Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
2.2. Samples for Molecular Study
2.3. RNA Extraction and Quantification
2.4. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
2.5. Identification of Genetic Biomarkers
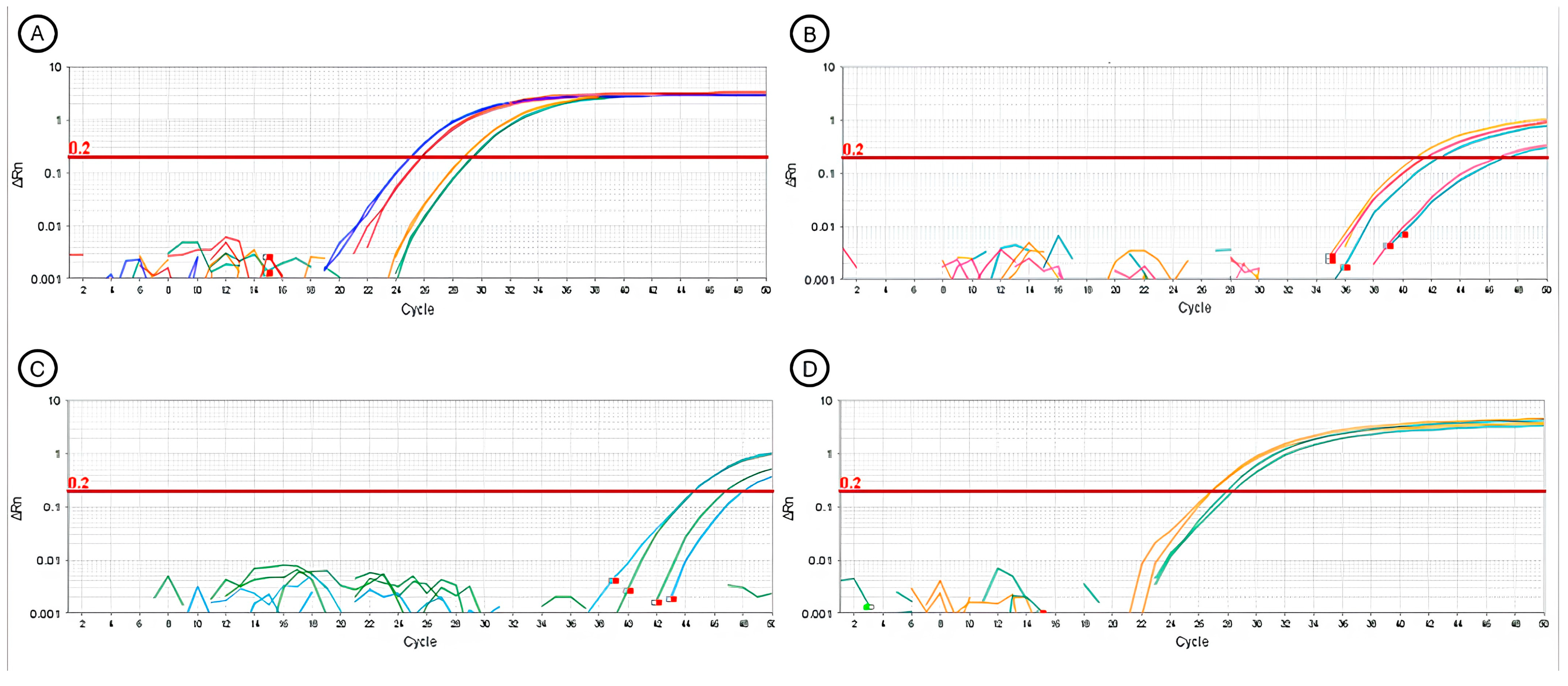
2.5.1. Nested Polymerase Chain Reaction (Nested-PCR)
2.5.2. Real-Time Polymerase Chain Reaction (RT-qPCR)
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Staudt, L.M. Molecular Diagnosis of the Hematologic Cancers. N. Engl. J. Med. 2003, 348, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; O’Brien, S.; Konopleva, M.; Kantarjian, H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer 2015, 121, 2517–2528. [Google Scholar] [CrossRef] [PubMed]
- Béné, M.C.; Grimwade, D.; Haferlach, C.; Haferlach, T.; Zini, G. Leukemia diagnosis: Today and tomorrow. Eur. J. Haematol. 2015, 95, 365–373. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Noronha, E.P.; Marinho, H.T.; Thomaz, E.B.A.F.; Silva, C.A.; Veras, G.L.R.; Oliveira, R.A.G. Caracterização imunofenotípica das leucemias agudas em um centro oncológico de referência público no Maranhão, Nordeste do Brasil. Sao Paulo Med. J. 2011, 129, 392–401. [Google Scholar] [CrossRef]
- Farias, M.G.; Castro, S.M.d. Diagnóstico laboratorial das leucemias linfóides agudas. J. Bras. Patol. e Med. Lab. 2004, 40, 91–98. [Google Scholar] [CrossRef][Green Version]
- Da Silva, G.C.; Pilger, D.A.; De Castro, S.M.; Wagner, S.C. Diagnóstico laboratorial das leucemias mielóides agudas. J. Bras. Patol. e Med. Lab. 2006, 42, 77–84. [Google Scholar] [CrossRef][Green Version]
- Rodrigues, C.A.; Gonçalves, M.V.; Ikoma, M.R.V.; Lorand-Metze, I.; Pereira, A.D.; Farias, D.L.C.d.; Chauffaille, M.d.L.L.F.; Schaffel, R.; Ribeiro, E.F.O.; Rocha, T.S.d.; et al. Diagnosis and treatment of chronic lymphocytic leukemia: Recommendations from the Brazilian Group of Chronic Lymphocytic Leukemia. Rev. Bras. Hematol. Hemoter. 2016, 38, 346–357. [Google Scholar] [CrossRef]
- White, T.J.; Arnheim, N.; Erlich, A.H. The polymerase chain reaction. Tech. Focus 1989, 5, 185–189. [Google Scholar] [CrossRef]
- Joshi, M.; Deshpande, J.D. Polymerase Chain Reaction: Methods, Principles and Application. Int. J. Biomed. Res. 2011, 2, 81–97. [Google Scholar] [CrossRef]
- Schochetman, G.; Ou, C.-Y.; Jones, W.K. Polymerase Chain Reaction. J. Infect. Dis. 1988, 158, 1154–1157. Available online: https://www.jstor.org/stable/30137034 (accessed on 15 January 2023). [CrossRef] [PubMed]
- Erlich, H.A. Polymerase chain reaction. J. Clin. Immunol. 1989, 9, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Kadri, K. Polymerase Chain Reaction (PCR): Principle and Applications. Synthetic Biology—New in Terdisciplinary Science. IntechOpen 2019, 19, 138–142. [Google Scholar]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR past, present and future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Wang, J.Y.J. The Capable ABL: What Is Its Biological Function? Mol. Cell. Biol. 2014, 34, 1188–1197. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Anthony, B.; Link, D.C. Regulation of Hematopoietic Stem Cells by Bone Marrow Stromal Cells. Trends Immunol. 2014, 35, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Sule, W.F.; Oluwayelu, D.O. Real-time RT-PCR for COVID-19 diagnosis: Challenges and prospects. Pan Afr. Med. J. 2020, 35, 121. [Google Scholar] [CrossRef]
- Van Dongen, J.; Macintyre, E.; Gabert, J.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Acute leukemia for detection of minimal residual disease Report of the BIOMED-I Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef]
- Pessoa, F.M.C.d.P.; Viana, V.B.d.J.; de Oliveira, M.B.; Nogueira, B.M.D.; Ribeiro, R.M.; Oliveira, D.d.S.; Lopes, G.S.; Vieira, R.P.G.; de Moraes Filho, M.O.; de Moraes, M.E.A.; et al. Validation of Endogenous Control Genes by Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction for Acute Leukemia Gene Expression Studies. Genes 2024, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Climent, J.A. Molecular cytogenetics of childhood hematological malignancies. Leukemia 1997, 11, 1999–2021. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dongen, J.J.M.; van Velden, V.H.J.; van der Brüggemann, M.; Orfao, A. Minimal residual disease diagnostics in acute lymphoblastic leukemia: Need for sensitive, fast, and standardized technologies. Blood 2015, 125, 3996–4009. [Google Scholar] [CrossRef]
- González, Á.; Hierro, N.; Poblet, M.; Mas, A.; Guillamón, J.M. Enumeration and detection of acetic acid bacteria by real-time PCR and nested PCR. FEMS Microbiol. Lett. 2006, 254, 123–128. [Google Scholar] [CrossRef]
- Gleißner, B.; Rieder, H.; Thiel, E.; Fonatsch, C.; Janssen, L.A.J.; Heinze, B.; Janssen, J.W.G.; Schoch, C.; Goekbuget, N.; Maurer, J.; et al. Prospective BCR-ABL analysis by polymerase chain reaction (RT-PCR) in adult acute B-lineage lymphoblastic leukemia: Reliability of RT-nested-PCR and comparison to cytogenetic data. Leukemia 2001, 15, 1834–1840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Janssen, J.W.; Fonatsch, C.; Ludwig, W.D.; Rieder, H.; Maurer, J.; Bartram, C.R. Polymerase chain reaction analysis of BCR-ABL sequences in adult Philadelphia chromosome-negative acute lymphoblastic leukemia patients. Leukemia 1992, 6, 463–464. [Google Scholar]
- Mason, J.; Griffiths, M. Molecular diagnosis of leukemia. Expert Rev. Mol. Diagn. 2012, 12, 511–526. [Google Scholar] [CrossRef]
- Bacher, U.; Schnittger, S.; Haferlach, C.; Haferlach, T. Molecular diagnostics in acute leukemias. Clin. Chem. Lab. Med. 2009, 47, 1333–1341. [Google Scholar] [CrossRef]
- van der Velden, V.H.J.; Hochhaus, A.; Cazzaniga, G.; Szczepanski, T.; Gabert, J.; van Dongen, J.J.M. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: Principles, approaches, and laboratory aspects. Leukemia 2003, 17, 1013–1034. [Google Scholar] [CrossRef]
- Van Der Velden, V.H.J.; Van Dongen, J.J.M. MRD Detection in Acute Lymphoblastic Leukemia Patients Using Ig/TCR Gene Rearrangements as Targets for Real-Time Quantitative PCR; Springer: Berlin/Heidelberg, Germany, 2009; Volume 538, ISBN 9781588299895. [Google Scholar]
- Scott, S.; Travis, D.; Whitby, L.; Bainbridge, J.; Cross, N.C.P.; Barnett, D. Measurement of BCR-ABL1 by RT-qPCR in chronic myeloid leukaemia: Findings from an International EQA Programme. Br. J. Haematol. 2017, 177, 414–422. [Google Scholar] [CrossRef]
- Watt, C.D.; Bagg, A. Molecular diagnosis of acute myeloid leukemia. Expert Rev. Mol. Diagn. 2010, 10, 993–1012. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, J.M.; Gomes, A.d.; de Oliveira, E.M.; Marques, G.; Fonseca, R.d.; Frota, S.d.; Aquino, P.E. DIAGNÓSTICO MOLECULAR DAS LEUCEMIAS. Rev. Arq. Científicos 2022, 5, 20–34. [Google Scholar]
- Chauhan, R.; Sazawal, S.; Pati, H.P. Laboratory Monitoring of Chronic Myeloid Leukemia in Patients on Tyrosine Kinase Inhibitors. Indian J. Hematol. Blood Transfus. 2018, 34, 197–203. [Google Scholar] [CrossRef]
- Lesieur, A.; Thomas, X.; Nibourel, O.; Boissel, N.; Fenwarth, L.; de Botton, S.; Fournier, E.; Celli-Lebras, K.; Raffoux, E.; Recher, C.; et al. Minimal residual disease monitoring in acute myeloid leukemia with non-A/B/D NPM1 mutations by digital polymerase chain reaction: Feasibility and clinical use. Haematologica 2021, 106, 1767–1769. [Google Scholar] [CrossRef]
- Pongers-Willemse, M.J.; Verhagen, O.J.H.M.; Tibbe, G.J.M.; Wijkhuijs, A.J.M.; De Haas, V.; Roovers, E.; Van Der School, C.E.; Van Dongen, J.J.M. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998, 12, 2006–2014. [Google Scholar] [CrossRef]
- Press, R.D.; Kamel-Reid, S.; Ang, D. BCR-ABL1 RT-qPCR for monitoring the molecular response to tyrosine kinase inhibitors in chronic myeloid leukemia. J. Mol. Diagn. 2013, 15, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Arthur, D.C.; Berger, R.; Golomb, H.M.; Swansbury, G.J.; Reeves, B.R.; Alimena, G.; Van Den Berghe, H.; Bloomfield, C.D.; de a Chapelle, A.; Dewald, G.W.; et al. The Clinical Significance of Karyotype in Acute Myelogenous Leukemia. Cancer Genet. Cytogenet. 1989, 40, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Mrozek, K.; Heeremab, N.A.; Bloomfielda, C.D. Cytogenetics in acute leukemia. Blood Rev. 2004, 18, 115–136. [Google Scholar] [CrossRef]
- Deepak, S.A.; Kottapalli, K.R.; Rakwal, R.; Oros, G.; Rangappa, K.S.; Iwahashi, H.Y.; Masuo, Y.; Agrawal, G.K. Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr. Genom. 2007, 8, 234–251. [Google Scholar] [CrossRef]
- Lin, M.T.; Tseng, L.H.; Rich, R.G.; Hafez, M.J.; Harada, S.; Murphy, K.M.; Eshleman, J.R.; Gocke, C.D. PCR, A Simple Method to Detect Translocations and Insertion/Deletion Mutations. J. Mol. Diagn. 2011, 13, 1. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Döhner, K.; Döhner, H. Molecular characterization of acute myeloid leukemia. Haematologica 2008, 93, 7. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S. Current findings for recurring mutations in acute myeloid leukemia. J. Hematol. Oncol. 2011, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Prassek, V.V.; Rothenberg-Thurley, M.; Sauerland, M.C.; Herold, T.; Janke, H.; Ksienzyk, B.; Konstandin, N.P.; Goerlich, D.; Krug, U.; Faldum, A. Genetics of acute myeloid leukemia in the elderly: Mutation spectrum and clinical impact in intensively treated patients aged 75 years or older. Haematologica 2018, 103, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Kamaneh, E.A.; Asenjan, K.S.; Akbari, A.M.; Laleh, P.A.; Chavoshi, H.; Ziaei, J.E.; Nikanfar, A.; Kermani, I.A.; Esfahani, A. Characterization of Common Chromosomal Translocations and Their Frequencies in Acute Myeloid Leukemia Patients of Northwest Iran. Cell J. 2016, 18, 37–45. [Google Scholar]
- Avet-loiseau, H. Fish Analysis at Diagnosis in Acute Lymphoblastic Leukemia. Leuk. Lymphoma 1999, 33, 441–449. [Google Scholar] [CrossRef]
- Gasparovic, L.; Weiler, S.; Higi, L.; Burden, A.M. Incidence of differentiation syndrome associated with treatment regimens in acute myeloid leukemia: A systematic review of the literature. J. Clin. Med. 2020, 9, 3342. [Google Scholar] [CrossRef]
- Grignani, F.; Ferrucci, P.F.; Testa, U.; Talamo, G.; Fagioli, M.; Alcalay, M.; Mencarelli, A.; Grignani, F.; Peschle, C.; Nicoletti, I.; et al. The acute promyelocytic leukemia-specific PML-RARα fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 1993, 74, 423–431. [Google Scholar] [CrossRef]
- Iaccarino, L.; Divona, M.; Ottone, T.; Cicconi, L.; Lavorgna, S.; Ciardi, C.; Alfonso, V.; Travaglini, S.; Facchini, L.; Cimino, G.; et al. Identification and monitoring of atypical PML/RARA fusion transcripts in acute promyelocytic leukemia. Genes Chromosom. Cancer 2019, 58, 60–65. [Google Scholar] [CrossRef]
- Tse, K.F.; Novelli, E.; Civin, C.I.; Bohmer, F.D.; Small, D. Inhibition of FLT3-mediated transformation by use of a tyrosine kinase inhibitor. Leukemia 2001, 15, 1001–1010. [Google Scholar] [CrossRef]
- Swart, L.E.; Heidenreich, O. The RUNX1/RUNX1T1 network: Translating insights into therapeutic options. Exp. Hematol. 2021, 94, 1–10. [Google Scholar] [CrossRef]
- Schwind, S.; Edwards, C.G.; Nicolet, D.; Mrózek, K.; Maharry, K.; Wu, Y.Z.; Paschka, P.; Eisfeld, A.K.; Hoellerbauer, P.; Becker, H.; et al. Inv(16)/t(16;16) acute myeloid leukemia with non-type A CBFB-MYH11 fusions associate with distinct clinical and genetic features and lack KIT mutations. Blood 2013, 121, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.E.; Luthra, R.; Jabbour, E.; Patel, K.P.; Khoury, J.D.; Tang, Z.; Alvarez, H.; Mallampati, S.; Garcia-Manero, G.; Montalban-Bravo, G.; et al. Incidental identification of inv(16)(p13.1q22)/CBFB-MYH11 variant transcript in a patient with therapy-related acute myeloid leukemia by routine leukemia translocation panel screen: Implications for diagnosis and therapy. Cold Spring Harb. Mol. Case Stud. 2021, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, F.M.C.d.P.; Machado, C.B.; Barreto, I.V.; Sampaio, G.F.; Oliveira, D.d.S.; Ribeiro, R.M.; Lopes, G.S.; de Moraes, M.E.A.; de Moraes Filho, M.O.; de Souza, L.E.B.; et al. Association between Immunophenotypic Parameters and Molecular Alterations in Acute Myeloid Leukemia. Biomedicines 2023, 11, 1098. [Google Scholar] [CrossRef]
- Meshinchi, S.; Alonzo, T.A.; Stirewalt, D.L.; Zwaan, M.; Zimmerman, M.; Reinhardt, D.; Kaspers, G.J.L.; Heerema, N.A.; Gerbing, R.; Lange, B.J.; et al. Clinical implications of FLT3 mutations in pediatric AML. Blood 2006, 108, 3654–3661. [Google Scholar] [CrossRef]
- Grinev, V.V.; Barneh, F.; Ilyushonak, I.M.; Nakjang, S.; Smink, J.; van Oort, A.; Clough, R.; Seyani, M.; McNeill, H.; Reza, M.; et al. RUNX1/RUNX1T1 mediates alternative splicing and reorganises the transcriptional landscape in leukemia. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Kindler, T.; Lipka, D.B.; Fischer, T. FLT3 as a therapeutic target in AML: Still challenging after all these years. Blood 2010, 116, 5089–5102. [Google Scholar] [CrossRef]
- Diakos, C.; Xiao, Y.; Zheng, S.; Kager, L.; Dworzak, M.; Wiemels, J.L. Direct and indirect targets of the E2A-PBX1 leukemia-specific fusion protein. PLoS ONE 2014, 9, e87602. [Google Scholar] [CrossRef]
- Faderl, S.; Kantarjian, H.M.; Thomas, D.A.; Cortes, J.; Giles, F.; Pierce, S.; Albitar, M.; Estrov, Z. Outcome of Philadelphia Chromosome-Positive Adult Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2000, 36, 263–273. [Google Scholar] [CrossRef]
- Felice, M.S.; Gallego, M.S.; Alonso, C.N.; Alfaro, E.M.; Guitter, M.R.; Bernasconi, A.R.; Rubio, P.L.; Zubizarreta, P.A.; Rossi, J.G. Prognostic impact of t(1;19)/ TCF3-PBX1 in childhood acute lymphoblastic leukemia in the context of Berlin-Frankfurt-Münster-based protocols. Leuk. Lymphoma 2011, 52, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Gestrich, C.K.; Lancy, S.J.D.; Kresak, A.; Sinno, M.G.; Yalley, A.; Pateva, I.; Meyerson, H.; Shetty, S.; Jr, K.A.O. Mucin 4 protein is expressed in B-acute lymphoblastic leukemia and is restricted to BCR::ABL1-positive and BCR::ABL-like subtypes. Hum. Pathol. 2023, 136, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Kager, L.; Lion, T.; Attarbaschi, A.; Koenig, M.; Strehl, S.; Haas, O.A.; Dworzak, M.N.; Schrappe, M.; Gadner, H.; Mann, G. Incidence and outcome of TCF3-PBX1-positive acute lymphoblastic leukemia in Austrian children. Haematologica 2007, 92, 1561–1564. [Google Scholar] [CrossRef]
- Maino, E.; Sancetta, R.; Viero, P.; Imbergamo, S.; Scattolin, A.M.; Vespignani, M.; Bassan, R. Current and future management of Ph/BCR-ABL positive ALL. Expert Rev. Anticancer Ther. 2014, 14, 723–740. [Google Scholar] [CrossRef]
- Reckel, S.; Hamelin, R.; Georgeon, S.; Armand, F.; Jolliet, Q.; Chiappe, D.; Moniatte, M.; Hantschel, O. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia 2017, 31, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Grote, D.; Olmos, A.; Kofoet, A.; Tuset, J.J.; Bertolini, E.; Cambra, M. Specific and Sensitive Detection of Phytophthora nicotianae By Simple and Nested-PCR. Eur. J. Plant Pathol. 2002, 108, 197–207. [Google Scholar]
- Lin, C.; Ying, F.; Lai, Y.; Li, X.; Xue, X.; Zhou, T.; Hu, D. Use of nested PCR for the detection of trichomonads in bronchoalveolar lavage fluid. BMC Infect. Dis. 2019, 19, 1–6. [Google Scholar] [CrossRef]
- Lan, J.; Ossewaarde, J.M.; Walboomers, J.M.M.; Meijer, C.J.L.M.; Van den Brule, A.J.C. Improved PCR sensitivity for direct genotyping of Chlamydia trachomatis serovars by using a nested PCR. J. Clin. Microbiol. 1994, 32, 528–530. [Google Scholar] [CrossRef]
- Strom, C.M.; Rechitsky, S. Use of nested PCR to identify charred human remains and minute amounts of blood. J. Forensic Sci. 1998, 43, 696–700. [Google Scholar] [CrossRef]
- Alvarez-Martínez, M.J.; Miró, J.M.; Valls, M.E.; Moreno, A.; Rivas, P.V.; Solé, M.; Benito, N.; Domingo, P.; Muñoz, C.; Rivera, E.; et al. Sensitivity and specificity of nested and real-time PCR for the detection of Pneumocystis jiroveci in clinical specimens. Diagn. Microbiol. Infect. Dis. 2006, 56, 153–160. [Google Scholar] [CrossRef]
- Hafez, H.M.; Hauck, R.; Lüschow, D.; McDougald, L. Comparison of the specificity and sensitivity of PCR, nested PCR, and real-time PCR for the diagnosis of histomoniasis. Avian Dis. 2005, 49, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Kortela, E.; Kirjavainen, V.; Ahava, M.J.; Jokiranta, S.T.; But, A.; Lindahl, A.; Jääskeläinen, A.E.; Jääskeläinen, A.J.; Järvinen, A.; Jokela, P.; et al. Real-life clinical sensitivity of SARS-CoV-2 RT-PCR test in symptomatic patients. PLoS ONE 2021, 16, 1–19. [Google Scholar] [CrossRef]
- Gregory, L.; Lara, M.C.C.S.H.; Hasegawa, M.Y.; Castro, R.S.; Rodrigues, J.N.M.; Araújo, J.; Keller, L.W.; Silva, L.K.F.; Durigon, E.L. Detecção Do Vírus Da Artrite Encefalite Caprina No Sêmen Através Das Técnicas De Pcr E Nested-Pcr. Arq. Inst. Biol. 2011, 78, 599–603. [Google Scholar] [CrossRef]
- Šeligová, B.; Lukáč, Ľ.; Bábelová, M.; Vávrová, S.; Sulo, P. Diagnostic reliability of nested PCR depends on the primer design and threshold abundance of Helicobacter pylori in biopsy, stool, and saliva samples. Helicobacter 2020, 25, e12680. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Fadrosh, D.; Goedert, J.J.; Ravel, J.; Goldstein, A.M. Nested PCR biases in interpreting microbial community structure in 16S rRNA gene sequence datasets. PLoS ONE 2015, 10, e0132253. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, G.H.; Gardner, S.N. Predicting the sensitivity and specificity of published real-time PCR assays. Ann. Clin. Microbiol. Antimicrob. 2008, 7, 18. [Google Scholar] [CrossRef]
- Trovato, L.; Domina, M.; Calvo, M.; De Pasquale, R.; Scalia, G.; Oliveri, S. Use of real time multiplex PCR for the diagnosis of dermatophytes onychomycosis in patients with empirical antifungal treatments. J. Infect. Public Health 2022, 15, 539–544. [Google Scholar] [CrossRef]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Am. J. Physiol.—Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef]
- Klein, D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- da Costa Lima, M.S.; Zorzenon, D.C.R.; Dorval, M.E.C.; Pontes, E.R.J.C.; Oshiro, E.T.; Cunha, R.; Andreotti, R.; de Fatima Cepa Matos, M. Sensitivity of PCR and real-time PCR for the diagnosis of human visceral leishmaniasis using peripheral blood. Asian Pacific J. Trop. Dis. 2013, 3, 10–15. [Google Scholar] [CrossRef]
- Nordkamp, L.O.; Mellink, C.; van der Schoot, E.; Berg, H. van den Karyotyping, FISH, and PCR in acute lymphoblastic leukemia: Competing or complementary diagnostics? J. Pediatr. Hematol. Oncol. 2009, 31, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.-Q.; Zhao, Z.-J.; Liu, B.; Bao, A.-Y.; Zheng, H.-Y.; Gu, J.; Xia, Y.; McGrath, M.; Dovat, S.; Song, C.-H.; et al. New rapid method to detect BCR-ABL fusion genes with multiplex RT-qPCR in one-tube at a time. Leuk. Res. 2018, 69, 47–53. [Google Scholar] [CrossRef] [PubMed]
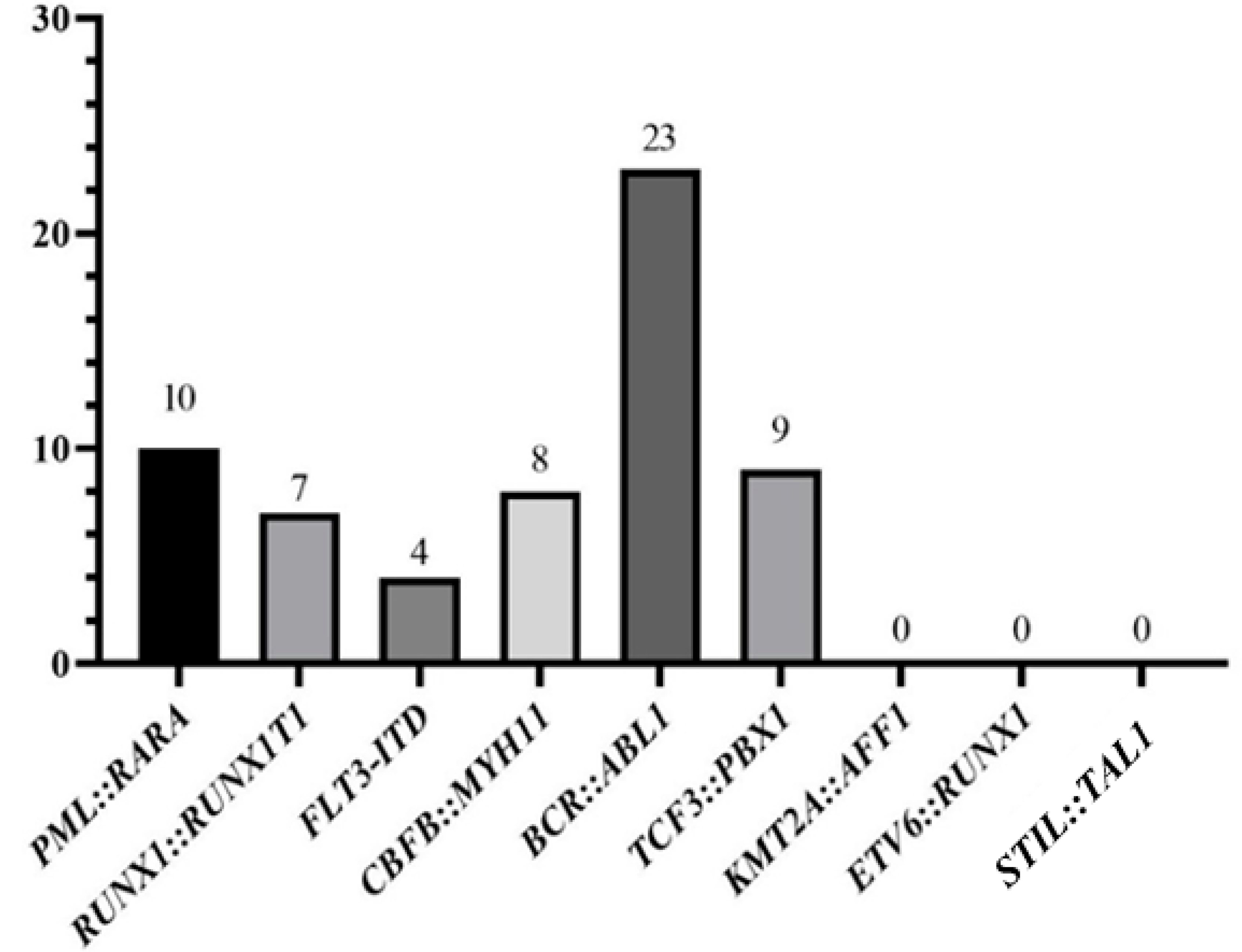


| N° of Patients | Gender | Age (Mean) | Hb | WBC | Blasts in PB | Karyotype | Deaths | |
|---|---|---|---|---|---|---|---|---|
| AML | ||||||||
| PML::RARA | 10 | Male: 8 | 40.5 years | Hb < 10: 9 | WBC < 10,000: 7 | Yes: 5 | Complex: 1 | 5 |
| Female: 2 | Hb > 10: 1 | WBC > 10,000: 3 | No: 5 | Classic translocation: 5 | ||||
| Normal: 1 | ||||||||
| NR: 3 | ||||||||
| RUNX1::RUNX1T1 | 7 | Male: 4 | 31 years | Hb < 10: 6 | WBC < 10,000: 7 | Yes: 3 | Complex: 2 | 2 |
| Female: 3 | Hb > 10: 1 | No: 4 | Classic translocation: 3 | |||||
| Normal: 1 | ||||||||
| Other alterations: 1 | ||||||||
| FLT3-ITD | 4 | Male: 2 | 50.2 years | Hb < 10: 4 | WBC < 10,000: 2 | Yes: 2 | Normal: 3 | 4 |
| Female: 2 | WBC > 10,000: 2 | No: 2 | Other alterations: 1 | |||||
| CBFB::MYH11 | 8 | Male: 6 Female: 2 | 41.9 years | Hb < 10: 8 | WBC < 10,000: 2 WBC > 10,000: 6 | Yes: 6 No: 2 | Complex: 1 Classic translocation: 1 Normal: 2 Other alterations: 3 NR: 1 | 5 |
| ALL | ||||||||
| BCR::ABL1 | 23 | Male: 14 | 42.2 years | Hb < 10: 19 | WBC < 10,000: 10 | Yes: 11 | Complex: 5 | 8 |
| Female: 9 | Hb > 10: 4 | WBC > 10,000: 13 | No: 12 | Normal: 7 | ||||
| Other alterations: 1 NR: 10 | ||||||||
| TCF3::PBX1 | 9 | Male: 5 Female: 4 | 36.9 years | Hb < 10: 8 Hb > 10: 1 | WBC < 10,000: 6 | Yes: 4 No: 5 | Complex: 4 | 3 |
| WBC > 10,000: 3 | Normal: 3 NR: 2 |
| N° of Patients | Gender | Age (Mean) | Hb | WBC | Blasts in PB | Karyotype | Deaths | |
|---|---|---|---|---|---|---|---|---|
| AML | 48 | Male: 26 | 55.5 years | Hb < 10: 47 | WBC < 10,000: 22 | Yes: 30 | Complex: 5 Normal: 16 | 23 |
| Female: 22 | Hb > 10: 1 | WBC > 10,000: 26 | No: 18 | Other alterations: 13 | ||||
| NR: 14 | ||||||||
| ALL | 8 | Male: 5 | 41.4 years | Hb < 10: 7 | WBC < 10,000: 3 | Yes: 4 | Complex: 2 | 5 |
| Female: 3 | Hb > 10: 1 | WBC > 10,000: 5 | No: 4 | Normal: 2 | ||||
| Other alterations: 1 | ||||||||
| NR: 3 |
| ALL Genetic Alterations | AML Genetic Alterations | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Type | Test Type | BCR::ABL1 | TCF3::PBX1 | Total | CBFB::MYH11 | FLT3-ITD | PML::RARA | RUNX1::RUNX1T1 | Total |
| BM | |||||||||
| Karyotype | 4 | 3 | 7 | 1 | 0 | 6 | 6 | 13 | |
| Nested-PCR † | 6 | 2 | 8 | 0 | 0 | 0 | 0 | 0 | |
| RT-qPCR | 14 | 4 | 18 | 1 | 4 | 3 | 6 | 14 | |
| Total | 24 | 9 | 33 | 2 | 4 | 9 | 12 | 27 | |
| PB | |||||||||
| Karyotype * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Nested-PCR † | 10 | 1 | 11 | 0 | 0 | 0 | 0 | 0 | |
| RT-qPCR | 22 | 7 | 29 | 7 | 4 | 7 | 7 | 25 | |
| Total | 32 | 8 | 40 | 7 | 4 | 7 | 7 | 25 | |
| Total | |||||||||
| Karyotype * | 4 | 3 | 7 | 1 | 0 | 6 | 6 | 13 | |
| Nested-PCR † | 16 | 3 | 19 | 0 | 0 | 0 | 0 | 0 | |
| RT-qPCR | 36 | 11 | 47 | 7 | 8 | 10 | 13 | 38 | |
| Total | 56 | 17 | 73 | 8 | 8 | 16 | 19 | 51 | |
| Chi-Square AML Patients | Chi-Square ALL Patients | |||||
|---|---|---|---|---|---|---|
| Sample Type | Value | p | Value | p | ||
| BM | Χ2 | 4.970 | 0.174 | Χ2 | 1.109 | 0.574 |
| N | 27 | N | 33 | |||
| PB | Χ2 | - | * | Χ2 | - | * |
| N | 25 | N | 40 | |||
| Total | Χ2 | 5.365 | 0.147 | Χ2 | 2.099 | 0.350 |
| N | 52 | N | 73 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pessoa, F.M.C.d.P.; de Oliveira, M.B.; Barreto, I.V.; Machado, A.K.d.C.; Oliveira, D.S.d.; Ribeiro, R.M.; Medeiros, J.C.; Maciel, A.d.R.; Silva, F.A.C.; Gurgel, L.A.; et al. Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias. DNA 2024, 4, 285-299. https://doi.org/10.3390/dna4030019
Pessoa FMCdP, de Oliveira MB, Barreto IV, Machado AKdC, Oliveira DSd, Ribeiro RM, Medeiros JC, Maciel AdR, Silva FAC, Gurgel LA, et al. Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias. DNA. 2024; 4(3):285-299. https://doi.org/10.3390/dna4030019
Chicago/Turabian StylePessoa, Flávia Melo Cunha de Pinho, Marcelo Braga de Oliveira, Igor Valentim Barreto, Anna Karolyna da Costa Machado, Deivide Sousa de Oliveira, Rodrigo Monteiro Ribeiro, Jaira Costa Medeiros, Aurélia da Rocha Maciel, Fabiana Aguiar Carneiro Silva, Lívia Andrade Gurgel, and et al. 2024. "Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias" DNA 4, no. 3: 285-299. https://doi.org/10.3390/dna4030019
APA StylePessoa, F. M. C. d. P., de Oliveira, M. B., Barreto, I. V., Machado, A. K. d. C., Oliveira, D. S. d., Ribeiro, R. M., Medeiros, J. C., Maciel, A. d. R., Silva, F. A. C., Gurgel, L. A., de Albuquerque, K. M. C., Lopes, G. S., Vieira, R. P. G., Arraes, J. A., Alencar Filho, M. S. d., Khayat, A. S., Moraes, M. E. A. d., de Moraes Filho, M. O., & Moreira-Nunes, C. A. (2024). Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias. DNA, 4(3), 285-299. https://doi.org/10.3390/dna4030019








