Of Short Interspersed Nuclear Elements, Long Interspersed Nuclear Elements and Leeches: Identification and Molecular Characterization of Transposable Elements in Leech Genomes
Abstract
1. Introduction
2. Methods and Materials
2.1. Genome and Transcriptome Data
2.2. Sources of Reference Sequences
| Domain | Reference |
| Apurinic endonuclease (APE) | [34,35] |
| Aspartic protease (AP) | [36,37] |
| Integrase (IN) | [38,39] |
| Restriction-like endonuclease (RLE) | [35] |
| Reverse Transkriptase (RT) | [38,40,41,42] |
| RNA recognition motif (RRM) | [12,43] |
| RNase H (RH) | [44,45,46] |
| Tyrosin recombinase (YR) | [47,48,49] |
| Zinc finger knuckle motif (CCHC) | [50] |
2.3. Bioinformatics Tools
3. Results
3.1. Identification and Characterization of HvSINE1
3.2. Identification and Characterization of HvSINE2–4
3.3. Tissue-Specific Expression of HvSINE Sequences
3.4. Presence of HvSINE1-like Elements in Leech and Annelid Species
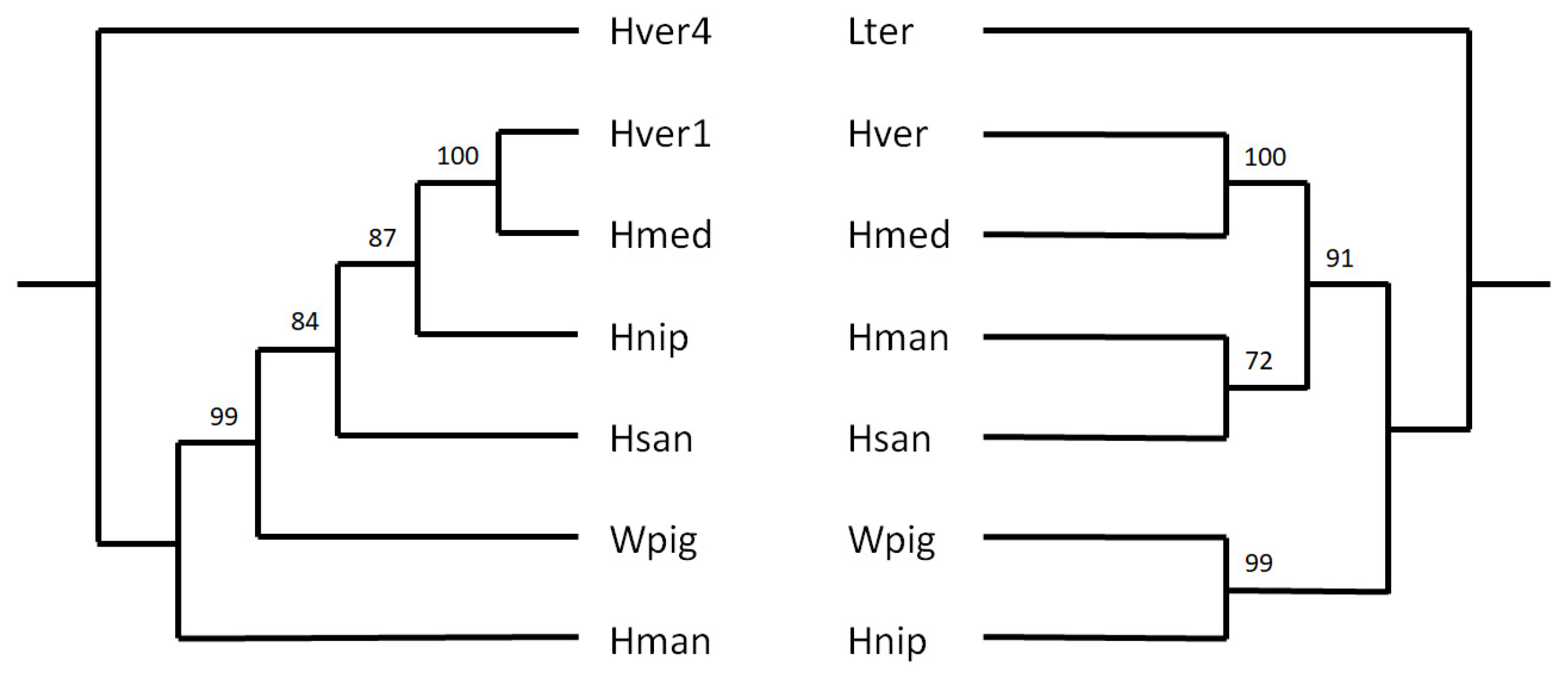
3.5. Phylogenetic Analyses Based on the HvSINE1 Sequence
3.6. Identification and Characterization of HmSINE_V2
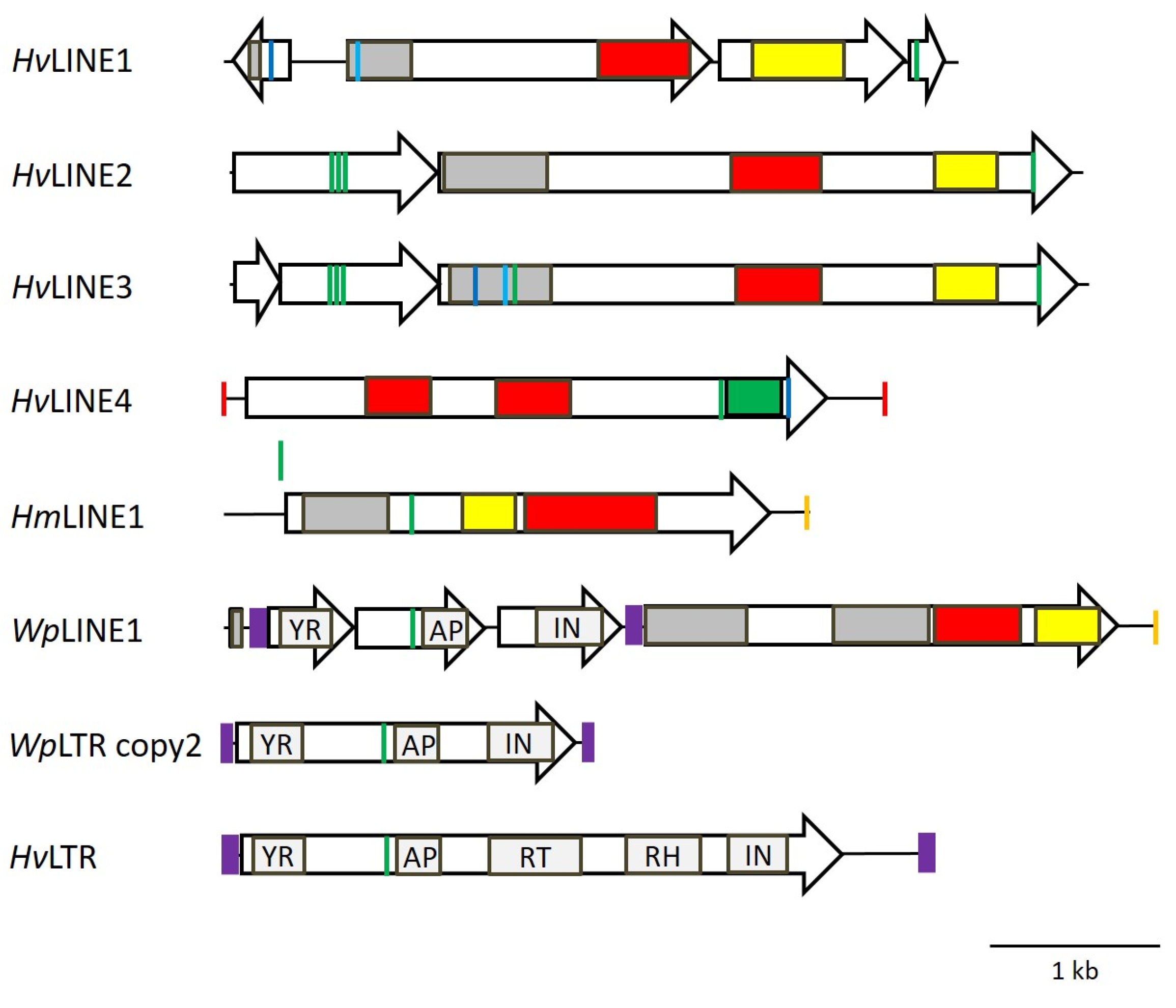
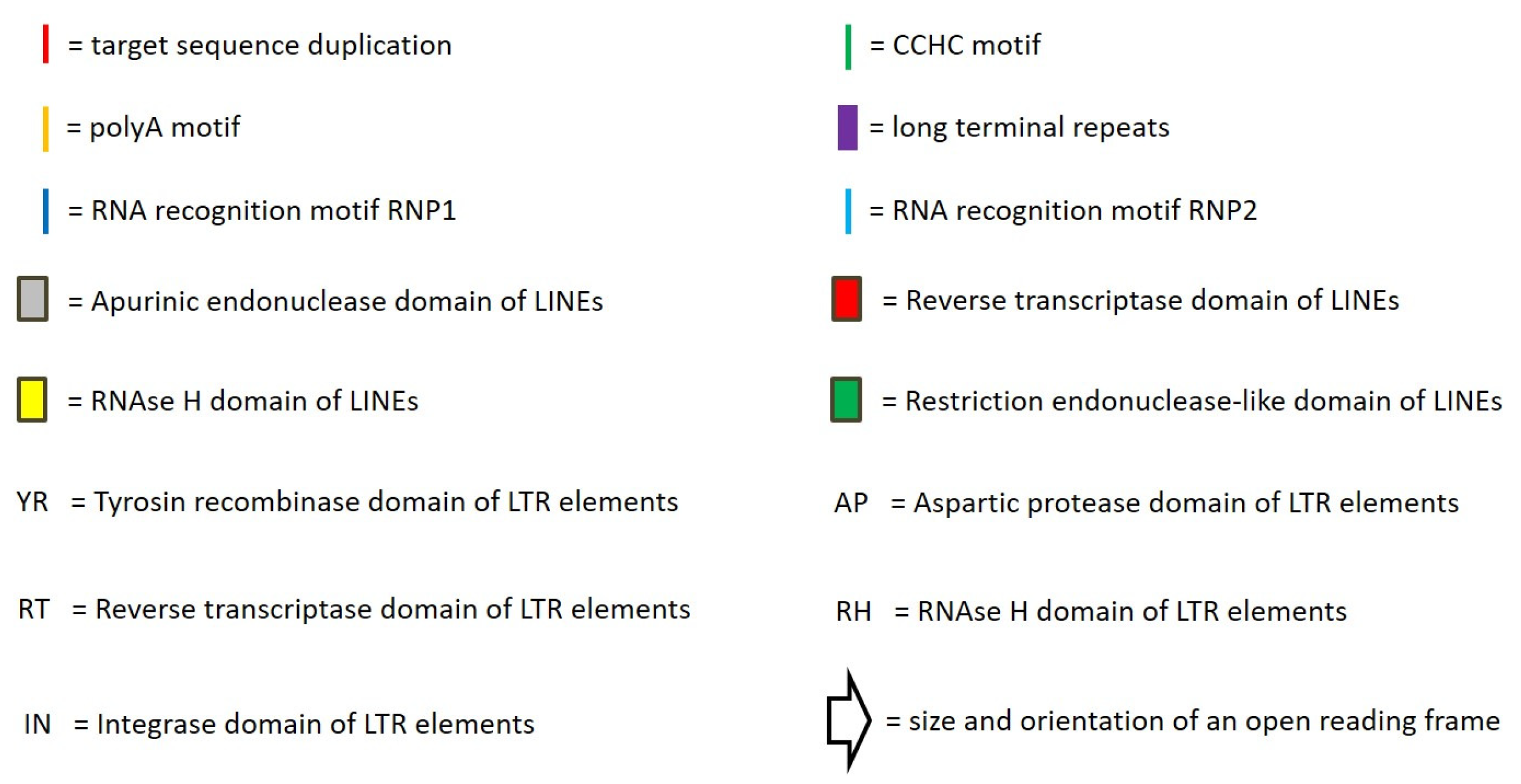
3.7. Identification of Corresponding LINEs
3.8. Abundance of WpLTRs
4. Discussion
4.1. Classification of SINEs
4.2. Classification of LINEs
4.3. Classification of LTR-Retrotransposons
4.4. Biological Significance
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H., Jr. Mobile elements: Drivers of genome evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef]
- Finnegan, D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Finnegan, D.L. Retrotransposons. Curr. Biol. 2012, 22, R432–R437. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Feschotte, C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Hamada, M.; Ogiwara, I.; Ohshima, K. SINEs and LINEs share common 3′ sequences: A review. Gene 1997, 205, 229–243. [Google Scholar] [CrossRef]
- Roy-Engel, A.M. A tale of an A-tail: The lifeline of a SINE. Mob. Genet. Elem. 2012, 2, 282–286. [Google Scholar] [CrossRef][Green Version]
- Borodulina, O.R.; Kramerov, D.A. Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner. RNA 2008, 14, 1865–1873. [Google Scholar] [CrossRef]
- Borodulina, O.R.; Golubchikova, J.S.; Ustyantsev, I.G.; Kramerov, D.A. Polyadenylation of RNA transcribed from mammalian SINEs by RNA polymerase III: Complex requirements for nucleotide sequences. Biochim. Biophys. Acta 2016, 1859, 355–365. [Google Scholar] [CrossRef]
- Borodulina, O.R.; Kramerov, D.A. Short interspersed elements (SINEs) from insectivores. Two classes of mammalian SINEs distinguished by A-rich tail structure. Mamm. Genome 2001, 12, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Khazina, E.; Weichenrieder, O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. USA 2009, 106, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C.J.; Casane, D. Modular organization and reticulate evolution of the ORF1 of Jockey superfamily transposable elements. Mob. DNA 2014, 5, 19. [Google Scholar] [CrossRef]
- SenGupta, D. RNA-Binding Domains in Proteins. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: New York, NY, USA, 2013; pp. 274–276. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. Origin and evolution of SINEs in eukaryotic genomes. Heredity 2011, 107, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.; Labuda, D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc. Natl. Acad. Sci. USA 1999, 96, 2869–2874. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, N.; Sun, Y. AnnoSINE: A short interspersed nuclear elements annotation tool for plant genomes. Plant Physiol. 2022, 188, 955–970. [Google Scholar] [CrossRef]
- Goubert, C.; Craig, R.J.; Bilat, A.F.; Peona, V.; Vogan, A.A.; Protasio, A.V. A beginner’s guide to manual curation of transposable elements. Mob. DNA 2022, 13, 7. [Google Scholar] [CrossRef]
- Kanhayuwa, L.; Coutts, R.H.A. Short Interspersed Nuclear Element (SINE) sequences in the genome of the human pathogenic fungus Aspergillus fumigatus Af293. PLoS ONE 2016, 11, e0163215. [Google Scholar] [CrossRef]
- Sket, B.; Trontelj, P. Global diversity of leeches (Hirudinea) in freshwater. Hydrobiologia 2008, 595, 129–137. [Google Scholar] [CrossRef]
- Kvist, S.; Utevsky, S.; Marrone, F.; Ben Ahmed, R.; Gajda, Ł.; Grosser, C.; Huseynov, M.; Jueg, U.; Khomenko, A.; Oceguera-Figueroa, A.; et al. Extensive sampling sheds light on species-level diversity in Palearctic Placobdella (Annelida: Clitellata: Glossiphoniiformes). Hydrobiologia 2022, 849, 1239–1259. [Google Scholar] [CrossRef]
- Sawyer, R.T. Leech Biology and Behaviour: Feeding, Biology, Ecology and Systematics; Clarendon Press: Oxford, UK, 1986; ISBN 978-019-857-622-8. [Google Scholar]
- Abdualkader, A.M.; Ghawi, A.M.; Alaama, M.; Awang, M.; Merzouk, A. Leech therapeutic applications. Indian. J. Pharm. Sci. 2013, 75, 127–137. [Google Scholar] [PubMed]
- Hildebrandt, J.-P.; Lemke, S. Small bite, large impact–saliva and salivary molecules in the medicinal leech, Hirudo medicinalis. Naturwissenschaften 2011, 98, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Lemke, S.; Vilcinskas, A. European Medicinal leeches-new roles in modern medicine. Biomedicines 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Greinacher, A.; Warkentin, T.E. The direct thrombin inhibitor hirudin. Thromb. Haemost. 2008, 99, 819–829. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Cho, S.-J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, D.-H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef]
- Guan, D.L.; Yang, J.; Liu, Y.K.; Li, Y.; Mi, D.; Ma, L.B.; Wang, Z.Z.; Xu, S.Q.; Qiu, Q. Draft Genome of the Asian buffalo leech Hirudinaria manillensis. Front. Genet. 2020, 10, 1321. [Google Scholar] [CrossRef]
- Kvist, S.; Manzano-Marín, A.; de Carle, D.; Trontelj, P.; Siddall, M.E. Draft genome of the European medicinal leech Hirudo medicinalis (Annelida, Clitellata, Hirudiniformes) with emphasis on anticoagulants. Sci. Rep. 2020, 10, 9885. [Google Scholar] [CrossRef]
- Babenko, V.V.; Podgorny, O.V.; Manuvera, V.A.; Kasianov, A.S.; Manolov, A.I.; Grafskaia, E.N.; Shirokov, D.A.; Kurdyumov, A.S.; Vinogradov, D.V.; Nikitina, A.S.; et al. Draft genome sequences of Hirudo medicinalis and salivary transcriptome of three closely related medicinal leeches. BMC Genomics 2020, 21, 331. [Google Scholar] [CrossRef]
- Tong, L.; Dai, S.-X.; Kong, D.-J.; Yang, P.-P.; Tong, X.; Tong, X.-R.; Bi, X.-X.; Su, Y.; Zhao, Y.-Q.; Liu, Z.-C. The genome of medicinal leech (Whitmania pigra) and comparative genomic study for exploration of bioactive ingredients. BMC Genomics 2022, 23, 76. [Google Scholar] [CrossRef]
- Paulsen, R.T.; Agany, D.D.M.; Petersen, J.; Davis, C.M.; Ehli, E.A.; Gnimpieba, E.; Burrell, B.S. A draft genome for Hirudo verbana, the Medicinal leech. BioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, F.; Huang, Z.; He, B.; Liu, K.; Li, J.; Liu, Z.; Lin, G. Comparative genomics of two Asian medicinal leeches Hirudo nipponia and Hirudo tianjinensis: With emphasis on antithrombotic genes and their corresponding proteins. Int. J. Biol. Macromol. 2024, 270, 132278. [Google Scholar] [CrossRef]
- Fillingham, J.S.; Thing, T.A.; Vythilingum, N.; Keuroghlian, A.; Bruno, D.; Golding, G.B.; Pearlman, R.E. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot. Cell 2004, 3, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K.; Fujiwara, H. An extraordinary retrotransposon family encoding dual endonucleases. Genome Res. 2005, 15, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J. Comparative studies on retroviral proteases: Substrate specificity. Viruses 2010, 2, 147–165. [Google Scholar] [CrossRef]
- Gazda, L.D.; Matúz, K.J.; Nagy, T.; Mótyán, J.A.; Tőzsér, J. Biochemical characterization of Ty1 retrotransposon protease. PLoS ONE 2020, 15, e0227062. [Google Scholar] [CrossRef]
- Evgen’ev, M.B.; Zelentsova, H.; Shostak, N.; Kozitsina, M.; Barskyi, V.; Lankenau, D.H.; Corces, V.G. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1997, 94, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Tsuchida, K.; Choi, S.; Sekine, Y.; Shiga, Y.; Ohtsubo, E. Presence of a characteristic D-D-E motif in IS1 transposase. J. Bacteriol. 2002, 184, 6146–6154. [Google Scholar] [CrossRef]
- Goodwin, T.J.D.; Poulter, R.T.M. The DIRS1 group of retrotransposons. Mol. Biol. Evol. 2001, 18, 2067–2082. [Google Scholar] [CrossRef][Green Version]
- Arkhipova, I.R. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst. Biol. 2006, 55, 875–885. [Google Scholar] [CrossRef]
- Meier, B.; Clejan, I.; Liu, Y.; Lowden, M.; Gartner, A.; Hodgkin, J.; Ahmed, S. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2006, 2, e18. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, X.; Han, J. A computational model for predicting RNase H domain of retrovirus. PLoS ONE 2016, 11, e0161913. [Google Scholar] [CrossRef]
- Moelling, K.; Broecker, F.; Russo, G.; Sunagawa, S. RNase H as gene modifier, driver of evolution and antiviral defense. Front. Microbiol. 2017, 8, 1745. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.J.D.; Poulter, R.T.M. A new group of tyrosine recombinase-encoding retrotransposons. Mol. Biol. Evol. 2004, 21, 746–759. [Google Scholar] [CrossRef]
- Poulter, R.T.M.; Goodwin, T.J.D. DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet. Genome Res. 2005, 110, 575–588. [Google Scholar] [CrossRef]
- Poulter, R.T.M.; Butler, M.I. Tyrosine recombinase retrotransposons and transposons. Microbiol. Spectr. 2015, 3, MDNA3-0036-2014. [Google Scholar] [CrossRef]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Kans. Univ. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar] [CrossRef]
- Müller, C.; Mescke, K.; Liebig, S.; Mahfoud, H.; Lemke, S.; Hildebrandt, J.-P. More than just one: Multiplicity of hirudins and hirudin-like factors in the medicinal leech Hirudo medicinalis. Mol. Genet. Genomics 2016, 291, 227–240. [Google Scholar] [CrossRef]
- Müller, C.; Haase, M.; Lemke, S.; Hildebrandt, J.-P. Hirudins and hirudin-like factors in Hirudinidae: Implications for function and phylogenetic relationships. Parasitol. Res. 2017, 116, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Geiduschek, E.P.; Tocchini-Valentini, G.P. Transcription by RNA polymerase III. Annu. Rev. Biochem. 1988, 57, 873–914. [Google Scholar] [CrossRef]
- Dieci, G.; Giuliodori, S.; Catellani, M.; Percudani, R.; Ottonello, S. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 6903–6914. [Google Scholar] [CrossRef]
- Traboni, C.; Ciliberto, G.; Cortese, R. A novel method for site-directed mutagenesis: Its application to an eukaryotic tRNAPro gene promoter. EMBO J. 1982, 1, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Siddall, M. Poly-paraphyly of Hirudinidae: Many lineages of medicinal leeches. BMC Evol. Biol. 2009, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Dornburg, A.; Zapfe, K.L.; Anderson, F.E.; James, S.W.; Erséus, C.; Lemmon, E.M.; Lemmon, A.R.; Wiliams, B.W. Phylogenomic analysis of a putative missing link sparks reinterpretation of leech evolution. Genome Biol. Evol. 2019, 11, 3082–3093. [Google Scholar] [CrossRef]
- Miyamoto, M.M. Molecular systematics: Perfect SINEs of evolutionary history? Curr. Biol. 1999, 9, R816–R819. [Google Scholar] [CrossRef]
- Ray, D.A.; Xing, J.; Salem, A.-H.; Batzer, M.A. SINEs of a nearly perfect character. Syst. Biol. 2006, 55, 928–935. [Google Scholar] [CrossRef]
- Korstian, J.M.; Paulat, N.S.; Platt, R.N., 2nd; Stevens, R.D.; Ray, D.A. SINE-based phylogenomics reveal extensive introgression and incomplete lineage sorting in Myotis. Genes 2022, 13, 399. [Google Scholar] [CrossRef]
- Martelossi, J.; Iannello, M.; Ghiselli, F.; Luchetti, A. Widespread HCD-tRNA derived SINEs in bivalves rely on multiple LINE partners and accumulate in genic regions. Mob. DNA 2024, 15, 22. [Google Scholar] [CrossRef]
- Lukas, P.; Melikian, G.; Hildebrandt, J.-P.; Müller, C. Make it double: Identification and characterization of a Tandem-Hirudin from the Asian medicinal leech Hirudinaria manillensis. Parasitol. Res. 2022, 121, 2995–3006. [Google Scholar] [CrossRef] [PubMed]
- Haren, L.; Ton-Hoang, B.; Chandler, M. Integrating DNA: Transposases and retroviral integrases. Annu. Rev. Microbiol. 1999, 53, 245–281. [Google Scholar] [CrossRef]
- Arkhipova, I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 2017, 8, 19. [Google Scholar] [CrossRef]
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2019, 94, 233–252. [Google Scholar] [CrossRef] [PubMed]
- McCullers, T.J.; Steiniger, M. Transposable elements in Drosophila. Mob. Genet. Elem. 2017, 7, 1–18. [Google Scholar] [CrossRef]
- Quesneville, H. Twenty years of transposable element analysis in the Arabidopsis thaliana genome. Mob. DNA 2020, 11, 28. [Google Scholar] [CrossRef]
- Han, G.; Zhang, N.; Jiang, H.; Meng, X.; Qian, K.; Zheng, Y.; Xu, J.; Wang, J. Diversity of short interspersed nuclear elements (SINEs) in lepidopteran insects and evidence of horizontal SINE transfer between baculovirus and lepidopteran hosts. BMC Genomics 2021, 22, 226. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 4.10.1–4.10.14. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Shi, J.; Liang, C. Generic Repeat Finder: A high-sensitivity tool for genome-wide de novo repeat detection. Plant Physiol. 2019, 180, 1803–1815. [Google Scholar] [CrossRef]
- Bell, E.A.; Butler, C.L.; Oliveira, C.; Marburger, S.; Yant, L.; Taylor, M.I. Transposable element annotation in non-model species: The benefits of species-specific repeat libraries using semi-automated EDTA and DeepTE de novo pipelines. Mol. Ecol. Resour. 2022, 22, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Deragon, J.-M.; Zhang, X. Short interspersed elements (SINEs) in plants: Origin, classification, and use as phylogenetic markers. Syst. Biol. 2006, 55, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, M.; Rooney, A.P.; Okada, N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: Hippopotamuses are the closest extant relatives of whales. Proc. Natl. Acad. Sci. USA 1999, 96, 10261–10266. [Google Scholar] [CrossRef]
- Trontelj, P.; Utevsky, S.Y. Phylogeny and phylogeography of medicinal leeches (genus Hirudo): Fast dispersal and shallow genetic structure. Mol. Phylogenet. Evol. 2012, 63, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Poulter, R.T.M.; Goodwin, T.J.D.; Butler, M.I. Vertebrate helentrons and other novel Helitrons. Gene 2003, 313, 201–212. [Google Scholar] [CrossRef]
- Karn, J. Retroviruses. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: New York, NY, USA, 2013; pp. 211–215. [Google Scholar] [CrossRef]
- Havecker, E.R.; Gao, X.; Voytas, D.F. The diversity of LTR retrotransposons. Genome Biol. 2004, 5, 225. [Google Scholar] [CrossRef][Green Version]
- Sabot, F.; Schulman, A.H. Parasitism and the retrotransposon life cycle in plants: A hitchhiker’s guide to the genome. Heredity 2006, 97, 381–388. [Google Scholar] [CrossRef]
- Sabot, F.; Sourdille, P.; Chantret, N.; Bernard, M. Morgane, a new LTR retrotransposon group, and its subfamilies in wheats. Genetica 2006, 128, 439–447. [Google Scholar] [CrossRef]
- Tanskanen, J.A.; Sabot, F.; Vicient, C.; Schulman, A.H. Life without GAG: The BARE-2 retrotransposon as a parasite’s parasite. Gene 2007, 390, 166–174. [Google Scholar] [CrossRef]
- Goodwin, T.J.D.; Butler, M.I.; Poulter, R.T.M. Cryptons: A group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology 2003, 149, 3099–3109. [Google Scholar] [CrossRef]
- Piednoël, M.; Gonçalves, I.R.; Higuet, D.; Bonnivard, E. Eukaryote DIRS1-like retrotransposons: An overview. BMC Genomics 2011, 12, 621. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Derbyshire, K.M. The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003, 4, 865–877. [Google Scholar] [CrossRef]
- Nefedova, L.; Kim, A. Mechanisms of LTR-retroelement transposition: Lessons from Drosophila melanogaster. Viruses 2017, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Warren, I.A.; Naville, M.; Chalopin, D.; Levin, P.; Berger, C.S.; Galiana, D.; Volff, J.-N. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 2015, 23, 505–531. [Google Scholar] [CrossRef]
- Nishihara, H. Transposable elements as genetic accelerators of evolution: Contribution to genome size, gene regulatory network rewiring and morphological innovation. Genes Genet. Syst. 2020, 94, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Serrato-Capuchina, A.; Matute, D.R. The role of transposable elements in speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef]
- Platt, R.N., 2nd; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Gross, U.; Roth, M. The biochemistry of leech saliva. In Medicinal Leech Therapy; Michalsen, A., Roth, M., Dobos, G., Eds.; Georg Thieme Verlag KG: Stuttgart, Germany, 2007. [Google Scholar] [CrossRef]
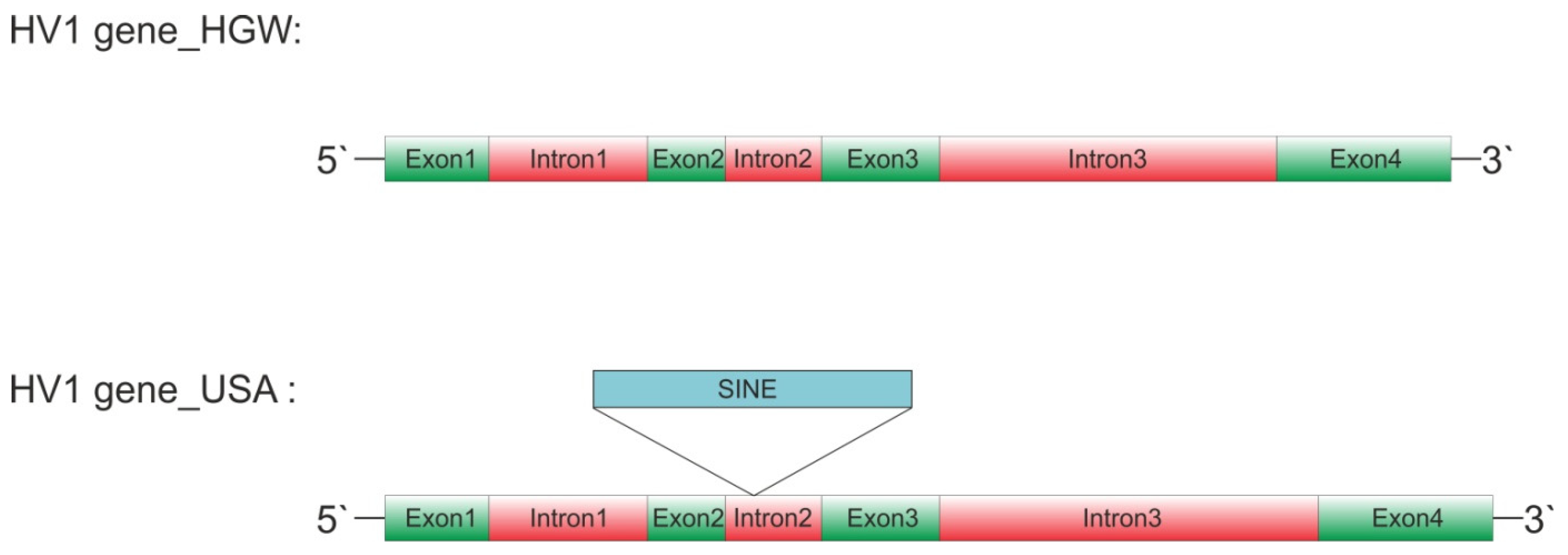
| Gene | Exon 1 | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | Exon 4 | GenBank |
|---|---|---|---|---|---|---|---|---|
| Hv_HGW1 | 61 | 103 | 50 | 62 | 76 | 199 | 71 | KX215734.1 |
| Hv_HGW2 | 61 | 103 | 50 | 62 | 76 | 199 | 71 | KX215735.1 |
| Hv_US | 61 | 103 | 50 | 272 | 76 | 224 | 71 | GCA_020137395.1 |
| JAIQDV010043103.1 | ||||||||
| Hv_contig_43718 | ||||||||
| position 3801-2954 | ||||||||
| Hm_HGW1 | 61 | 103 | 50 | 62 | 76 | 199 | 71 | KR066930.1 |
| Hm_HGW2 | 61 | 103 | 50 | 62 | 76 | 199 | 71 | KR066931.1 |
| Hm_Kvist | 61 | 103 | 50 | 62 | 76 | 199 | 71 | GCA_903470615.1 |
| CAGKPE010009153.1 | ||||||||
| contig SCF_090848 | ||||||||
| position 35665-36286 |
| 5S rRNA genes | ||||||
|---|---|---|---|---|---|---|
| Box A | 16 (13) bp | Box B/IE 4 bp | Box C | |||
| Homo sapiens: | TTGGAAGCTAAGCAGGGTCAGGCCTGGTTGGTACCT-GATGGGAGAGAG | |||||
| Plutella xylostella: | ACCGAAGTCAAGCAACGTCGGGC----GTAGTCATTGGATGGGTGACCG | |||||
| Urechis unicinctus: | ACTGAAGTTAAGCAACGTCGGGCCCGGTTAGTACTTGGATGGGTGACCG | |||||
| Hirudo medicinalis: | ACCGAAGTTAAGCAACGTCGAGCCCGGTTAGTACTTGGATGGGTGACCG | |||||
| Hirudo verbana: | ACCGAAGTTAAGCAACGTCGAGCCCGGTTAGTACTTGGATGGGTGACCG | |||||
| tRNA and 7SL genes | ||||||
| Box 1 | spacer | Box 2 | ||||
| tRNA consensus | TRGYBYAGTGG | 33 bp | RGTTCGADYCY + | |||
| TRGCNNAGYGG | 33 bp | GGTTCGANTCC * | ||||
| Human tRNAPro | TGGTCTAGTGG | 31 bp | GGTTCAA_TCC # | |||
| Human 7SL RNA | GGGCGCGGTGG | 47 bp | GCTTGAG_TCC | |||
| D. melanogaster 7SL RNA | TGGAAGGTTGG | 49 bp | GGCTGGGATCT | |||
| H. medicinalis 7SL RNA | TGGAGTCGTAG | 44 bp | GTTTGAGGTCG | |||
| H. verbana 7SL RNA | TGGAGTCGTAG | 44 bp | GTTTGAGGTCG | |||
| putative HvSINE1–4 promoters | ||||||
| Box 1 | spacer | Box 2 | ||||
| Hirudo sp. tRNA promoter | TGGTCTAATGG | 29–32 bp | GAATCGAATCC | |||
| HvSINE1 | TATCCCAATGG | 31 bp | TATATAGCGCC | |||
| HvSINE2 | GATCCGGGTTGG | 30 bp | TATATAGCACC | |||
| HvSINE3 | TGGATGCGAAGG | 31 bp | TGTGTGGATCA | |||
| HvSINE4 | TGCGCGGAGGG | 29 bp | TGTTTTAATCG | |||
| Hirudo verbana | Hirudo medicinalis | |
|---|---|---|
| HvSINE1 | >1000 copies | >1000 copies |
| HvSINE2 | 1 copy | 1 copy |
| HvSINE3 | 1 copy | 1 copy |
| HvSINE4 | 21 (about 200) copies | 14 (about 200) copies |
| HvSINE1 | HvSINE2 | HvSINE3 | HvSINE4 | |
|---|---|---|---|---|
| salivary gland | ✓ | - | - | ✓ |
| muscle | ✓ | - | - | ✓ |
| ganglion | ✓ | ✓ | - | ✓ |
| central nervous system | ✓ | ✓ | ✓ | ✓ |
| head | ✓ | ✓ | ✓ | ✓ |
| Hirudo medicinalis | + | Hirudinidae | Europe |
| Hirudinaria manillensis | + | Hirudinidae | Southeast |
| Whitmania pigra | + | Hirudinidae | Asia |
| Hirudo nipponia | + | Hirudinidae | East Asia |
| Haemopis sanguisuga | + | Hirudinidae | East Asia |
| Limnobdella mexicana | − | Hirudinidae | Europe, North Africa |
| Macrobdella decora | − | Hirudinidae | North America |
| Asiaticobdella fenestrata | − | Hirudinidae | North America |
| Haemadipsa interrupta | − | Haemadipsidae | Southern Africa |
| Haementeria vizzotoi | − | Glossiphoniidae | Southern Asia |
| Helobdella robusta | − | Glossiphoniidae | South America |
| Erpobdella octoculata | − | Erpobdellidae | North America |
| Piscicola geometra | − | Piscicolidae | Europe |
| Enchytraeus crypticus | − | Enchytraeidae | Europe |
| Lumbricus terrestris | − | Lumbricidae | globally |
| Eisenia fetida | − | Lumbricidae | Europe |
| Capitella teleta | − | Capitellidae | Europe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, C. Of Short Interspersed Nuclear Elements, Long Interspersed Nuclear Elements and Leeches: Identification and Molecular Characterization of Transposable Elements in Leech Genomes. DNA 2025, 5, 30. https://doi.org/10.3390/dna5020030
Müller C. Of Short Interspersed Nuclear Elements, Long Interspersed Nuclear Elements and Leeches: Identification and Molecular Characterization of Transposable Elements in Leech Genomes. DNA. 2025; 5(2):30. https://doi.org/10.3390/dna5020030
Chicago/Turabian StyleMüller, Christian. 2025. "Of Short Interspersed Nuclear Elements, Long Interspersed Nuclear Elements and Leeches: Identification and Molecular Characterization of Transposable Elements in Leech Genomes" DNA 5, no. 2: 30. https://doi.org/10.3390/dna5020030
APA StyleMüller, C. (2025). Of Short Interspersed Nuclear Elements, Long Interspersed Nuclear Elements and Leeches: Identification and Molecular Characterization of Transposable Elements in Leech Genomes. DNA, 5(2), 30. https://doi.org/10.3390/dna5020030