Mutational Characterization of Astrocytoma, IDH-Mutant, CNS WHO Grade III in the AACR GENIE Database
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genetic Differences by Race and Sex
3.2. Co-Occurrence and Exclusivity
3.3. Primary vs. Metastatic Samples
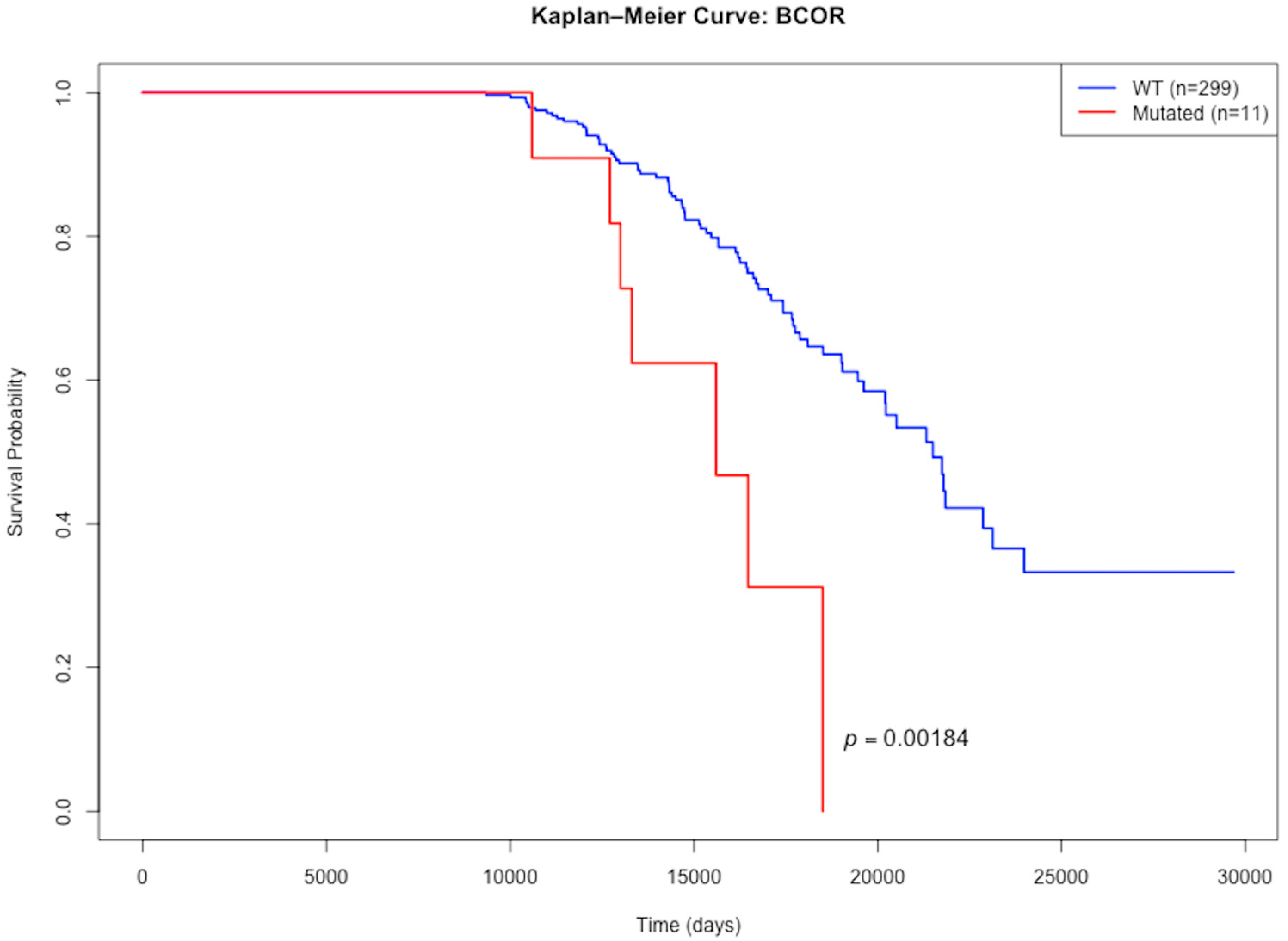
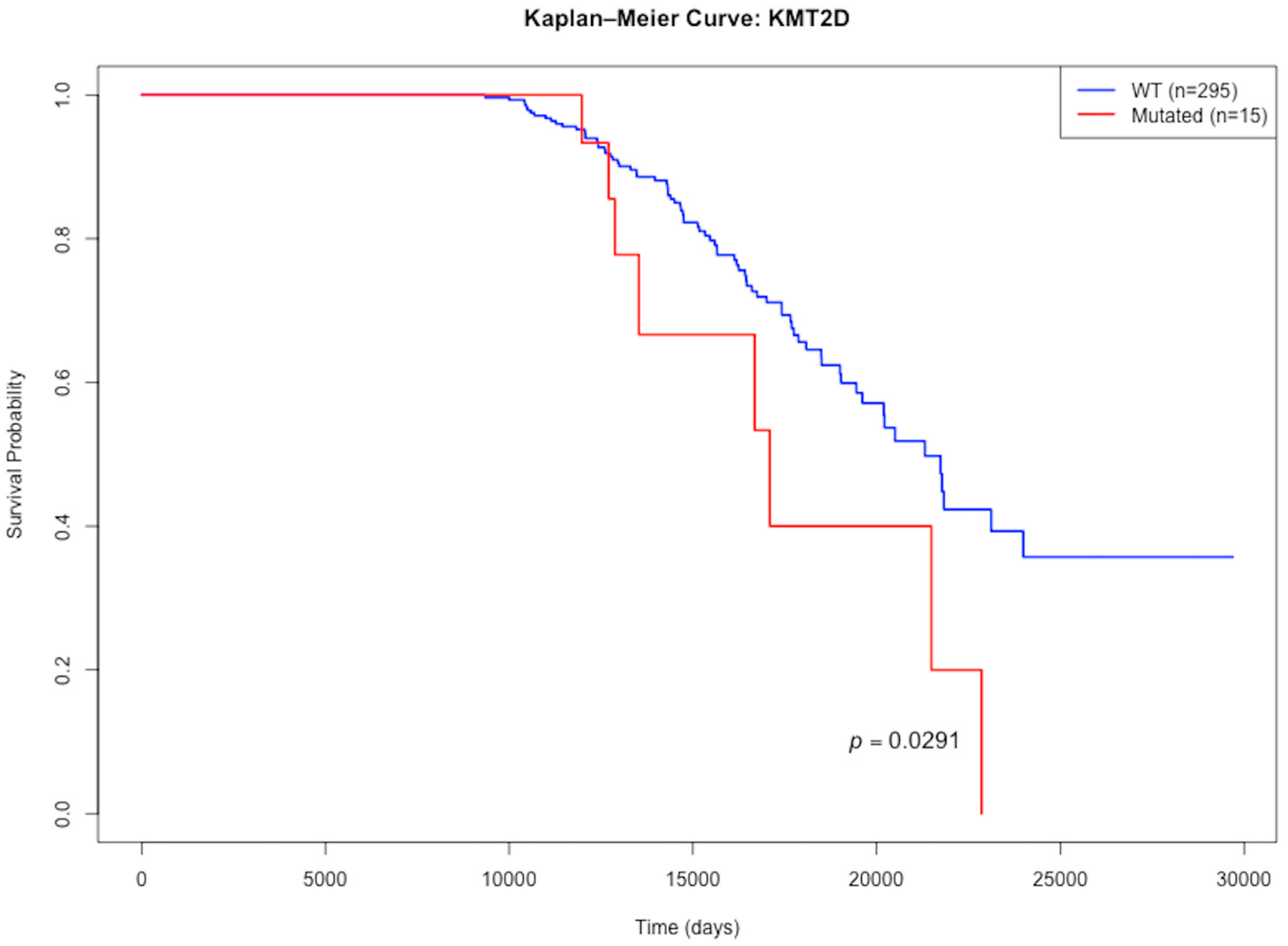
3.4. Mutations with Differential Effects on Lifespan
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GENIE | AACR Project Genomics Evidence Neoplasia Information Exchange |
| AACR | American Association for Cancer Research |
| 2-HG | (R)-2-hydroxyglutarate |
| 2OG | Alpha-ketoglutarate |
| IDH-mut A3 | Astrocytoma, IDH-mutant, CNS WHO grade 3 |
| ALT | Alternative lengthening of telomeres |
| ATR | ATR serine/threonine kinase |
| ATRX | ATP-dependent helicase ATRX |
| AXIN2 | Axin 2 |
| BCOR | BCL6 corepressor |
| BRAF | B-Raf Proto-oncogene, serine/threonine kinase |
| BRD4 | Bromodomain-containing protein 4 |
| CDK4 | Cyclin-dependent kinase 4 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CNS | Central nervous system |
| CREBBP | CREB binding protein |
| EGFR | Epidermal growth factor receptor |
| FAT1 | FAT atypical cadherin 1 |
| FDR | False discovery rate |
| G-CIMP | Glioma CpG island methylator phenotype |
| GLI1 | GLI family zinc finger 1 |
| IDH | Isocitrate dehydrogenase |
| KIT | KIT proto-oncogene, receptor tyrosine kinase |
| KMT2D | Lysine methyltransferase 2D |
| MAP2K2 | Mitogen-activated protein kinase 2 |
| MLH1 | MutL homolog 1 |
| MTAP | Methylthioadenosine phosphorylase |
| MTOR | Mechanistic target of rapamycin kinase |
| NF1 | Neurofibromin 1 |
| NOTCH1 | Notch receptor 1 |
| PDGFRA | Platelet-derived growth factor receptor alpha |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 |
| PRKDC | Protein kinase, DNA-activated, catalytic subunit |
| RET | Ret proto-oncogene |
| RICTOR | RPTOR independent companion of MTOR complex 2 |
| ROS1 | ROS proto-oncogene 1, receptor tyrosine kinase |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A |
| SMARCA4 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 |
| SMO | Smoothened, frizzled class receptor |
| SV | Structural variant |
| TERT | Telomerase reverse transcriptase |
| TET1 | Tet methylcytosine dioxygenase 1 |
| TP53 | Tumor protein p53 |
| WHO | World Health Organization |
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Alshiekh Nasany, R.; de la Fuente, M.I. Therapies for IDH-Mutant Gliomas. Curr. Neurol. Neurosci. Rep. 2023, 23, 225–233. [Google Scholar] [CrossRef]
- Anvari, K.; Seilanian Toussi, M.; Shahidsales, S.; Motlagh, F.; Reza Ehsaee, M.; Afshari, F. Treatment Outcomes and Prognostic Factors in Adult Astrocytoma: In North East of Iran. Iran. J. Cancer Prev. 2016, 9, e4099. [Google Scholar] [CrossRef] [PubMed]
- Smoll, N.R.; Hamilton, B. Incidence and Relative Survival of Anaplastic Astrocytomas. Neuro Oncol. 2014, 16, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Brandner, S.; McAleenan, A.; Jones, H.E.; Kernohan, A.; Robinson, T.; Schmidt, L.; Dawson, S.; Kelly, C.; Leal, E.S.; Faulkner, C.L.; et al. Diagnostic accuracy of 1p/19q codeletion tests in oligodendroglioma: A comprehensive meta-analysis based on a Cochrane systematic review. Neuropathol. Appl. Neurobiol. 2022, 48, e12790. [Google Scholar] [CrossRef]
- Lin, M.D.; Tsai, A.C.-Y.; Abdullah, K.G.; McBrayer, S.K.; Shi, D.D. Treatment of IDH-Mutant Glioma in the INDIGO Era. npj Precis. Oncol. 2024, 8, 149. [Google Scholar] [CrossRef]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 Mutation Is Sufficient to Establish the Glioma Hypermethylator Phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Malta, T.M.; de Souza, C.F.; Sabedot, T.S.; Silva, T.C.; Mosella, M.S.; Kalkanis, S.N.; Snyder, J.; Castro, A.V.B.; Noushmehr, H. Glioma CpG Island Methylator Phenotype (G-CIMP): Biological and Clinical Implications. Neuro Oncol. 2018, 20, 608–620. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH Mutation in Glioma: Molecular Mechanisms and Potential Therapeutic Targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Byun, Y.H.; Park, C.-K. Classification and Diagnosis of Adult Glioma: A Scoping Review. Brain Neurorehabilit. 2022, 15, e23. [Google Scholar] [CrossRef]
- Haase, S.; Garcia-Fabiani, M.B.; Carney, S.; Altshuler, D.; Núñez, F.J.; Méndez, F.M.; Núñez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a New Therapeutic Target for Glioma. Expert Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef]
- Katzendobler, S.; Niedermeyer, S.; Blobner, J.; Trumm, C.; Harter, P.N.; von Baumgarten, L.; Stoecklein, V.M.; Tonn, J.-C.; Weller, M.; Thon, N.; et al. Determinants of Long-Term Survival in Patients with IDH-Mutant Gliomas. J. Neurooncol 2024, 170, 655–664. [Google Scholar] [CrossRef]
- Mohile, N.A.; Messersmith, H.; Gatson, N.T.; Hottinger, A.F.; Lassman, A.; Morton, J.; Ney, D.; Nghiemphu, P.L.; Olar, A.; Olson, J.; et al. Therapy for Diffuse Astrocytic and Oligodendroglial Tumors in Adults: ASCO-SNO Guideline. J. Clin. Oncol. 2022, 40, 403–426. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef]
- Childhood Astrocytomas and Other Gliomas Treatment (PDQ®)-NCI. Available online: https://www.cancer.gov/types/brain/hp/child-astrocytoma-glioma-treatment-pdq (accessed on 8 August 2025).
- AACR Project GENIE Consortium; André, F.; Arnedos, M.; Baras, A.S.; Baselga, J.; Bedard, P.L.; Berger, M.F.; Bierkens, M.; Calvo, F.; Cerami, E. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Reuss, D.E. Updates on the WHO diagnosis of IDH-mutant glioma. J. Neurooncol. 2023, 162, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Whiting, K. Cbioportalr: Browse and Query Clinical and Genomic Data from Cbioportal, R package version 1.1.1. 2024. Available online: https://CRAN.R-project.org/package=cbioportalR (accessed on 1 May 2025).
- Wickham, H.; Vaughan, D.; Girlich, M. Tidyr: Tidy Messy Data, R package version 1.3.1. 2024. Available online: https://CRAN.R-project.org/package=tidyr (accessed on 1 May 2025).
- Xie, Y. Knitr: A General-Purpose Package for Dynamic Report Generation in R, R Package Version 1.49. 2024. Available online: https://yihui.org/knitr/ (accessed on 1 May 2025).
- Xie, Y. Dynamic Documents with R and Knitr, 2nd ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2015; ISBN 978-1498716963. Available online: https://github.com/yihui/knitr-book (accessed on 1 May 2025).
- Xie, Y. knitr: A Comprehensive Tool for Reproducible Research in R. In Implementing Reproducible Computational Research; Stodden, V., Leisch, F., Peng, R.D., Eds.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014; ISBN 978-1466561595. [Google Scholar]
- Therneau, T. A Package for Survival Analysis in R, R Package Version 3.7-0. 2024. Available online: https://CRAN.R-project.org/package=survival (accessed on 1 May 2025).
- Terry, M.T.; Patricia, M. Grambsch. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation, R package version 1.1.4. 2023. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 1 May 2025).
- Arora, S.; Morgan, M.; Carlson, M.; Pagès, H. GenomeInfoDb: Utilities for Manipulating Chromosome Names, Including Modifying Them to Follow a Particular Naming Style, R package version 1.42.3. 2025. Available online: https://bioconductor.org/packages/GenomeInfoDb (accessed on 1 May 2025).
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.; Carey, V. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Pagès, H.; Lawrence, M.; Aboyoun, P. S4Vectors: Foundation of Vector-Like and List-Like Containers in Bioconductor, R Package Version 0.44.0. 2024. Available online: https://bioconductor.org/packages/S4Vectors (accessed on 8 August 2025).
- Huber, W.; Carey, J.V.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, S.B.; Bravo, C.H.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Hocking, T.; Schwendinger, B. Data.Table: Extension of ‘data.frame’, R Package Version 1.16.2. 2024. Available online: https://CRAN.R-project.org/package=data.table (accessed on 8 August 2025).
- Williams, E.A.; Sharaf, R.; Decker, B.; Werth, A.J.; Toma, H.; Montesion, M.; Sokol, E.S.; Pavlick, D.C.; Shah, N.; Williams, K.J.; et al. CDKN2C-Null Leiomyosarcoma: A Novel, Genomically Distinct Class of TP53/RB1–Wild-Type Tumor with Frequent CIC Genomic Alterations and 1p/19q-Codeletion. JCO Precis. Oncol. 2020, 4, 955–971. [Google Scholar] [CrossRef]
- Perez, G.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res. 2025, 53, D1243–D1249. [Google Scholar] [CrossRef]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Li, Z.; Deng, Z.; Liu, F.; Li, C.; Yang, K.; Gong, X.; Feng, S.; Zeng, Y.; Zhou, H.; Fan, F.; et al. Clinical Sequencing Reveals Diagnostic, Therapeutic, and Prognostic Biomarkers for Adult-Type Diffuse Gliomas. Heliyon 2024, 10, e37712. [Google Scholar] [CrossRef] [PubMed]
- Mayr, L.; Neyazi, S.; Schwark, K.; Trissal, M.; Beck, A.; Labelle, J.; Eder, S.K.; Weiler-Wichtl, L.; Marques, J.G.; de Biagi-Junior, C.A.O.; et al. Effective Targeting of PDGFRA-Altered High-Grade Glioma with Avapritinib. Cancer Cell 2025, 43, 740–756.e8. [Google Scholar] [CrossRef]
- Guan, B.; Wang, T.-L.; Shih, I.-M. ARID1A, a Factor That Promotes Formation of SWI/SNF-Mediated Chromatin Remodeling, Is a Tumor Suppressor in Gynecologic Cancers. Cancer Res. 2011, 71, 6718–6727. [Google Scholar] [CrossRef]
- Lee, K.; Kim, S.-I.; Kim, E.E.; Shim, Y.-M.; Won, J.-K.; Park, C.-K.; Choi, S.H.; Yun, H.; Lee, H.; Park, S.-H. Genomic Profiles of IDH-Mutant Gliomas: MYCN-Amplified IDH-Mutant Astrocytoma Had the Worst Prognosis. Sci. Rep. 2023, 13, 6761. [Google Scholar] [CrossRef]
- Hatanpaa, K.J.; Burma, S.; Zhao, D.; Habib, A.A. Epidermal Growth Factor Receptor in Glioma: Signal Transduction, Neuropathology, Imaging, and Radioresistance. Neoplasia 2010, 12, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liu, G.; Zhang, Y.; Zhou, Y.; Pan, Y.; Zhang, Q.; Yu, R.; Gao, S. Gender Differences in Gliomas: From Epidemiological Trends to Changes at the Hormonal and Molecular Levels. Cancer Lett. 2024, 598, 217114. [Google Scholar] [CrossRef] [PubMed]
| Demographics | Category | N (%) |
|---|---|---|
| Sex | Male | 201 (58.4) |
| Female | 142 (41.3) | |
| Unknown | 1 (0.3) | |
| Age category | Adult | 347 (99.7) |
| Unknown | 1 (0.3) | |
| Pediatric | 0 (0) | |
| Ethnicity | Non-Hispanic | 273 (79.4) |
| Unknown/Not Collected | 51 (14.8) | |
| Spanish/Hispanic | 20 (5.8) | |
| Race | White | 260 (75.6) |
| Unknown/Not Collected | 29 (8.4) | |
| Asian | 24 (7.0) | |
| Other | 17 (4.9) | |
| Black | 11 (3.2) | |
| Pacific Islander | 1 (0.3) | |
| Sample Type | Primary | 303 (87.1) |
| Metastatic | 40 (11.5) | |
| Not Collected/Unspecified | 5 (1.4) |
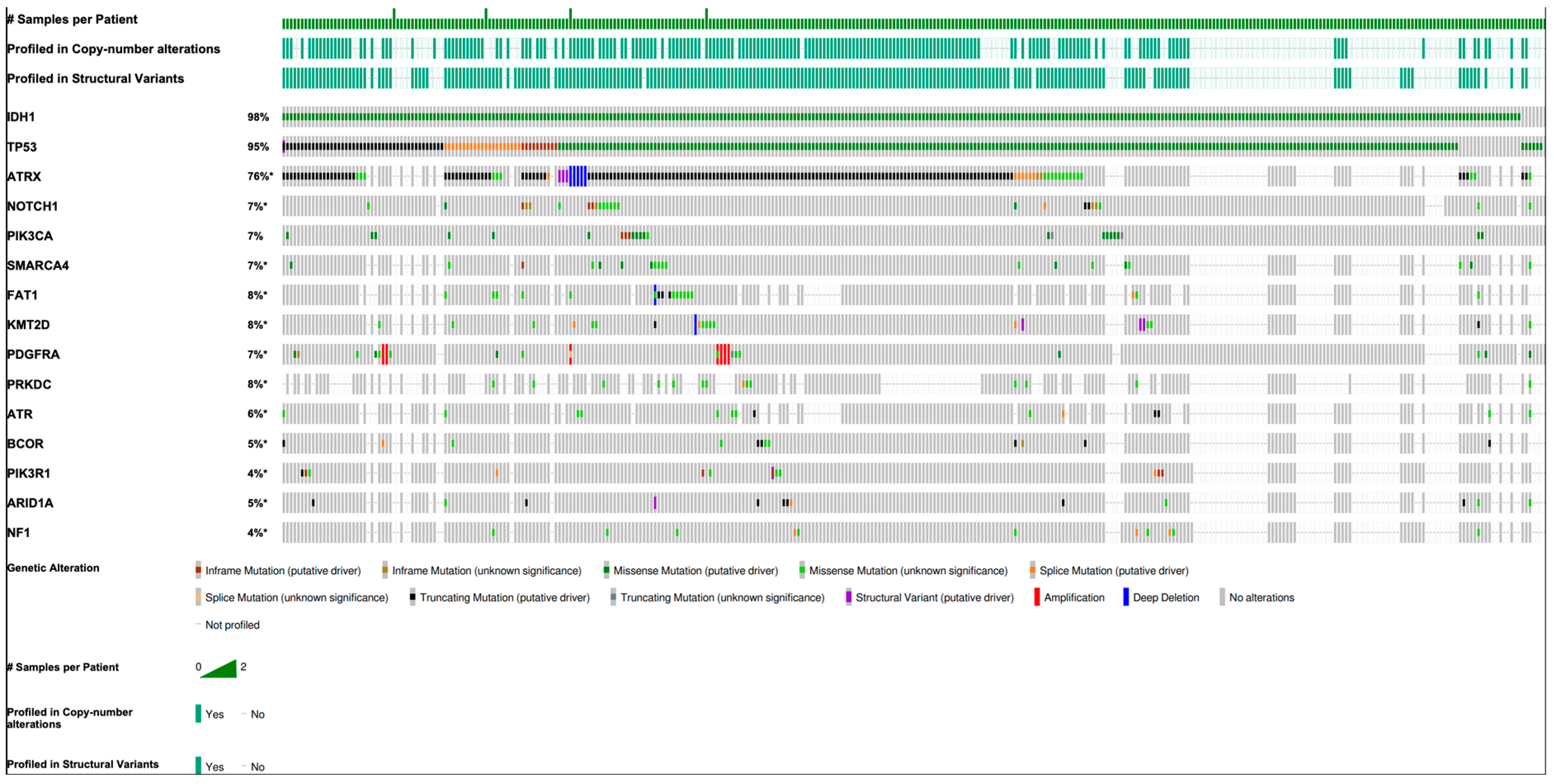
| Gene | N (%) | Function |
|---|---|---|
| IDH1 | 341 (98.0) | Converts alpha-ketoglutarate (2OG) into the oncometabolite (R)-2-hydroxyglutarate |
| IDH2 | 8 (2.3) | |
| TP53 | 330 (94.8) | Tumor suppressor |
| ATRX | 192 (55.2) | Chromatin remodeler |
| NOTCH1 | 24 (6.9) | Cell surface receptor in NOTCH pathway |
| PIK3CA | 24 (6.9) | PI3K/AKT/mTOR signaling pathway |
| SMARCA4 | 19 (5.5) | Chromatin remodeler |
| FAT1 | 18 (5.2) | Tumor suppressor |
| KMT2D | 18 (5.2) | Histone methyltransferase |
| PDGFRA | 16 (4.3) | AKT1/MAP signaling pathway |
| PRKDC | 15 (4.2) | Non-homologous end joining DNA repair |
| ATR | 14 (4.0) | DNA damage sensor |
| BCOR | 12 (3.4) | Transcriptional corepressor |
| PIK3R1 | 13 (3.7) | Regulatory subunit of PI3K |
| ARID1A | 12 (3.4) | Chromatin remodeler |
| NF1 | 11 (3.2) | Tumor suppressor of Ras pathway |
| Type | Gene Name | N (%) | Function |
|---|---|---|---|
| Amplification | CCND2 | 8 (4.0) | Cyclin D2 cell cycle regulator |
| CDK4 | 8 (4.0) | Cyclin-dependent kinase 4 cell cycle regulator | |
| PDGFRA | 7 (3.5) | AKT1/MAP signaling pathway | |
| MYCN | 5 (2.5) | Cellular proliferation transcription factor | |
| KRAS | 5 (2.5) | Oncogene GTPase | |
| Homozygous Deletion | ATRX | 6 (3.0) | Chromatin remodeler |
| CDKN1B | 3 (1.5) | Cell cycle inhibitor | |
| EPHA7 | 3 (1.5) | Receptor tyrosine kinase | |
| PDCD1 | 3 (1.5) | Immune checkpoint receptor | |
| TERT | 3 (1.5) | Telomerase reverse transcriptase |
| Gene Name | N | Deletions | Inversions | Duplications | Translocation | NA |
|---|---|---|---|---|---|---|
| KMT2D | 4 | 0 | 0 | 1 | 1 | 2 |
| ATRX | 3 | 0 | 0 | 0 | 0 | 3 |
| SDHA | 2 | 0 | 1 | 0 | 0 | 1 |
| ABL1 | 2 | 0 | 0 | 0 | 0 | 2 |
| BRCA1 | 1 | 0 | 0 | 0 | 0 | 1 |
| Gene | Effect on Lifespan | Median Lifespan Mutated | Median Lifespan WT | N_Mutated | N_WT | p Value |
|---|---|---|---|---|---|---|
| BCOR | Lessens | 15,595 | 21,488 | 11 | 299 | 0.00184 |
| KMT2D | Lessens | 17,094 | 21,310 | 15 | 295 | 0.0291 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torbenson, E.; Hsia, B.; Lang, N.; Silberstein, P. Mutational Characterization of Astrocytoma, IDH-Mutant, CNS WHO Grade III in the AACR GENIE Database. DNA 2025, 5, 43. https://doi.org/10.3390/dna5030043
Torbenson E, Hsia B, Lang N, Silberstein P. Mutational Characterization of Astrocytoma, IDH-Mutant, CNS WHO Grade III in the AACR GENIE Database. DNA. 2025; 5(3):43. https://doi.org/10.3390/dna5030043
Chicago/Turabian StyleTorbenson, Elijah, Beau Hsia, Nigel Lang, and Peter Silberstein. 2025. "Mutational Characterization of Astrocytoma, IDH-Mutant, CNS WHO Grade III in the AACR GENIE Database" DNA 5, no. 3: 43. https://doi.org/10.3390/dna5030043
APA StyleTorbenson, E., Hsia, B., Lang, N., & Silberstein, P. (2025). Mutational Characterization of Astrocytoma, IDH-Mutant, CNS WHO Grade III in the AACR GENIE Database. DNA, 5(3), 43. https://doi.org/10.3390/dna5030043






