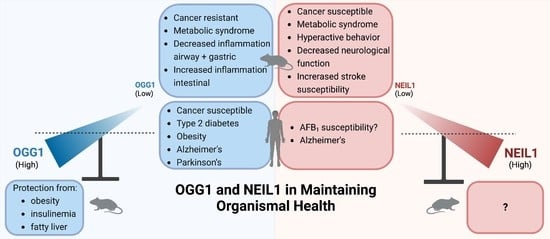
Complex Roles of NEIL1 and OGG1: Insights Gained from Murine Knockouts and Human Polymorphic Variants
Abstract
1. Introduction
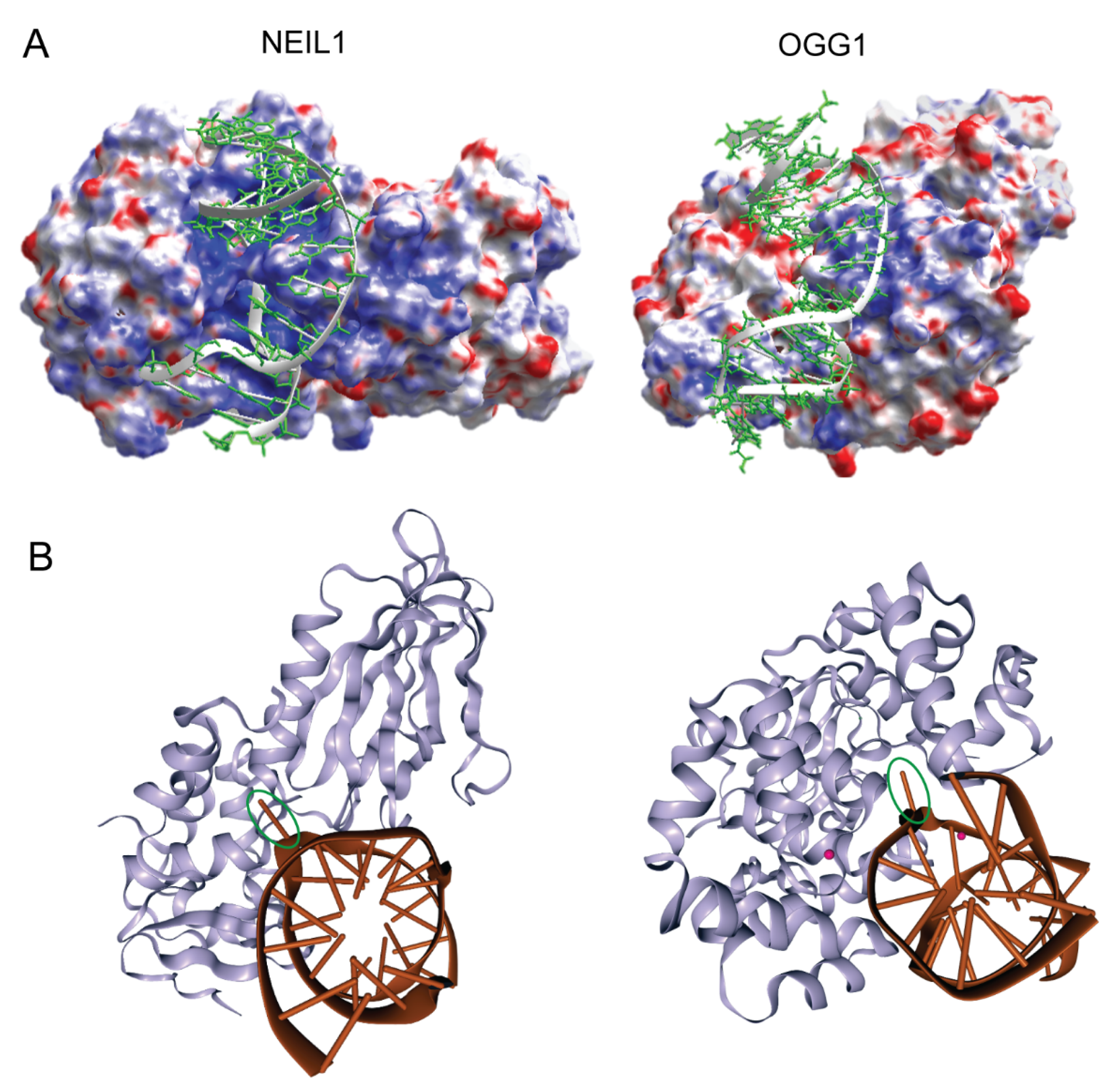
2. Target Site Location and Substrate Recognition
3. Mechanisms of Chemical Catalysis with Implications for the Completion of the BER Pathway
4. Cell-Cycle Specificity and Protein-Binding Partners
5. Phenotypes in Knockout Mice
5.1. Cancers
5.2. Metabolic Syndrome
5.3. Inflammation: The Role of OGG1 as a Regulator of Transcriptional Responses to Oxidatively Induced Base Damage
5.4. Roles of OGG1 and NEIL1 in Huntington’s Disease
5.5. Neurodegeneration in Neil1 Deficiency
6. Extrapolation to Human Investigations
6.1. OGG1-Associated Cancer Variants
6.2. Low-Level Expression of OGG1 in Therapeutically Responsive Subtypes of Human Acute Myeloid Leukemia (AML)
6.3. NEIL1-Associated Cancers
6.4. Potential Applications of Agonists of OGG1 in Disease Prevention
7. Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kow, Y.W. Base excision repair in E. coli—An overview. Ann. N. Y. Acad. Sci. 1994, 726, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Laval, J. Role of DNA repair enzymes in the cellular resistance to oxidative stress. Pathol. Biol. 1996, 44, 14–24. [Google Scholar] [PubMed]
- Beard, W.A.; Horton, J.K.; Prasad, R.; Wilson, S.H. Eukaryotic base excision repair: New approaches shine light on mechanism. Annu. Rev. Biochem. 2019, 88, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Bandaru, V.; Sunkara, S.; Wallace, S.S.; Bond, J.P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair 2002, 1, 517–529. [Google Scholar] [CrossRef]
- Hazra, T.K.; Izumi, T.; Boldogh, I.; Imhoff, B.; Kow, Y.W.; Jaruga, P.; Dizdaroglu, M.; Mitra, S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 3523–3528. [Google Scholar] [CrossRef]
- Morland, I.; Rolseth, V.; Luna, L.; Rognes, T.; Bjoras, M.; Seeberg, E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002, 30, 4926–4936. [Google Scholar] [CrossRef]
- Rosenquist, T.A.; Zaika, E.; Fernandes, A.S.; Zharkov, D.O.; Miller, H.; Grollman, A.P. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair 2003, 2, 581–591. [Google Scholar] [CrossRef]
- van der Kemp, P.A.; Thomas, D.; Barbey, R.; de Oliveira, R.; Boiteux, S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 1996, 93, 5197–5202. [Google Scholar] [CrossRef]
- Aburatani, H.; Hippo, Y.; Ishida, T.; Takashima, R.; Matsuba, C.; Kodama, T.; Takao, M.; Yasui, A.; Yamamoto, K.; Asano, M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997, 57, 2151–2156. [Google Scholar]
- Arai, K.; Morishita, K.; Shinmura, K.; Kohno, T.; Kim, S.R.; Nohmi, T.; Taniwaki, M.; Ohwada, S.; Yokota, J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene 1997, 14, 2857–2861. [Google Scholar] [CrossRef]
- Bjoras, M.; Luna, L.; Johnsen, B.; Hoff, E.; Haug, T.; Rognes, T.; Seeberg, E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997, 16, 6314–6322. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Nash, H.M.; Verdine, G.L. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997, 7, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Radicella, J.P.; Dherin, C.; Desmaze, C.; Fox, M.S.; Boiteux, S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 8010–8015. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Arjona, T.; Wei, Y.F.; Carter, K.C.; Klungland, A.; Anselmino, C.; Wang, R.P.; Augustus, M.; Lindahl, T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 1997, 94, 8016–8020. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, T.A.; Zharkov, D.O.; Grollman, A.P. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 1997, 94, 7429–7434. [Google Scholar] [CrossRef] [PubMed]
- Doublie, S.; Bandaru, V.; Bond, J.P.; Wallace, S.S. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10284–10289. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Yang, W.; Karplus, M.; Verdine, G.L. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 2005, 434, 612–618. [Google Scholar] [CrossRef]
- Imamura, K.; Wallace, S.S.; Doublie, S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J. Biol. Chem. 2009, 284, 26174–26183. [Google Scholar] [CrossRef]
- Imamura, K.; Averill, A.; Wallace, S.S.; Doublie, S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J. Biol. Chem. 2012, 287, 4288–4298. [Google Scholar] [CrossRef]
- Zhu, C.; Lu, L.; Zhang, J.; Yue, Z.; Song, J.; Zong, S.; Liu, M.; Stovicek, O.; Gao, Y.Q.; Yi, C. Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair. Proc. Natl. Acad. Sci. USA 2016, 113, 7792–7797. [Google Scholar] [CrossRef]
- Li, H.; Endutkin, A.V.; Bergonzo, C.; Fu, L.; Grollman, A.; Zharkov, D.O.; Simmerling, C. DNA deformation-coupled recognition of 8-oxoguanine: Conformational kinetic gating in human DNA glycosylase. J. Am. Chem. Soc. 2017, 139, 2682–2692. [Google Scholar] [CrossRef]
- Shigdel, U.K.; Ovchinnikov, V.; Lee, S.J.; Shih, J.A.; Karplus, M.; Nam, K.; Verdine, G.L. The trajectory of intrahelical lesion recognition and extrusion by the human 8-oxoguanine DNA glycosylase. Nat. Commun. 2020, 11, 4437. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, J.; Zhu, C.; Zhang, X.; Xiao, W.; Yan, Y.; Liu, L.; Zeng, H.; Gao, Y.Q.; Yi, C. DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes. Nat. Commun. 2021, 12, 4108. [Google Scholar] [CrossRef]
- Lloyd, R.S.; Hanawalt, P.C.; Dodson, M.L. Processive action of T4 endonuclease V on ultraviolet-irradiated DNA. Nucleic Acids Res. 1980, 8, 5113–5127. [Google Scholar] [CrossRef]
- Gruskin, E.A.; Lloyd, R.S. The DNA scanning mechanism of T4 endonuclease V. Effect of NaCl concentration on processive nicking activity. J. Biol. Chem. 1986, 261, 9607–9613. [Google Scholar] [CrossRef]
- Dowd, D.R.; Lloyd, R.S. Site-directed mutagenesis of the T4 endonuclease V gene: The role of arginine-3 in the target search. Biochemistry 1989, 28, 8699–8705. [Google Scholar] [CrossRef]
- Dowd, D.R.; Lloyd, R.S. Biological significance of facilitated diffusion in protein-DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J. Biol. Chem. 1990, 265, 3424–3431. [Google Scholar] [CrossRef]
- Zharkov, D.O.; Grollman, A.P. The DNA trackwalkers: Principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005, 577, 24–54. [Google Scholar] [CrossRef]
- Prakash, A.; Doublie, S.; Wallace, S.S. The Fpg/Nei family of DNA glycosylases: Substrates, structures, and search for damage. Prog. Mol. Biol. Transl. Sci. 2012, 110, 71–91. [Google Scholar] [CrossRef]
- Lee, A.J.; Wallace, S.S. Hide and seek: How do DNA glycosylases locate oxidatively damaged DNA bases amidst a sea of undamaged bases? Free Radic. Biol. Med. 2017, 107, 170–178. [Google Scholar] [CrossRef]
- D’Augustin, O.; Huet, S.; Campalans, A.; Radicella, J.P. Lost in the crowd: How does human 8-oxoguanine DNA glycosylase 1 (OGG1) find 8-oxoguanine in the genome? Int. J. Mol. Sci. 2020, 21, 8360. [Google Scholar] [CrossRef]
- Kong, M.; Beckwitt, E.C.; Van Houten, B. Dynamic action of DNA repair proteins as revealed by single molecule techniques: Seeing is believing. DNA Repair 2020, 93, 102909. [Google Scholar] [CrossRef]
- Rose, A.S.; Bradley, A.R.; Valasatava, Y.; Duarte, J.M.; Prlic, A.; Rose, P.W. NGL viewer: Web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755–3758. [Google Scholar] [CrossRef]
- McCullough, A.K.; Minko, I.G.; Nilsen, A.; Nagarajan, S.; Lloyd, R.S. Modulation of DNA glycosylase activities via small molecules. In DNA Damage, DNA Repair and Disease; Dizdaroglu, M., Lloyd, R.S., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2021; Volume 1, pp. 323–347. [Google Scholar]
- Hegde, M.L.; Hegde, P.M.; Bellot, L.J.; Mandal, S.M.; Hazra, T.K.; Li, G.M.; Boldogh, I.; Tomkinson, A.E.; Mitra, S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E3090–E3099. [Google Scholar] [CrossRef]
- Hegde, P.M.; Dutta, A.; Sengupta, S.; Mitra, J.; Adhikari, S.; Tomkinson, A.E.; Li, G.M.; Boldogh, I.; Hazra, T.K.; Mitra, S.; et al. The C-terminal domain (CTD) of human DNA glycosylase NEIL1 is required for forming BERosome repair complex with DNA replication proteins at the replicating genome: Dominant negative function of the CTD. J. Biol. Chem. 2015, 290, 20919–20933. [Google Scholar] [CrossRef]
- Dou, H.; Mitra, S.; Hazra, T.K. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003, 278, 49679–49684. [Google Scholar] [CrossRef]
- Das, A.; Boldogh, I.; Lee, J.W.; Harrigan, J.A.; Hegde, M.L.; Piotrowski, J.; de Souza Pinto, N.; Ramos, W.; Greenberg, M.M.; Hazra, T.K.; et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007, 282, 26591–26602. [Google Scholar] [CrossRef]
- Hegde, M.L.; Theriot, C.A.; Das, A.; Hegde, P.M.; Guo, Z.; Gary, R.K.; Hazra, T.K.; Shen, B.; Mitra, S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J. Biol. Chem. 2008, 283, 27028–27037. [Google Scholar] [CrossRef]
- Yeo, J.; Goodman, R.A.; Schirle, N.T.; David, S.S.; Beal, P.A. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. USA 2010, 107, 20715–20719. [Google Scholar] [CrossRef]
- Zhao, X.; Krishnamurthy, N.; Burrows, C.J.; David, S.S. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry 2010, 49, 1658–1666. [Google Scholar] [CrossRef]
- Hegde, M.L.; Banerjee, S.; Hegde, P.M.; Bellot, L.J.; Hazra, T.K.; Boldogh, I.; Mitra, S. Enhancement of NEIL1 protein-initiated oxidized DNA base excision repair by heterogeneous nuclear ribonucleoprotein U (hnRNP-U) via direct interaction. J. Biol. Chem. 2012, 287, 34202–34211. [Google Scholar] [CrossRef]
- Vartanian, V.; Minko, I.G.; Chawanthayatham, S.; Egner, P.A.; Lin, Y.C.; Earley, L.F.; Makar, R.; Eng, J.R.; Camp, M.T.; Li, L.; et al. NEIL1 protects against aflatoxin-induced hepatocellular carcinoma in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 4207–4212. [Google Scholar] [CrossRef]
- Roy, L.M.; Jaruga, P.; Wood, T.G.; McCullough, A.K.; Dizdaroglu, M.; Lloyd, R.S. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 2007, 282, 15790–15798. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat. Res. Rev. Mutat. Res. 2017, 771, 99–127. [Google Scholar] [CrossRef]
- Minko, I.G.; Vartanian, V.L.; Tozaki, N.N.; Coskun, E.; Coskun, S.H.; Jaruga, P.; Yeo, J.; David, S.S.; Stone, M.P.; Egli, M.; et al. Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1. DNA Repair 2020, 85, 102741. [Google Scholar] [CrossRef]
- Prakash, A.; Carroll, B.L.; Sweasy, J.B.; Wallace, S.S.; Doublie, S. Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing. DNA Repair 2014, 14, 17–26. [Google Scholar] [CrossRef]
- Minko, I.G.; Christov, P.P.; Li, L.; Stone, M.P.; McCullough, A.K.; Lloyd, R.S. Processing of N5-substituted formamidopyrimidine DNA adducts by DNA glycosylases NEIL1 and NEIL3. DNA Repair 2019, 73, 49–54. [Google Scholar] [CrossRef]
- Tomar, R.; Minko, I.G.; Kellum, A.H., Jr.; Voehler, M.W.; Stone, M.P.; McCullough, A.K.; Lloyd, R.S. DNA sequence modulates the efficiency of NEIL1-catalyzed excision of the aflatoxin B1-induced formamidopyrimidine guanine adduct. Chem. Res. Toxicol. 2021, 34, 901–911. [Google Scholar] [CrossRef]
- Hailer, M.K.; Slade, P.G.; Martin, B.D.; Rosenquist, T.A.; Sugden, K.D. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair 2005, 4, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Zhao, X.; Burrows, C.J.; David, S.S. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 2008, 47, 7137–7146. [Google Scholar] [CrossRef]
- McKibbin, P.L.; Fleming, A.M.; Towheed, M.A.; Van Houten, B.; Burrows, C.J.; David, S.S. Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 2013, 135, 13851–13861. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef] [PubMed]
- Slyvka, A.; Mierzejewska, K.; Bochtler, M. Nei-like 1 (NEIL1) excises 5-carboxylcytosine directly and stimulates TDG-mediated 5-formyl and 5-carboxylcytosine excision. Sci. Rep. 2017, 7, 9001. [Google Scholar] [CrossRef] [PubMed]
- Couve-Privat, S.; Mace, G.; Rosselli, F.; Saparbaev, M.K. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007, 35, 5672–5682. [Google Scholar] [CrossRef] [PubMed]
- Couve, S.; Mace-Aime, G.; Rosselli, F.; Saparbaev, M.K. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J. Biol. Chem. 2009, 284, 11963–11970. [Google Scholar] [CrossRef]
- Martin, P.R.; Couve, S.; Zutterling, C.; Albelazi, M.S.; Groisman, R.; Matkarimov, B.T.; Parsons, J.L.; Elder, R.H.; Saparbaev, M.K. The human DNA glycosylases NEIL1 and NEIL3 excise psoralen-induced DNA-DNA cross-links in a four-stranded DNA structure. Sci. Rep. 2017, 7, 17438. [Google Scholar] [CrossRef]
- Hu, J.; de Souza-Pinto, N.C.; Haraguchi, K.; Hogue, B.A.; Jaruga, P.; Greenberg, M.M.; Dizdaroglu, M.; Bohr, V.A. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 2005, 280, 40544–40551. [Google Scholar] [CrossRef]
- Sidorenko, V.S.; Grollman, A.P.; Jaruga, P.; Dizdaroglu, M.; Zharkov, D.O. Substrate specificity and excision kinetics of natural polymorphic variants and phosphomimetic mutants of human 8-oxoguanine-DNA glycosylase. FEBS J. 2009, 276, 5149–5162. [Google Scholar] [CrossRef]
- Sassa, A.; Beard, W.A.; Prasad, R.; Wilson, S.H. DNA sequence context effects on the glycosylase activity of human 8-oxoguanine DNA glycosylase. J. Biol. Chem. 2012, 287, 36702–36710. [Google Scholar] [CrossRef]
- Sidorenko, V.S.; Nevinsky, G.A.; Zharkov, D.O. Specificity of stimulation of human 8-oxoguanine-DNA glycosylase by AP endonuclease. Biochem. Biophys. Res. Commun. 2008, 368, 175–179. [Google Scholar] [CrossRef]
- Dherin, C.; Radicella, J.P.; Dizdaroglu, M.; Boiteux, S. Excision of oxidatively damaged DNA bases by the human a-hOgg1 protein and the polymorphic a-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999, 27, 4001–4007. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 2005, 591, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Schrock, R.D., 3rd; Lloyd, R.S. Reductive methylation of the amino terminus of endonuclease V eradicates catalytic activities. Evidence for an essential role of the amino terminus in the chemical mechanisms of catalysis. J. Biol. Chem. 1991, 266, 17631–17639. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.L.; Schrock, R.D., 3rd; Lloyd, R.S. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry 1993, 32, 8284–8290. [Google Scholar] [CrossRef]
- Schrock, R.D., 3rd; Lloyd, R.S. Site-directed mutagenesis of the NH2 terminus of T4 endonuclease V. The position of the aNH2 moiety affects catalytic activity. J. Biol. Chem. 1993, 268, 880–886. [Google Scholar] [CrossRef]
- Golan, G.; Zharkov, D.O.; Grollman, A.P.; Dodson, M.L.; McCullough, A.K.; Lloyd, R.S.; Shoham, G. Structure of T4 pyrimidine dimer glycosylase in a reduced imine covalent complex with abasic site-containing DNA. J. Mol. Biol. 2006, 362, 241–258. [Google Scholar] [CrossRef]
- Dodson, M.L.; Michaels, M.L.; Lloyd, R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994, 269, 32709–32712. [Google Scholar] [CrossRef]
- Sun, B.; Latham, K.A.; Dodson, M.L.; Lloyd, R.S. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J. Biol. Chem. 1995, 270, 19501–19508. [Google Scholar] [CrossRef]
- Vik, E.S.; Alseth, I.; Forsbring, M.; Helle, I.H.; Morland, I.; Luna, L.; Bjoras, M.; Dalhus, B. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair 2012, 11, 766–773. [Google Scholar] [CrossRef]
- Forsbring, M.; Vik, E.S.; Dalhus, B.; Karlsen, T.H.; Bergquist, A.; Schrumpf, E.; Bjoras, M.; Boberg, K.M.; Alseth, I. Catalytically impaired hMYH and NEIL1 mutant proteins identified in patients with primary sclerosing cholangitis and cholangiocarcinoma. Carcinogenesis 2009, 30, 1147–1154. [Google Scholar] [CrossRef]
- Dalhus, B.; Forsbring, M.; Helle, I.H.; Vik, E.S.; Forstrom, R.J.; Backe, P.H.; Alseth, I.; Bjoras, M. Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure 2011, 19, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Allgayer, J.; Kitsera, N.; Bartelt, S.; Epe, B.; Khobta, A. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016, 44, 7267–7280. [Google Scholar] [CrossRef] [PubMed]
- Morland, I.; Luna, L.; Gustad, E.; Seeberg, E.; Bjoras, M. Product inhibition and magnesium modulate the dual reaction mode of hOgg1. DNA Repair 2005, 4, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Kidane, D.; Murphy, D.L.; Sweasy, J.B. Accumulation of abasic sites induces genomic instability in normal human gastric epithelial cells during Helicobacter pylori infection. Oncogenesis 2014, 3, e128. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Maayah, M.; Sweasy, J.B.; Alnajjar, K.S. The role of cysteines in the structure and function of OGG1. J. Biol. Chem. 2021, 296, 100093. [Google Scholar] [CrossRef]
- Popov, A.V.; Endutkin, A.V.; Yatsenko, D.D.; Yudkina, A.V.; Barmatov, A.E.; Makasheva, K.A.; Raspopova, D.Y.; Diatlova, E.A.; Zharkov, D.O. Molecular dynamics approach to identification of new OGG1 cancer-associated somatic variants with impaired activity. J. Biol. Chem. 2021, 296, 100229. [Google Scholar] [CrossRef]
- Wiederhold, L.; Leppard, J.B.; Kedar, P.; Karimi-Busheri, F.; Rasouli-Nia, A.; Weinfeld, M.; Tomkinson, A.E.; Izumi, T.; Prasad, R.; Wilson, S.H.; et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 2004, 15, 209–220. [Google Scholar] [CrossRef]
- Whitehouse, C.J.; Taylor, R.M.; Thistlethwaite, A.; Zhang, H.; Karimi-Busheri, F.; Lasko, D.D.; Weinfeld, M.; Caldecott, K.W. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 2001, 104, 107–117. [Google Scholar] [CrossRef]
- Guan, X.; Bai, H.; Shi, G.; Theriot, C.A.; Hazra, T.K.; Mitra, S.; Lu, A.L. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007, 35, 2463–2472. [Google Scholar] [CrossRef]
- Dou, H.; Theriot, C.A.; Das, A.; Hegde, M.L.; Matsumoto, Y.; Boldogh, I.; Hazra, T.K.; Bhakat, K.K.; Mitra, S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 2008, 283, 3130–3140. [Google Scholar] [CrossRef]
- Theriot, C.A.; Hegde, M.L.; Hazra, T.K.; Mitra, S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair 2010, 9, 643–652. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Fitzpatrick, M.; Kompaniez, K.; Jacob, K.D.; Moore, B.R.; Nagle, J.; Barnes, J.; Lohani, A.; Evans, M.K. Coordination of DNA repair by NEIL1 and PARP-1: A possible link to aging. Aging 2012, 4, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Dhenaut, A.; Boiteux, S.; Radicella, J.P. Characterization of the hOGG1 promoter and its expression during the cell cycle. Mutat. Res. 2000, 461, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Boldogh, I.; Milligan, D.; Lee, M.S.; Bassett, H.; Lloyd, R.S.; McCullough, A.K. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: Evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001, 29, 2802–2809. [Google Scholar] [CrossRef] [PubMed]
- Luna, L.; Rolseth, V.; Hildrestrand, G.A.; Otterlei, M.; Dantzer, F.; Bjoras, M.; Seeberg, E. Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res. 2005, 33, 1813–1824. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.W.; Hazra, T.K.; Izumi, T.; Mitra, S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: Potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001, 29, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.E.; Hickson, I.D.; Boiteux, S.; Radicella, J.P. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: Bypass of the AP lyase activity step. Nucleic Acids Res. 2001, 29, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Marsin, S.; Vidal, A.E.; Sossou, M.; Menissier-de Murcia, J.; Le Page, F.; Boiteux, S.; de Murcia, G.; Radicella, J.P. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003, 278, 44068–44074. [Google Scholar] [CrossRef] [PubMed]
- Mokkapati, S.K.; Wiederhold, L.; Hazra, T.K.; Mitra, S. Stimulation of DNA glycosylase activity of OGG1 by NEIL1: Functional collaboration between two human DNA glycosylases. Biochemistry 2004, 43, 11596–11604. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Park, J.H.; Hahm, S.H.; Ko, S.I.; Lee, Y.R.; Chung, J.H.; Sohn, S.Y.; Cho, Y.; Kang, L.W.; Han, Y.S. Repair activities of human 8-oxoguanine DNA glycosylase are stimulated by the interaction with human checkpoint sensor Rad9-Rad1-Hus1 complex. DNA Repair 2009, 8, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Kompaniez, K.; Barnes, J.; Lohani, A.; Evans, M.K. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1). J. Biol. Chem. 2011, 286, 44679–44690. [Google Scholar] [CrossRef] [PubMed]
- Ramdzan, Z.M.; Vadnais, C.; Pal, R.; Vandal, G.; Cadieux, C.; Leduy, L.; Davoudi, S.; Hulea, L.; Yao, L.; Karnezis, A.N.; et al. RAS transformation requires CUX1-dependent repair of oxidative DNA damage. PLoS Biol. 2014, 12, e1001807. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Ramdzan, Z.M.; Kaur, S.; Duquette, P.M.; Marcotte, R.; Leduy, L.; Davoudi, S.; Lamarche-Vane, N.; Iulianella, A.; Nepveu, A. CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage. J. Biol. Chem. 2015, 290, 22520–22531. [Google Scholar] [CrossRef]
- Ramdzan, Z.M.; Pal, R.; Kaur, S.; Leduy, L.; Berube, G.; Davoudi, S.; Vadnais, C.; Nepveu, A. The function of CUX1 in oxidative DNA damage repair is needed to prevent premature senescence of mouse embryo fibroblasts. Oncotarget 2015, 6, 3613–3626. [Google Scholar] [CrossRef]
- Kaur, S.; Coulombe, Y.; Ramdzan, Z.M.; Leduy, L.; Masson, J.Y.; Nepveu, A. Special AT-rich sequence-binding protein 1 (SATB1) functions as an accessory factor in base excision repair. J. Biol. Chem. 2016, 291, 22769–22780. [Google Scholar] [CrossRef]
- Jang, S.; Kumar, N.; Beckwitt, E.C.; Kong, M.; Fouquerel, E.; Rapic-Otrin, V.; Prasad, R.; Watkins, S.C.; Khuu, C.; Majumdar, C.; et al. Damage sensor role of UV-DDB during base excision repair. Nat. Struct. Mol. Biol. 2019, 26, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Han, X.; Akbari, M.; Croteau, D.L.; Rasmussen, L.J.; Bohr, V.A. Interaction between RECQL4 and OGG1 promotes repair of oxidative base lesion 8-oxoG and is regulated by SIRT1 deacetylase. Nucleic Acids Res. 2020, 48, 6530–6546. [Google Scholar] [CrossRef]
- Kumar, N.; Theil, A.F.; Roginskaya, V.; Ali, Y.; Calderon, M.; Watkins, S.C.; Barnes, R.P.; Opresko, P.L.; Pines, A.; Lans, H.; et al. Global and transcription-coupled repair of 8-oxoG is initiated by nucleotide excision repair proteins. Nat. Commun. 2022, 13, 974. [Google Scholar] [CrossRef]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef]
- Bangalore, D.M.; Tessmer, I. Direct hOGG1-Myc interactions inhibit hOGG1 catalytic activity and recruit Myc to its promoters under oxidative stress. Nucleic Acids Res. 2022, 50, 10385–10398. [Google Scholar] [CrossRef]
- Sharma, N.; Chakravarthy, S.; Longley, M.J.; Copeland, W.C.; Prakash, A. The C-terminal tail of the NEIL1 DNA glycosylase interacts with the human mitochondrial single-stranded DNA binding protein. DNA Repair 2018, 65, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Thompson, M.K.; Arrington, J.F.; Terry, D.M.; Chakravarthy, S.; Prevelige, P.E.; Prakash, A. Novel interaction interfaces mediate the interaction between the NEIL1 DNA glycosylase and mitochondrial transcription factor A. Front. Cell Dev. Biol. 2022, 10, 893806. [Google Scholar] [CrossRef] [PubMed]
- Kamenisch, Y.; Fousteri, M.; Knoch, J.; von Thaler, A.K.; Fehrenbacher, B.; Kato, H.; Becker, T.; Dolle, M.E.; Kuiper, R.; Majora, M.; et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010, 207, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.T.; De Luca, G.; Degan, P.; Parlanti, E.; Dogliotti, E.; Barnes, D.E.; Lindahl, T.; Yang, H.; Miller, J.H.; Bignami, M. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004, 64, 4411–4414. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yang, H.; Cunanan, C.; Okamoto, K.; Shibata, D.; Pan, J.; Barnes, D.E.; Lindahl, T.; McIlhatton, M.; Fishel, R.; et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004, 64, 3096–3102. [Google Scholar] [CrossRef]
- Arai, T.; Kelly, V.P.; Minowa, O.; Noda, T.; Nishimura, S. The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology 2006, 221, 179–186. [Google Scholar] [CrossRef]
- Kakehashi, A.; Ishii, N.; Okuno, T.; Fujioka, M.; Gi, M.; Wanibuchi, H. Enhanced susceptibility of Ogg1 mutant mice to multiorgan carcinogenesis. Int. J. Mol. Sci. 2017, 18, 1801. [Google Scholar] [CrossRef]
- Kunisada, M.; Sakumi, K.; Tominaga, Y.; Budiyanto, A.; Ueda, M.; Ichihashi, M.; Nakabeppu, Y.; Nishigori, C. 8-Oxoguanine formation induced by chronic UVB exposure makes Ogg1 knockout mice susceptible to skin carcinogenesis. Cancer Res. 2005, 65, 6006–6010. [Google Scholar] [CrossRef]
- Kakehashi, A.; Ishii, N.; Okuno, T.; Fujioka, M.; Gi, M.; Fukushima, S.; Wanibuchi, H. Progression of hepatic adenoma to carcinoma in Ogg1 mutant mice induced by phenobarbital. Oxid. Med. Cell. Longev. 2017, 2017, 8541064. [Google Scholar] [CrossRef]
- Vartanian, V.; Lowell, B.; Minko, I.G.; Wood, T.G.; Ceci, J.D.; George, S.; Ballinger, S.W.; Corless, C.L.; McCullough, A.K.; Lloyd, R.S. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. USA 2006, 103, 1864–1869. [Google Scholar] [CrossRef]
- Chan, M.K.; Ocampo-Hafalla, M.T.; Vartanian, V.; Jaruga, P.; Kirkali, G.; Koenig, K.L.; Brown, S.; Lloyd, R.S.; Dizdaroglu, M.; Teebor, G.W. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair 2009, 8, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Rolseth, V.; Luna, L.; Olsen, A.K.; Suganthan, R.; Scheffler, K.; Neurauter, C.G.; Esbensen, Y.; Kusnierczyk, A.; Hildrestrand, G.A.; Graupner, A.; et al. No cancer predisposition or increased spontaneous mutation frequencies in NEIL DNA glycosylases-deficient mice. Sci. Rep. 2017, 7, 4384. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakatsuru, Y.; Zhang, S.; Shimizu, Y.; Kume, H.; Tanaka, K.; Ide, F.; Ishikawa, T. Enhanced spontaneous and aflatoxin-induced liver tumorigenesis in xeroderma pigmentosum group A gene-deficient mice. Carcinogenesis 2002, 23, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Calkins, M.J.; Vartanian, V.; Owen, N.; Kirkali, G.; Jaruga, P.; Dizdaroglu, M.; McCullough, A.K.; Lloyd, R.S. Enhanced sensitivity of Neil1−/− mice to chronic UVB exposure. DNA Repair 2016, 48, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Sampath, H.; Batra, A.K.; Vartanian, V.; Carmical, J.R.; Prusak, D.; King, I.B.; Lowell, B.; Earley, L.F.; Wood, T.G.; Marks, D.L.; et al. Variable penetrance of metabolic phenotypes and development of high-fat diet-induced adiposity in NEIL1-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E724–E734. [Google Scholar] [CrossRef] [PubMed]
- Sampath, H.; Vartanian, V.; Rollins, M.R.; Sakumi, K.; Nakabeppu, Y.; Lloyd, R.S. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS ONE 2012, 7, e51697. [Google Scholar] [CrossRef]
- Vartanian, V.; Tumova, J.; Dobrzyn, P.; Dobrzyn, A.; Nakabeppu, Y.; Lloyd, R.S.; Sampath, H. 8-oxoguanine DNA glycosylase (OGG1) deficiency elicits coordinated changes in lipid and mitochondrial metabolism in muscle. PLoS ONE 2017, 12, e0181687. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Warmbrunn, M.V.; Nieuwdorp, M.; Clement, K. Metabolism and metabolic disorders and the microbiome: The intestinal microbiota associated with obesity, lipid metabolism, and metabolic health-pathophysiology and therapeutic strategies. Gastroenterology 2021, 160, 573–599. [Google Scholar] [CrossRef]
- Simon, H.; Vartanian, V.; Wong, M.H.; Nakabeppu, Y.; Sharma, P.; Lloyd, R.S.; Sampath, H. OGG1 deficiency alters the intestinal microbiome and increases intestinal inflammation in a mouse model. PLoS ONE 2020, 15, e0227501. [Google Scholar] [CrossRef]
- Komakula, S.S.B.; Tumova, J.; Kumaraswamy, D.; Burchat, N.; Vartanian, V.; Ye, H.; Dobrzyn, A.; Lloyd, R.S.; Sampath, H. The DNA repair protein OGG1 protects against obesity by altering mitochondrial energetics in white adipose tissue. Sci. Rep. 2018, 8, 14886. [Google Scholar] [CrossRef]
- Burchat, N.; Sharma, P.; Ye, H.; Komakula, S.S.B.; Dobrzyn, A.; Vartanian, V.; Lloyd, R.S.; Sampath, H. Maternal transmission of human OGG1 protects mice against genetically- and diet-Induced obesity through increased tissue mitochondrial content. Front. Cell Dev. Biol. 2021, 9, 718962. [Google Scholar] [CrossRef] [PubMed]
- Komakula, S.S.B.; Blaze, B.; Ye, H.; Dobrzyn, A.; Sampath, H. A Novel Role for the DNA Repair Enzyme 8-Oxoguanine DNA Glycosylase in Adipogenesis. Int. J. Mol. Sci. 2021, 22, 1152. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Galick, H.; Wallace, S.S. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair 2004, 3, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chaudhry, M.A.; Wallace, S.S. Base excision repair by hNTH1 and hOGG1: A two edged sword in the processing of DNA damage in g-irradiated human cells. DNA Repair 2006, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yuan, K.; Yan, C.; Fox, J., 3rd; Gaid, M.; Breitwieser, W.; Bansal, A.K.; Zeng, H.; Gao, H.; Wu, M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic. Biol. Med. 2012, 52, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Ba, X.; Aguilera-Aguirre, L.; Rashid, Q.T.; Bacsi, A.; Radak, Z.; Sur, S.; Hosoki, K.; Hegde, M.L.; Boldogh, I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int. J. Mol. Sci. 2014, 15, 16975–16997. [Google Scholar] [CrossRef] [PubMed]
- Mabley, J.G.; Pacher, P.; Deb, A.; Wallace, R.; Elder, R.H.; Szabo, C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB J. 2005, 19, 290–292. [Google Scholar] [CrossRef]
- Touati, E.; Michel, V.; Thiberge, J.M.; Ave, P.; Huerre, M.; Bourgade, F.; Klungland, A.; Labigne, A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter 2006, 11, 494–505. [Google Scholar] [CrossRef]
- Yang, J.; Mitra, A.; Dojer, N.; Fu, S.; Rowicka, M.; Brasier, A.R. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-kB/RelA regulatory network using ChIP-Seq. Nucleic Acids Res. 2013, 41, 7240–7259. [Google Scholar] [CrossRef][Green Version]
- Pan, L.; Zhu, B.; Hao, W.; Zeng, X.; Vlahopoulos, S.A.; Hazra, T.K.; Hegde, M.L.; Radak, Z.; Bacsi, A.; Brasier, A.R.; et al. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor kB-driven gene expression. J. Biol. Chem. 2016, 291, 25553–25566. [Google Scholar] [CrossRef]
- Park, J.W.; Han, Y.I.; Kim, S.W.; Kim, T.M.; Yeom, S.C.; Kang, J.; Park, J. 8-OxoG in GC-rich Sp1 binding sites enhances gene transcription in adipose tissue of juvenile mice. Sci. Rep. 2019, 9, 15618. [Google Scholar] [CrossRef]
- Perillo, B.; Ombra, M.N.; Bertoni, A.; Cuozzo, C.; Sacchetti, S.; Sasso, A.; Chiariotti, L.; Malorni, A.; Abbondanza, C.; Avvedimento, E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 2008, 319, 202–206. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013, 26, 593–607. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Visser, J.A.; Zhu, J.; Burrows, C.J. Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5′-untranslated regions. Biochemistry 2018, 57, 991–1002. [Google Scholar] [CrossRef]
- Rogers, R.A.; Fleming, A.M.; Burrows, C.J. Rapid screen of potential i-motif forming sequences in DNA repair gene promoters. ACS Omega 2018, 3, 9630–9635. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Esders, S.; Burrows, C.J. Oxidative modification of guanine in a potential Z-DNA-forming sequence of a gene promoter impacts gene expression. Chem. Res. Toxicol. 2019, 32, 899–909. [Google Scholar] [CrossRef]
- Donley, N.; Jaruga, P.; Coskun, E.; Dizdaroglu, M.; McCullough, A.K.; Lloyd, R.S. Small molecule inhibitors of 8-oxoguanine DNA glycosylase-1 (OGG1). ACS Chem. Biol. 2015, 10, 2334–2343. [Google Scholar] [CrossRef]
- Tahara, Y.K.; Auld, D.; Ji, D.; Beharry, A.A.; Kietrys, A.M.; Wilson, D.L.; Jimenez, M.; King, D.; Nguyen, Z.; Kool, E.T. Potent and selective inhibitors of 8-oxoguanine DNA glycosylase. J. Am. Chem. Soc. 2018, 140, 2105–2114. [Google Scholar] [CrossRef]
- Visnes, T.; Cazares-Korner, A.; Hao, W.; Wallner, O.; Masuyer, G.; Loseva, O.; Mortusewicz, O.; Wiita, E.; Sarno, A.; Manoilov, A.; et al. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science 2018, 362, 834–839. [Google Scholar] [CrossRef]
- Tahara, Y.K.; Kietrys, A.M.; Hebenbrock, M.; Lee, Y.; Wilson, D.L.; Kool, E.T. Dual inhibitors of 8-oxoguanine surveillance by OGG1 and NUDT1. ACS Chem. Biol. 2019, 14, 2606–2615. [Google Scholar] [CrossRef]
- Ling, H.; Song, C.; Fang, Y.; Yin, Y.; Wu, Z.; Wang, Y.; Xu, Z.; Gao, S.; Li, A.; Liu, G. TH5487, a small molecule inhibitor of OGG1, attenuates pulmonary fibrosis by NEDD4L-mediated OGG1 degradation. Chem. Biol. Interact. 2022, 362, 109999. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wu, S.; Wu, G.; Yang, J. Inhibition of 8-oxoguanine DNA glycosylase (OGG1) expression suppresses polycystic ovarian syndrome via the NF-kB signaling pathway. Reprod. Biol. 2022, 22, 100679. [Google Scholar] [CrossRef] [PubMed]
- Baquero, J.M.; Benitez-Buelga, C.; Rajagopal, V.; Zhenjun, Z.; Torres-Ruiz, R.; Muller, S.; Hanna, B.M.F.; Loseva, O.; Wallner, O.; Michel, M.; et al. Small molecule inhibitor of OGG1 blocks oxidative DNA damage repair at telomeres and potentiates methotrexate anticancer effects. Sci. Rep. 2021, 11, 3490. [Google Scholar] [CrossRef] [PubMed]
- Baquero, J.M.; Marchena-Perea, E.; Mirabet, R.; Torres-Ruiz, R.; Blanco-Aparicio, C.; Rodriguez-Perales, S.; Helleday, T.; Benitez-Buelga, C.; Benitez, J.; Osorio, A. OGG1 inhibition triggers synthetic lethality and enhances the effect of PARP inhibitor olaparib in BRCA1-deficient TNBC cells. Front. Oncol. 2022, 12, 888810. [Google Scholar] [CrossRef]
- Karsten, S. Targeting the DNA repair enzymes MTH1 and OGG1 as a novel approach to treat inflammatory diseases. Basic Clin. Pharmacol. Toxicol. 2022, 131, 95–103. [Google Scholar] [CrossRef]
- Gatto, E.M.; Rojas, N.G.; Persi, G.; Etcheverry, J.L.; Cesarini, M.E.; Perandones, C. Huntington disease: Advances in the understanding of its mechanisms. Clin. Park. Relat. Disord. 2020, 3, 100056. [Google Scholar] [CrossRef]
- Monckton, D.G. The contribution of somatic expansion of the CAG repeat to symptomatic development in Huntington’s disease: A historical perspective. J. Huntingtons. Dis. 2021, 10, 7–33. [Google Scholar] [CrossRef]
- Kovtun, I.V.; Liu, Y.; Bjoras, M.; Klungland, A.; Wilson, S.H.; McMurray, C.T. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 2007, 447, 447–452. [Google Scholar] [CrossRef]
- Goula, A.V.; Berquist, B.R.; Wilson, D.M., 3rd; Wheeler, V.C.; Trottier, Y.; Merienne, K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009, 5, e1000749. [Google Scholar] [CrossRef]
- Kovtun, I.V.; Johnson, K.O.; McMurray, C.T. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging 2011, 3, 509–514. [Google Scholar] [CrossRef]
- Iyer, R.R.; Pluciennik, A. DNA mismatch repair and its role in Huntington’s disease. J. Huntingtons. Dis. 2021, 10, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Budworth, H.; Harris, F.R.; Williams, P.; Lee, D.Y.; Holt, A.; Pahnke, J.; Szczesny, B.; Acevedo-Torres, K.; Ayala-Pena, S.; McMurray, C.T. Suppression of somatic expansion delays the onset of pathophysiology in a mouse model of Huntington’s disease. PLoS Genet. 2015, 11, e1005267. [Google Scholar] [CrossRef] [PubMed]
- Mollersen, L.; Rowe, A.D.; Illuzzi, J.L.; Hildrestrand, G.A.; Gerhold, K.J.; Tveteras, L.; Bjolgerud, A.; Wilson, D.M., 3rd; Bjoras, M.; Klungland, A. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum. Mol. Genet. 2012, 21, 4939–4947. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Canugovi, C.; Yoon, J.S.; Feldman, N.H.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proc. Natl. Acad. Sci. USA 2012, 109, 14948–14953. [Google Scholar] [CrossRef] [PubMed]
- Canugovi, C.; Misiak, M.; Scheibye-Knudsen, M.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. Loss of NEIL1 causes defects in olfactory function in mice. Neurobiol. Aging 2015, 36, 1007–1012. [Google Scholar] [CrossRef]
- Yang, B.; Figueroa, D.M.; Hou, Y.; Babbar, M.; Baringer, S.L.; Croteau, D.L.; Bohr, V.A. NEIL1 stimulates neurogenesis and suppresses neuroinflammation after stress. Free Radic. Biol. Med. 2019, 141, 47–58. [Google Scholar] [CrossRef]
- Hildrestrand, G.A.; Rolseth, V.; Kunath, N.; Suganthan, R.; Jensen, V.; Bugaj, A.M.; Fernandez-Berrocal, M.S.; Sikko, S.B.; Vetlesen, S.; Kusnierczyk, A.; et al. NEIL1 and NEIL2 DNA glycosylases modulate anxiety and learning in a cooperative manner in mice. Commun. Biol. 2021, 4, 1354. [Google Scholar] [CrossRef]
- Hung, R.J.; Hall, J.; Brennan, P.; Boffetta, P. Genetic polymorphisms in the base excision repair pathway and cancer risk: A HuGE review. Am. J. Epidemiol. 2005, 162, 925–942. [Google Scholar] [CrossRef]
- Hill, J.W.; Evans, M.K. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006, 34, 1620–1632. [Google Scholar] [CrossRef]
- Bravard, A.; Vacher, M.; Moritz, E.; Vaslin, L.; Hall, J.; Epe, B.; Radicella, J.P. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009, 69, 3642–3649. [Google Scholar] [CrossRef]
- Baptiste, B.A.; Katchur, S.R.; Fivenson, E.M.; Croteau, D.L.; Rumsey, W.L.; Bohr, V.A. Enhanced mitochondrial DNA repair of the common disease-associated variant, Ser326Cys, of hOGG1 through small molecule intervention. Free Radic. Biol. Med. 2018, 124, 149–162. [Google Scholar] [CrossRef]
- Kabzinski, J.; Walczak, A.; Dziki, A.; Mik, M.; Majsterek, I. Impact of the Ser326Cys polymorphism of the OGG1 gene on the level of oxidative DNA damage in patients with colorectal cancer. Pol. Przegl. Chir. 2018, 90, 13–15. [Google Scholar] [CrossRef]
- Alanazi, M.; Pathan, A.A.K.; Shaik, J.P.; Alhadheq, A.; Khan, Z.; Khan, W.; Al Naeem, A.; Parine, N.R. The hOGG1 Ser326Cys gene polymorphism and breast cancer risk in Saudi population. Pathol. Oncol. Res. 2017, 23, 525–535. [Google Scholar] [CrossRef]
- Costa, E.F.; Santos, E.S.; Liutti, V.T.; Leal, F.; Santos, V.C.; Rinck-Junior, J.A.; Mariano, F.V.; Coutinho-Camillo, C.M.; Altemani, A.; Lima, C.S.; et al. Association between polymorphisms in genes related to DNA base-excision repair with risk and prognosis of oropharyngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2016, 142, 1917–1926. [Google Scholar] [CrossRef]
- Gotoh, N.; Saitoh, T.; Takahashi, N.; Kasamatsu, T.; Minato, Y.; Lobna, A.; Oda, T.; Hoshino, T.; Sakura, T.; Shimizu, H.; et al. Association between OGG1 S326C CC genotype and elevated relapse risk in acute myeloid leukemia. Int. J. Hematol. 2018, 108, 246–253. [Google Scholar] [CrossRef]
- Smolarz, B.; Michalska, M.M.; Samulak, D.; Wojcik, L.; Romanowicz, H. Studies of correlations between single nucleotide polymorphisms of DNA repair genes and endometrial cancer in Polish women. Anticancer Res. 2018, 38, 5223–5229. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, S.L.; Turner, A.; Isaacs, S.D.; Wiley, K.E.; Hawkins, G.A.; Chang, B.L.; Bleecker, E.R.; Walsh, P.C.; Meyers, D.A.; et al. Associations between hOGG1 sequence variants and prostate cancer susceptibility. Cancer Res. 2002, 62, 2253–2257. [Google Scholar]
- Chen, L.; Elahi, A.; Pow-Sang, J.; Lazarus, P.; Park, J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J. Urol. 2003, 170, 2471–2474. [Google Scholar] [CrossRef]
- Le Marchand, L.; Donlon, T.; Lum-Jones, A.; Seifried, A.; Wilkens, L.R. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol. Biomark. Prev. 2002, 11, 409–412. [Google Scholar]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef]
- Hyun, J.W.; Choi, J.Y.; Zeng, H.H.; Lee, Y.S.; Kim, H.S.; Yoon, S.H.; Chung, M.H. Leukemic cell line, KG-1 has a functional loss of hOGG1 enzyme due to a point mutation and 8-hydroxydeoxyguanosine can kill KG-1. Oncogene 2000, 19, 4476–4479. [Google Scholar] [CrossRef]
- Hyun, J.W.; Cheon, G.J.; Kim, H.S.; Lee, Y.S.; Choi, E.Y.; Yoon, B.H.; Kim, J.S.; Chung, M.H. Radiation sensitivity depends on OGG1 activity status in human leukemia cell lines. Free Radic. Biol. Med. 2002, 32, 212–220. [Google Scholar] [CrossRef]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Owen, N.; Minko, I.G.; Moellmer, S.A.; Cammann, S.K.; Lloyd, R.S.; McCullough, A.K. Enhanced cytarabine-induced killing in OGG1-deficient acute myeloid leukemia cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2016833118. [Google Scholar] [CrossRef]
- Galick, H.A.; Marsden, C.G.; Kathe, S.; Dragon, J.A.; Volk, L.; Nemec, A.A.; Wallace, S.S.; Prakash, A.; Doublie, S.; Sweasy, J.B. The NEIL1 G83D germline DNA glycosylase variant induces genomic instability and cellular transformation. Oncotarget 2017, 8, 85883–85895. [Google Scholar] [CrossRef]
- Minko, I.G.; Vartanian, V.L.; Tozaki, N.N.; Linde, O.K.; Jaruga, P.; Coskun, S.H.; Coskun, E.; Qu, C.; He, H.; Xu, C.; et al. Characterization of rare NEIL1 variants found in East Asian populations. DNA Repair 2019, 79, 32–39. [Google Scholar] [CrossRef]
- Thameem, F.; Puppala, S.; Lehman, D.M.; Stern, M.P.; Blangero, J.; Abboud, H.E.; Duggirala, R.; Habib, S.L. The Ser(326)Cys polymorphism of 8-oxoguanine glycosylase 1 (OGG1) is associated with type 2 diabetes in Mexican Americans. Hum. Hered. 2010, 70, 97–101. [Google Scholar] [CrossRef]
- Anderson, S.M.; Naidoo, R.N.; Ramkaran, P.; Asharam, K.; Muttoo, S.; Chuturgoon, A.A. OGG1 Ser326Cys polymorphism, HIV, obesity and air pollution exposure influences adverse birth outcome susceptibility, within South African Women. Reprod. Toxicol. 2018, 79, 8–15. [Google Scholar] [CrossRef]
- Sliwinska, A.; Kwiatkowski, D.; Czarny, P.; Toma, M.; Wigner, P.; Drzewoski, J.; Fabianowska-Majewska, K.; Szemraj, J.; Maes, M.; Galecki, P.; et al. The levels of 7,8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1)—A potential diagnostic biomarkers of Alzheimer’s disease. J. Neurol. Sci. 2016, 368, 155–159. [Google Scholar] [CrossRef]
- Sanders, L.H.; Paul, K.C.; Howlett, E.H.; Lawal, H.; Boppana, S.; Bronstein, J.M.; Ritz, B.; Greenamyre, J.T. Editor’s highlight: Base excision repair variants and pesticide exposure increase Parkinson’s disease risk. Toxicol. Sci. 2017, 158, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Ramirez-Sabio, J.B.; Coltell, O.; Ortega-Azorin, C.; Estruch, R.; Martinez-Gonzalez, M.A.; Salas-Salvado, J.; Sorli, J.V.; Castaner, O.; Aros, F.; et al. Effects of the Ser326Cys polymorphism in the DNA repair OGG1 gene on cancer, cardiovascular, and all-cause mortality in the PREDIMED study: Modulation by diet. J. Acad. Nutr. Diet. 2018, 118, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Fromme, J.C.; Bruner, S.D.; Yang, W.; Karplus, M.; Verdine, G.L. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol. 2003, 10, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Katchur, S.R.; Jiang, Y.; Briand, J.; Schaber, M.; Kreatsoulas, C.; Schwartz, B.; Thrall, S.; Davis, A.M.; Duvall, S.; et al. Small molecule-mediated allosteric activation of the base excision repair enzyme 8-oxoguanine DNA glycosylase and its impact on mitochondrial function. Sci. Rep. 2022, 12, 14685. [Google Scholar] [CrossRef]
- Michel, M.; Benitez-Buelga, C.; Calvo, P.A.; Hanna, B.M.F.; Mortusewicz, O.; Masuyer, G.; Davies, J.; Wallner, O.; Sanjiv, K.; Albers, J.J.; et al. Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function. Science 2022, 376, 1471–1476. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lloyd, R.S. Complex Roles of NEIL1 and OGG1: Insights Gained from Murine Knockouts and Human Polymorphic Variants. DNA 2022, 2, 279-301. https://doi.org/10.3390/dna2040020
Lloyd RS. Complex Roles of NEIL1 and OGG1: Insights Gained from Murine Knockouts and Human Polymorphic Variants. DNA. 2022; 2(4):279-301. https://doi.org/10.3390/dna2040020
Chicago/Turabian StyleLloyd, R. Stephen. 2022. "Complex Roles of NEIL1 and OGG1: Insights Gained from Murine Knockouts and Human Polymorphic Variants" DNA 2, no. 4: 279-301. https://doi.org/10.3390/dna2040020
APA StyleLloyd, R. S. (2022). Complex Roles of NEIL1 and OGG1: Insights Gained from Murine Knockouts and Human Polymorphic Variants. DNA, 2(4), 279-301. https://doi.org/10.3390/dna2040020