Multiplexed Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq) to Study DNA Methylation Using Low Amounts of DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Peripheral Blood Mononuclear Cells (PBMCs) Isolation
2.2. Micrococcal Nuclease Digestion
2.3. End-Repair, A-Tailing, and Adapter Ligation
2.4. Multiplex Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq)
2.5. Size Selection
2.6. Sequencing
2.7. Bioinformatics Analysis
- cutadapt --cores=40 --no-trim --quality-cutoff=15,10 -o .fastq.gz fastq.gz > .fastq.gz.txt
- methylQA medip -o file -m file.bed hg38_lite.size file
- macs2 callpeak \
- -B \
- -t trimmed_M192-A18_S11_ME_L001.extended.bed \
- -c trimmed_M206-A18_S11_ME_L001.extended.bed \
- -f BEDPE \
- -g hs \
- -n M192-A18_vs_M206-A18_q01_macs2 \
- -q 0.01 > M192-A18_q01_macs2.log
2.8. Statistical Analyses
3. Results
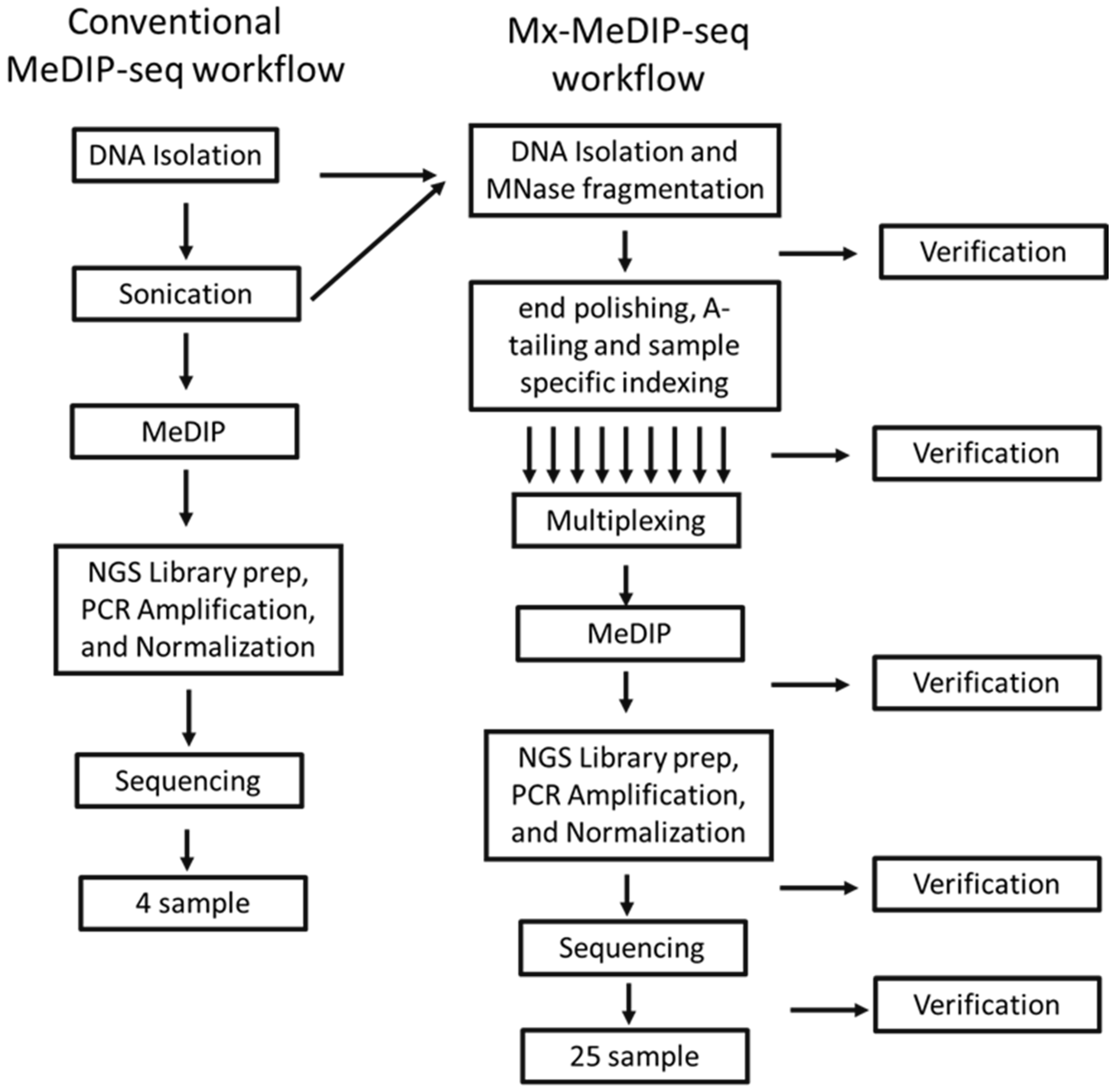
3.1. Development of Mx-MeDIP-Seq
3.2. Sonication vs. Enzyme Fragmentation on Methylation Data
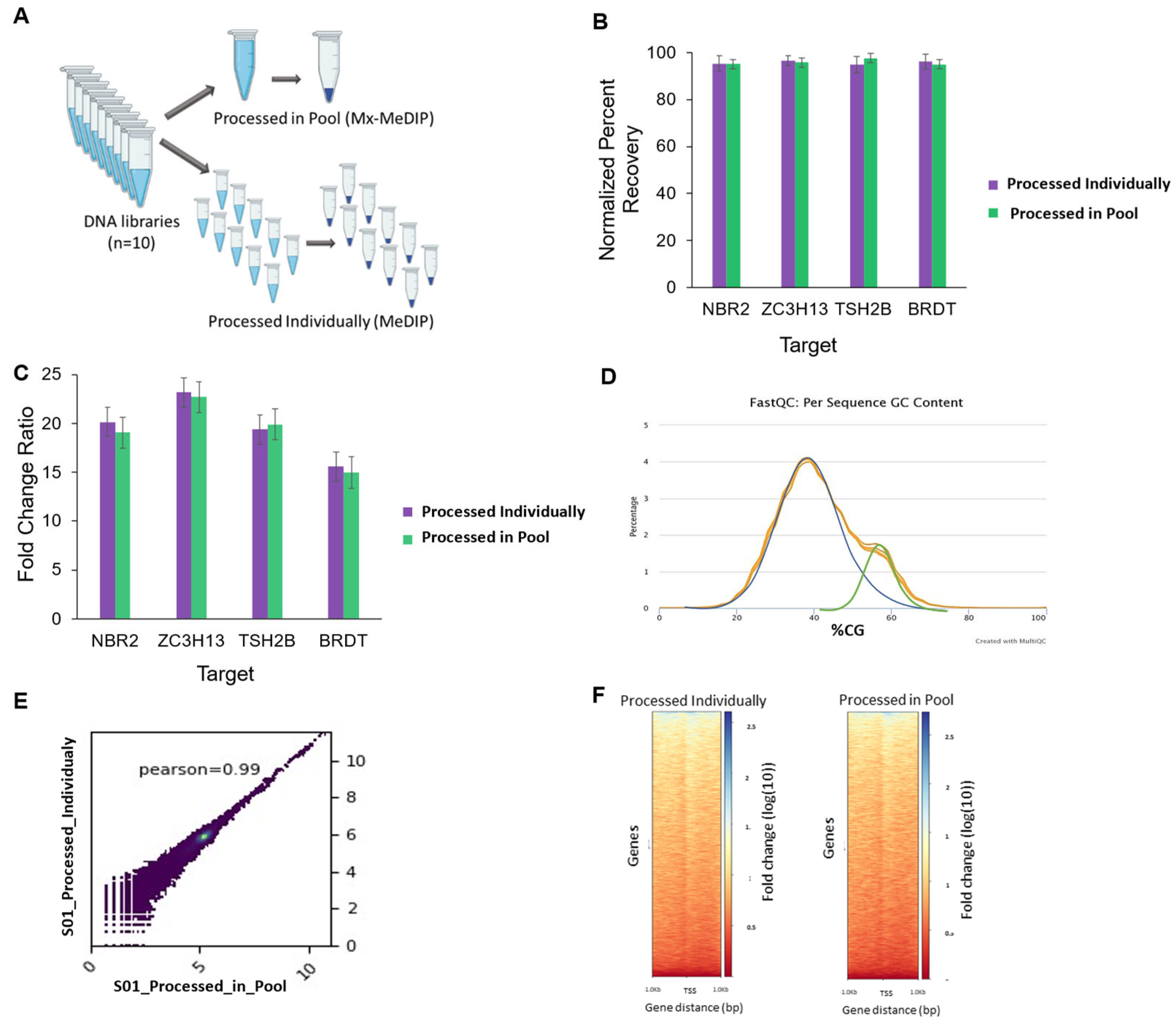
3.3. Individual MeDIP and Mx-MeDIP Resulted in Similar Enrichment and Correlation
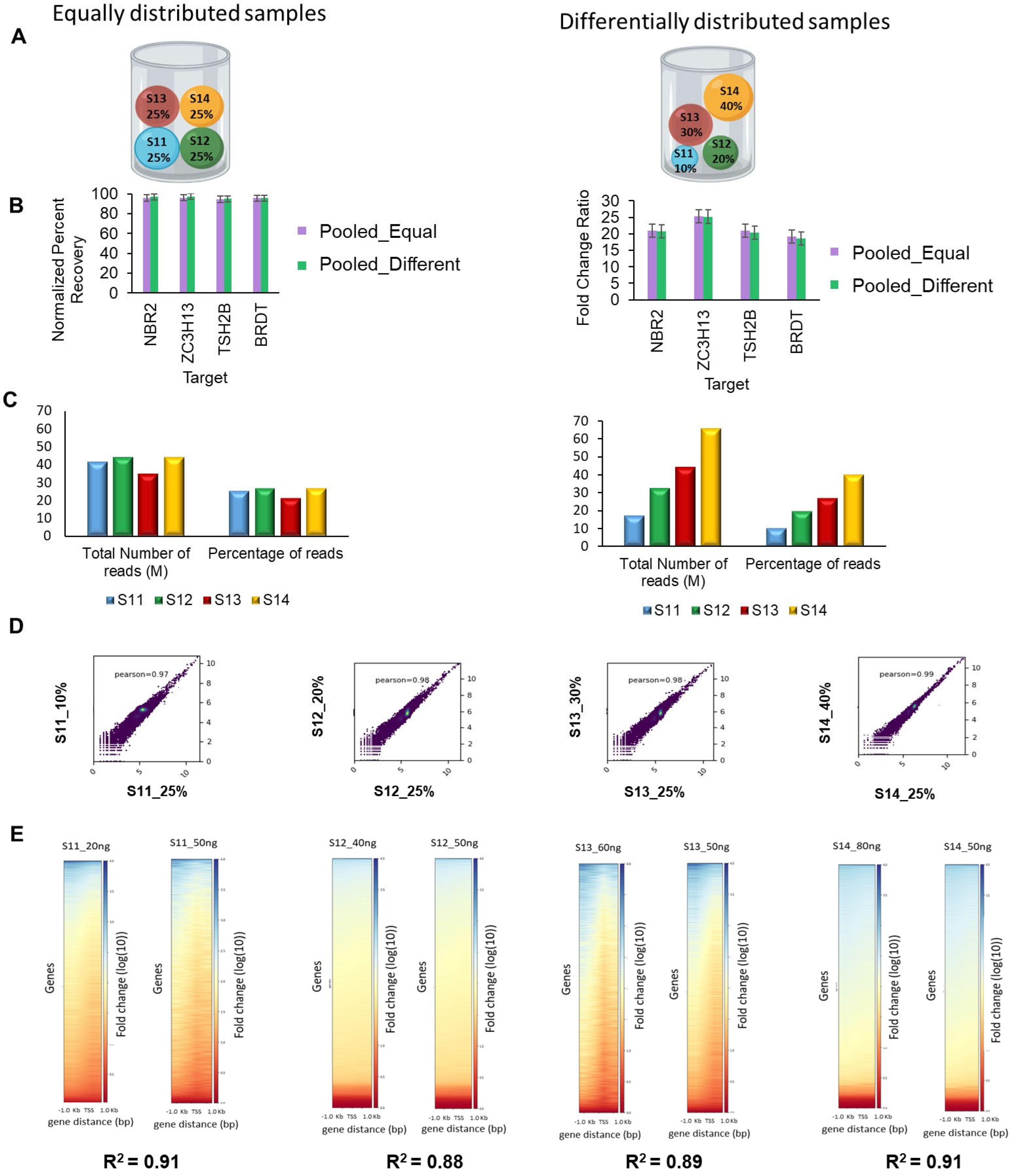
3.4. Optimizing Sample Needs for Mx-MeDIP-Seq
3.5. Mx-MeDIP-Seq Can Be Performed Using Small Amounts of DNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyko, A.; Kovalchuk, I. Genome instability and epigenetic modification—Heritable responses to environmental stress? Curr. Opin. Plant Biol. 2011, 14, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005, 2 (Suppl. S1), S4–S11. [Google Scholar] [CrossRef]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes si-lencing. Oncotarget 2017, 8, 5629–5637. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Zhang, X.; Chen, P.Y.; Bostick, M.; Goll, M.G.; Hetzel, J.; Jain, J.; Strauss, S.H.; Halpern, M.E.; et al. Con-servation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA 2010, 107, 8689–8694. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Fuks, F.; Hurd, P.J.; Wolf, D.; Nan, X.; Bird, A.P.; Kouzarides, T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003, 278, 4035–4040. [Google Scholar] [CrossRef]
- Della Ragione, F.; Filosa, S.; Scalabri, F.; D’Esposito, M. MeCP2 as a genome-wide modulator: The renewal of an old story. Front. Genet. 2012, 3, 181. [Google Scholar] [CrossRef]
- Ludwig, A.K.; Zhang, P.; Hastert, F.D.; Meyer, S.; Rausch, C.; Herce, H.D.; Muller, U.; Lehmkuhl, A.; Hellmann, I.; Trummer, C.; et al. Binding of MBD proteins to DNA blocks Tet1 function thereby modulating transcriptional noise. Nucleic Acids Res. 2017, 45, 2438–2457. [Google Scholar] [CrossRef]
- Chowdhury, B.; Cho, I.H.; Hahn, N.; Irudayaraj, J. Quantification of 5-methylcytosine, 5-hydroxymethylcytosine and 5-carboxylcytosine from the blood of cancer patients by an enzyme-based immunoassay. Anal. Chim. Acta 2014, 852, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.R.; Hubler, S.L.; Nelson, C.D.; Nashold, F.E.; Spanier, J.A.; Hayes, C.E. 1,25-Dihydroxyvitamin D3 increases the me-thionine cycle, CD4+ T cell DNA methylation and Helios+ Foxp3+ T regulatory cells to reverse autoimmune neurodegenerative disease. J. Neuroimmunol. 2018, 324, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B. DNA methylation and autoimmune disease. Clin. Immunol. 2003, 109, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Heyn, H.; Vidal, E.; Ferreira, H.J.; Vizoso, M.; Sayols, S.; Gomez, A.; Moran, S.; Boque-Sastre, R.; Guil, S.; Martinez-Cardus, A.; et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016, 17, 11. [Google Scholar] [CrossRef]
- Cronin, P.; McCarthy, M.J.; Lim, A.S.P.; Salmon, D.P.; Galasko, D.; Masliah, E.; De Jager, P.L.; Bennett, D.A.; Desplats, P. Circadian alterations during early stages of Alzheimer’s disease are associated with aberrant cycles of DNA methylation in BMAL1. Alzheimers Dement. 2017, 13, 689–700. [Google Scholar] [CrossRef]
- Mossman, D.; Scott, R.J. Epimutations, inheritance and causes of aberrant DNA methylation in cancer. Hered. Cancer Clin. Pract. 2006, 4, 75–80. [Google Scholar] [CrossRef]
- Liu, L.; Wylie, R.C.; Andrews, L.G.; Tollefsbol, T.O. Aging, cancer and nutrition: The DNA methylation connection. Mech. Ageing Dev. 2003, 124, 989–998. [Google Scholar] [CrossRef]
- McCabe, D.C.; Caudill, M.A. DNA methylation, genomic silencing, and links to nutrition and cancer. Nutr. Rev. 2005, 63, 183–195. [Google Scholar] [CrossRef]
- Herceg, Z. Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007, 22, 91–103. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.B.; Conway, K.; Wang, X.W.; Bhamra, R.K.; Lin, X.H.; Cohen, M.D.; Annab, L.; Barrett, J.C.; Costa, M. Senescence of nickel-transformed cells by an X chromosome: Possible epigenetic control. Science 1991, 251, 796–799. [Google Scholar] [CrossRef]
- Illingworth, R.; Kerr, A.; Desousa, D.; Jorgensen, H.; Ellis, P.; Stalker, J.; Jackson, D.; Clee, C.; Plumb, R.; Rogers, J.; et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008, 6, e22. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.I.; Huang, T.H.; Yan, P.S. Methylated DNA immunoprecipitation and microarray-based analysis: Detection of DNA methylation in breast cancer cell lines. Methods Mol. Biol. 2009, 590, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Thu, K.L.; Pikor, L.A.; Kennett, J.Y.; Alvarez, C.E.; Lam, W.L. Methylation analysis by DNA immunoprecipitation. J. Cell Physiol. 2010, 222, 522–531. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Ji, L.; Sasaki, T.; Sun, X.; Ma, P.; Lewis, Z.A.; Schmitz, R.J. Methylated DNA is over-represented in whole-genome bisulfite se-quencing data. Front. Genet. 2014, 5, 341. [Google Scholar] [CrossRef]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef]
- Gu, H.; Smith, Z.D.; Bock, C.; Boyle, P.; Gnirke, A.; Meissner, A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011, 6, 468–481. [Google Scholar] [CrossRef]
- Libertini, E.; Heath, S.C.; Hamoudi, R.A.; Gut, M.; Ziller, M.J.; Czyz, A.; Ruotti, V.; Stunnenberg, H.G.; Frontini, M.; Ouwehand, W.H.; et al. Information recovery from low coverage whole-genome bisulfite sequencing. Nat. Commun. 2016, 7, 11306. [Google Scholar] [CrossRef]
- Cao, B.; Luo, H.; Luo, T.; Li, N.; Shao, K.; Wu, K.; Sahu, S.K.; Li, F.; Lin, C. The performance of whole genome bisulfite sequencing on DNBSEQ-Tx platform examined by different library preparation strategies. Heliyon 2023, 9, e16571. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Hall, M.L.; Lee, E.; Kim, S.-C.; Ramesh, N.; Lee, S.H.; Jang, J.-Y.; Bold, R.J.; Ku, J.-L.; Hwang, C.-I. Whole-genome bisulfite sequencing identifies stage- and subtype-specific DNA methylation signatures in pancreatic cancer. iScience 2024, 27, 109414. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Pastor, W.A.; Shen, Y.; Tahiliani, M.; Liu, D.R.; Rao, A. The behaviour of 5-hydroxymethylcytosine in bisulfite se-quencing. PLoS ONE 2010, 5, e8888. [Google Scholar] [CrossRef]
- Cerrizuela, S.; Kaya, O.; Kremer, L.P.M.; Sarvari, A.; Ellinger, T.; Straub, J.; Brunken, J.; Sanz-Morejón, A.; Korkmaz, A.; Mar-tín-Villalba, A. High-throughput scNMT protocol for multiomics profiling of single cells from mouse brain and pancreatic or-ganoids. STAR Protoc. 2022, 3, 101555. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.J.; Argelaguet, R.; Kapourani, C.A.; Stubbs, T.M.; Lee, H.J.; Alda-Catalinas, C.; Krueger, F.; Sanguinetti, G.; Kelsey, G.; Marioni, J.C.; et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018, 9, 781. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, K.; An, Q.; Du, G.; Hu, G.; Xue, J.; Zhu, X.; Wang, C.-Y.; Xue, Z.; Fan, G. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016, 17, 88. [Google Scholar] [CrossRef]
- Kremer, L.P.M.; Braun, M.M.; Ovchinnikova, S.; Küchenhoff, L.; Cerrizuela, S.; Martin-Villalba, A.; Anders, S. Analyzing sin-gle-cell bisulfite sequencing data with MethSCAn. Nat. Methods 2024, 21, 1616–1623. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Drablos, F.; Rye, M.B. DNA methylation data by sequencing: Experimental approaches and recommendations for tools and pipelines for data analysis. Clin. Epigenet. 2019, 11, 193. [Google Scholar] [CrossRef]
- Branco, M.R.; Ficz, G.; Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2011, 13, 7–13. [Google Scholar] [CrossRef]
- Kubiura, M.; Okano, M.; Kimura, H.; Kawamura, F.; Tada, M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosome Res. 2012, 20, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Wardenaar, R.; Liu, H.; Colot, V.; Colome-Tatche, M.; Johannes, F. Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in Arabidopsis. Methods Mol. Biol. 2013, 1067, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Hagmann, J.; Muller, J.; Koenig, D.; Stegle, O.; Borgwardt, K.; Weigel, D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011, 480, 245–249. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes. Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Shu, C.; Zhang, X.; Aouizerat, B.E.; Xu, K. Comparison of methylation capture sequencing and Infinium MethylationEPIC array in peripheral blood mononuclear cells. Epigenet. Chromatin 2020, 13, 51. [Google Scholar] [CrossRef]
- Mohn, F.; Weber, M.; Schubeler, D.; Roloff, T.C. Methylated DNA immunoprecipitation (MeDIP). Methods Mol. Biol. 2009, 507, 55–64. [Google Scholar] [CrossRef]
- Borgel, J.; Guibert, S.; Weber, M. Methylated DNA immunoprecipitation (MeDIP) from low amounts of cells. Methods Mol. Biol. 2012, 925, 149–158. [Google Scholar] [CrossRef]
- Taiwo, O.; Wilson, G.A.; Morris, T.; Seisenberger, S.; Reik, W.; Pearce, D.; Beck, S.; Butcher, L.M. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat. Protoc. 2012, 7, 617–636. [Google Scholar] [CrossRef]
- Zhao, M.T.; Whyte, J.J.; Hopkins, G.M.; Kirk, M.D.; Prather, R.S. Methylated DNA immunoprecipitation and high-throughput sequencing (MeDIP-seq) using low amounts of genomic DNA. Cell. Reprogram. 2014, 16, 175–184. [Google Scholar] [CrossRef]
- Thu, K.L.; Vucic, E.A.; Kennett, J.Y.; Heryet, C.; Brown, C.J.; Lam, W.L.; Wilson, I.M. Methylated DNA immunoprecipitation. J. Vis. Exp. 2009, 23, e935. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Hein, A.; Damiani, R. Nuclear DNA Content Varies with Cell Size across Human Cell Types. Cold Spring Harb. Perspect. Biol. 2015, 7, a019091. [Google Scholar] [CrossRef] [PubMed]
- Brind’Amour, J.; Liu, S.; Hudson, M.; Chen, C.; Karimi, M.M.; Lorincz, M.C. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 2015, 6, 6033. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Cao, Z.; Lu, C. Microfluidic MeDIP-seq for low-input methylomic analysis of mammary tumorigenesis in mice. Analyst 2019, 144, 1904–1915. [Google Scholar] [CrossRef]
- Auriol, E.; Billard, L.M.; Magdinier, F.; Dante, R. Specific binding of the methyl binding domain protein 2 at the BRCA1-NBR2 locus. Nucleic Acids Res. 2005, 33, 4243–4254. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, D.; Zhang, B.; Xing, X.; Wang, T. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods 2015, 72, 29–40. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Bronner, I.F.; Quail, M.A.; Turner, D.J.; Swerdlow, H. Improved Protocols for Illumina Sequencing. Curr. Protoc. Hum. Genet. 2014, 79, 18.2.1–18.2.42. [Google Scholar] [CrossRef]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schübeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Lisanti, S.; von Zglinicki, T.; Mathers, J.C. Standardization and quality controls for the methylated DNA immunoprecipitation technique. Epigenetics 2012, 7, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.; Palta, P.; Joyce, C.J.; Scott, C.; Grundberg, E.; Deloukas, P.; Palotie, A.; Coffey, A.J. A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation450 BeadChip((R)) for methylome profiling. PLoS ONE 2012, 7, e50233. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, C.; Kato, A.; Tempel, W.; Abreu, J.G.; Bian, C.; Hu, Y.; Hu, D.; Zhao, B.; Cerovina, T.; et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 2012, 151, 1200–1213. [Google Scholar] [CrossRef]
- Kim, M.C.; Kim, N.Y.; Seo, Y.R.; Kim, Y. An Integrated Analysis of the Genome-Wide Profiles of DNA Methylation and mRNA Expression Defining the Side Population of a Human Malignant Mesothelioma Cell Line. J. Cancer 2016, 7, 1668–1679. [Google Scholar] [CrossRef]
- Serre, D.; Lee, B.H.; Ting, A.H. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010, 38, 391–399. [Google Scholar] [CrossRef]
- Butcher, L.M.; Beck, S. AutoMeDIP-seq: A high-throughput, whole genome, DNA methylation assay. Methods 2010, 52, 223–231. [Google Scholar] [CrossRef]
- Shen, H.; Qiu, C.; Li, J.; Tian, Q.; Deng, H.W. Characterization of the DNA methylome and its interindividual variation in human peripheral blood monocytes. Epigenomics 2013, 5, 255–269. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Ballestar, E.; Esteller, M. Methyl-DNA immunoprecipitation (MeDIP): Hunting down the DNA methylome. Biotechniques 2008, 44, 35–43. [Google Scholar] [CrossRef]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenet. 2020, 12, 156. [Google Scholar] [CrossRef]
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Kandi, V.; Vadakedath, S. Effect of DNA Methylation in Various Diseases and the Probable Protective Role of Nutrition: A Mini-Review. Cureus 2015, 7, e309. [Google Scholar] [CrossRef]
- Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef]
- Hsu, H.K.; Weng, Y.I.; Hsu, P.Y.; Huang, T.H.; Huang, Y.W. Detection of DNA methylation by MeDIP and MBDCap assays: An overview of techniques. Methods Mol. Biol. 2014, 1105, 61–70. [Google Scholar] [CrossRef]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Aberg, K.A.; Chan, R.F.; Xie, L.; Shabalin, A.A.; van den Oord, E. Methyl-CpG-Binding Domain Sequencing: MBD-seq. Methods Mol. Biol. 2018, 1708, 171–189. [Google Scholar] [CrossRef]
- Jørgensen, H.F.; Adie, K.; Chaubert, P.; Bird, A.P. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006, 34, e96. [Google Scholar] [CrossRef]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Collings, C.K.; Anderson, J.N. Links between DNA methylation and nucleosome occupancy in the human genome. Epigenet. Chromatin 2017, 10, 18. [Google Scholar] [CrossRef]
- Fenouil, R.; Cauchy, P.; Koch, F.; Descostes, N.; Cabeza, J.Z.; Innocenti, C.; Ferrier, P.; Spicuglia, S.; Gut, M.; Gut, I.; et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012, 22, 2399–2408. [Google Scholar] [CrossRef] [PubMed]
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 2016, 7, 11485. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Liz, J.; Nogales, V.; Setién, F.; Villanueva, A.; Esteller, M. DNA methylation determines nucleosome occupancy in the 5′-CpG islands of tumor suppressor genes. Oncogene 2013, 32, 5421–5428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, W.; Jiang, J. Genome-Wide Nucleosome Occupancy and Positioning and Their Impact on Gene Expression and Evolution in Plants. Plant Physiol. 2015, 168, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, S.; Uffenorde, J.; Gimm, O.; Hosseinpour Feizi, M.A.; Miemczyk, S.; Coutinho, L.L.; Jensen, P.; Guerrero-Bosagna, C.; Pértille, F. GBS-MeDIP: A protocol for parallel identification of genetic and epigenetic variation in the same reduced fraction of genomes across individuals. STAR Protoc. 2022, 3, 101202. [Google Scholar] [CrossRef]
- Pértille, F.; Alvarez-Rodriguez, M.; da Silva, A.N.; Barranco, I.; Roca, J.; Guerrero-Bosagna, C.; Rodriguez-Martinez, H. Sperm Methylome Profiling Can Discern Fertility Levels in the Porcine Biomedical Model. Int. J. Mol. Sci. 2021, 22, 2679. [Google Scholar] [CrossRef]
- Nunes, J.R.S.; Pértille, F.; Andrade, S.C.S.; Perazza, C.A.; Villela, P.M.S.; Almeida-Val, V.M.F.; Gao, Z.-X.; Coutinho, L.L.; Hilsdorf, A.W.S. Genome-wide association study reveals genes associated with the absence of intermuscular bones in tambaqui (Colossoma macropomum). Anim. Genet. 2020, 51, 899–909. [Google Scholar] [CrossRef]
- Pértille, F.; Da Silva, V.H.; Johansson, A.M.; Lindström, T.; Wright, D.; Coutinho, L.L.; Jensen, P.; Guerrero-Bosagna, C. Mutation dynamics of CpG dinucleotides during a recent event of vertebrate diversification. Epigenetics 2019, 14, 685–707. [Google Scholar] [CrossRef]



| Reagents | Volume (mL) |
|---|---|
| 100 mM Tris-HCl (1 M, pH 8.0) | 20 |
| 300 mM NaCl (5 M) | 12 |
| 2% Triton X-100 (25%) | 16 |
| 0.2% sodium deoxycholate (DOC, 12.5%) | 3.2 |
| 10 mM CaCl2 (1 M) | 2 |
| 10 mM sodium butyrate (800 mM) | 2.5 |
| H2O | 144.3 |
| 1 U/µL MNase stock solution | 4 |
| Total volume | 200 |
| Target Name | Oligonucleotide Sequence |
|---|---|
| Human Testis/Sperm-Specific Histone H2B (TSH2B) | Forward: CAGACATCTCCTCGCATCAA Reverse: GGAGGATGAAAGATGCGGTA |
| Bromodomain Testis Associated (BRDT) | Forward: CCCTTTGGCCTTACCAACTT Reverse: GCCCTCCCTTGAAGAAAAAC |
| Zinc Finger CCCH-Type Containing 13 (ZC3H13) | Forward: TCTCGGTCCACTCGTGATG Reverse: CCGGGATTCTTCTGGATATG |
| Neighbor of BRCA1 Gene 2 (NBR2) | Forward: TGTTATTTTTCGGGTTCAGCTT Reverse: GATTGGCTCTTACCACTTGTCC |
| Glyceraldehyde-3-Phosphate Dehydrogenase Peptidyl-Cysteine S-Nitrosylase (GAPDH) | Forward: - TCGACAGTCAGCCGCATCT Reverse: CTAGCCTCCCGGGTTTCTCT |
| Samples | Shifted Peak in %CG (Processed Individually) | Shifted Peak in %CG (Processed in Pool) | Pearson Correlation Coefficient (of BAM Files Between Two Groups) | Pearson Correlation Coefficient (of Peaks Called on CpG Island) |
|---|---|---|---|---|
| S1 | 59 | 59 | 0.99 | 0.998 |
| S2 | 60 | 59 | 0.99 | 0.996 |
| S3 | 59 | 59 | 0.90 | 0.996 |
| S4 | 59 | 60 | 0.89 | 0.994 |
| S5 | 59 | 60 | 0.89 | 0.998 |
| S6 | 58 | 59 | 0.88 | 0.996 |
| S7 | 60 | 59 | 0.87 | 0.999 |
| S8 | 59 | 58 | 0.87 | 0.995 |
| S9 | 58 | 59 | 0.89 | 0.993 |
| S10 | 60 | 59 | 0.99 | 0.993 |
| Mean | 59.10 | 59.10 | 0.91 | 0.995 |
| standard deviation (SD) | 0.70 | 0.53 | 0.05 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ridha, I.; Xu, C.; Zhang, Y.; Chung, Y.; Park, J.G.; LaBaer, J.; Murugan, V. Multiplexed Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq) to Study DNA Methylation Using Low Amounts of DNA. DNA 2024, 4, 397-416. https://doi.org/10.3390/dna4040028
Ridha I, Xu C, Zhang Y, Chung Y, Park JG, LaBaer J, Murugan V. Multiplexed Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq) to Study DNA Methylation Using Low Amounts of DNA. DNA. 2024; 4(4):397-416. https://doi.org/10.3390/dna4040028
Chicago/Turabian StyleRidha, Inam, Chenxi Xu, Yining Zhang, Yunro Chung, Jin G Park, Joshua LaBaer, and Vel Murugan. 2024. "Multiplexed Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq) to Study DNA Methylation Using Low Amounts of DNA" DNA 4, no. 4: 397-416. https://doi.org/10.3390/dna4040028
APA StyleRidha, I., Xu, C., Zhang, Y., Chung, Y., Park, J. G., LaBaer, J., & Murugan, V. (2024). Multiplexed Methylated DNA Immunoprecipitation Sequencing (Mx-MeDIP-Seq) to Study DNA Methylation Using Low Amounts of DNA. DNA, 4(4), 397-416. https://doi.org/10.3390/dna4040028








