Novel Bacterial-Based Therapeutic Strategies | |||
|---|---|---|---|
| Beneficial Features | Clinical Applications | Limitations | |
| CRISPR-Cas9 | Able to remove the dominant allele from the cell via non-homologous end joining (NHEJ) to correct errors during mitosis, avoiding subsequent potential genetic mutations and mitotic catastrophe | Limitations in the direct editing of a gene into the right cell type | Can be used for the genomic editing of diseases in which there is a need for editing a selective allele |
| Poor selectivity | Can be used for antibacterial therapies | ||
| Riboswitches | Identified riboswitches respond to ubiquitous and important metabolites and second messengers related with mRNAs-encoding proteins, fundamental for survival or against pathogens | Exhibit a limited selectivity for their target genes | |
| Riboswitches-mediated small molecules recognition is through different mechanisms compared to eukaryotes, avoiding cross-species reactivity | Analog ligands still unknonw | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltani-Fard, E.; Taghvimi, S.; Abedi Kichi, Z.; Weber, C.; Shabaninejad, Z.; Taheri-Anganeh, M.; Hossein Khatami, S.; Mousavi, P.; Movahedpour, A.; Natarelli, L. Insights into the Function of Regulatory RNAs in Bacteria and Archaea. Int. J. Transl. Med. 2021, 1, 403-423. https://doi.org/10.3390/ijtm1030024
Soltani-Fard E, Taghvimi S, Abedi Kichi Z, Weber C, Shabaninejad Z, Taheri-Anganeh M, Hossein Khatami S, Mousavi P, Movahedpour A, Natarelli L. Insights into the Function of Regulatory RNAs in Bacteria and Archaea. International Journal of Translational Medicine. 2021; 1(3):403-423. https://doi.org/10.3390/ijtm1030024
Chicago/Turabian StyleSoltani-Fard, Elahe, Sina Taghvimi, Zahra Abedi Kichi, Christian Weber, Zahra Shabaninejad, Mortaza Taheri-Anganeh, Seyyed Hossein Khatami, Pegah Mousavi, Ahmad Movahedpour, and Lucia Natarelli. 2021. "Insights into the Function of Regulatory RNAs in Bacteria and Archaea" International Journal of Translational Medicine 1, no. 3: 403-423. https://doi.org/10.3390/ijtm1030024
APA StyleSoltani-Fard, E., Taghvimi, S., Abedi Kichi, Z., Weber, C., Shabaninejad, Z., Taheri-Anganeh, M., Hossein Khatami, S., Mousavi, P., Movahedpour, A., & Natarelli, L. (2021). Insights into the Function of Regulatory RNAs in Bacteria and Archaea. International Journal of Translational Medicine, 1(3), 403-423. https://doi.org/10.3390/ijtm1030024






