Classification Problems of Repetitive DNA Sequences
Abstract
:1. Introduction
2. Repetitive Sequences in Bivalve Genomes
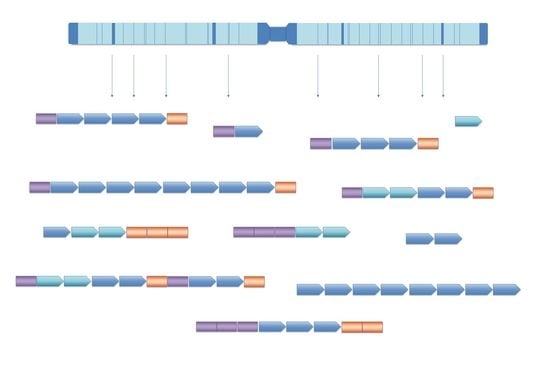
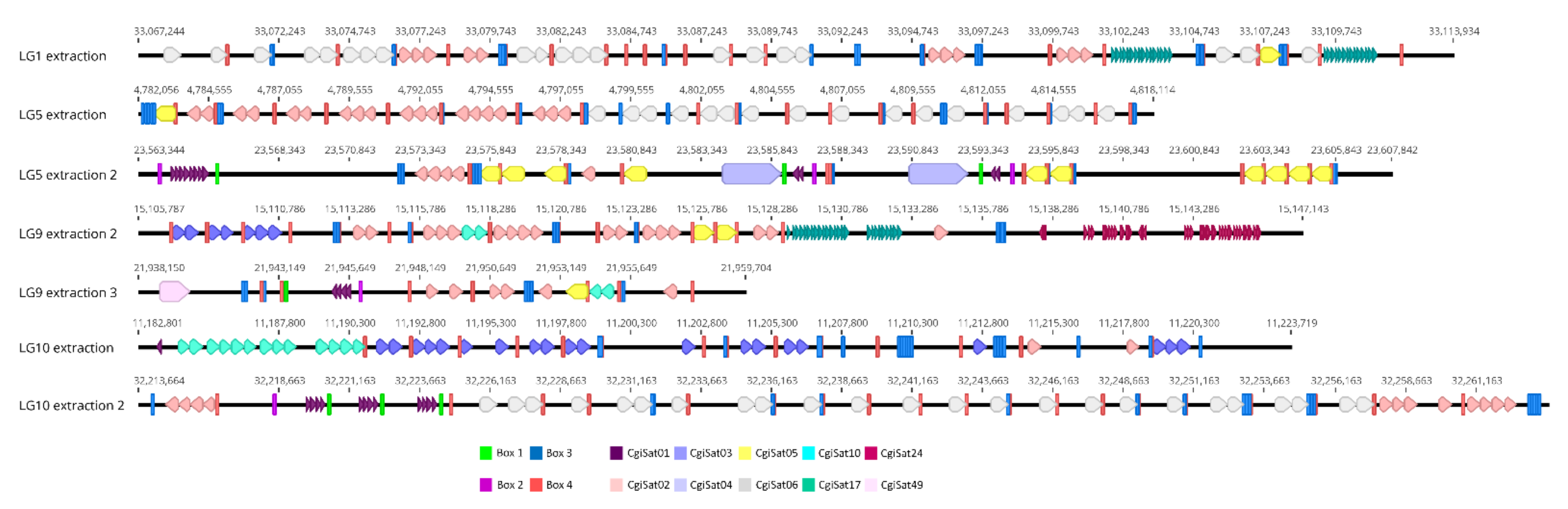
3. Problems in Classification of Repetitive DNA Sequences: The Case of the Pacific Oyster Crassostrea gigas
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA Evolution: Old Ideas, New Approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The Evolutionary Dynamics of Repetitive DNA in Eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, P.M.; Pierce, J.C.; Mackinley, A.G.; Titchen, D.A.; Glenn, W.K. Pearl, a Novel Family of Putative Transposable Elements in Bivalve Mollusks. J. Mol. Evol. 2003, 56, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Brajković, J.; Feliciello, I.; Bruvo-Mađaric, B.; Ugarković, D. Satellite DNA-Like Elements Associated With Genes Within Euchromatin of the Beetle Tribolium castaneum. G3—Genes Genom. Genet. 2012, 2, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Dias, G.B.; Svartman, M.; Delprat, A.; Ruiz, A.; Kuhn, G.C.S.S. Tetris Is a Foldback Transposon That Provided the Building Blocks for an Emerging Satellite DNA of Drosophila virilis. Genome Biol. Evol. 2014, 6, 1302–1313. [Google Scholar] [CrossRef] [Green Version]
- Dias, G.B.; Heringer, P.; Svartman, M.; Kuhn, G.C.S.S. Helitrons Shaping the Genomic Architecture of Drosophila: Enrichment of DINE-TR1 in α- and β-Heterochromatin, Satellite DNA Emergence, and PiRNA Expression. Chromosom. Res. 2015, 23, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Luchetti, A. TerMITEs: Miniature Inverted-Repeat Transposable Elements (MITEs) in the Termite Genome (Blattodea: Termitoidae). Mol. Genet. Genom. 2015, 290, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Šatović, E.; Vojvoda Zeljko, T.; Luchetti, A.; Mantovani, B.; Plohl, M. Adjacent Sequences Disclose Potential for Intra-Genomic Dispersal of Satellite DNA Repeats and Suggest a Complex Network with Transposable Elements. BMC Genom. 2016, 17, 997. [Google Scholar] [CrossRef] [Green Version]
- Feliciello, I.; Pezer, Ž.; Kordiš, D.; Mađarić, B.B.; Ugarković, Đ. Evolutionary History of Alpha Satellite DNA Repeats Dispersed within Human Genome Euchromatin. Genome Biol. Evol. 2020, 14561197, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Meštrović, N.; Mravinac, B.; Pavlek, M.; Vojvoda-Zeljko, T.; Šatović, E.; Plohl, M. Structural and Functional Liaisons between Transposable Elements and Satellite DNAs. Chromosom. Res. 2015, 23, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Tørresen, O.K.; Star, B.; Mier, P.; Andrade-Navarro, M.A.; Bateman, A.; Jarnot, P.; Gruca, A.; Grynberg, M.; Kajava, A.V.; Promponas, V.J.; et al. Tandem Repeats Lead to Sequence Assembly Errors and Impose Multi-Level Challenges for Genome and Protein Databases. Nucleic Acids Res. 2019, 47, 10994–11006. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the Dark Matter: Bioinformatics of Long-Range Sequencing and Mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Šatović, E.; Tunjić Cvitanić, M.; Plohl, M. Tools and Databases for Solving Problems in Detection and Identification of Repetitive DNA Sequences. Period. Biol. 2020, 121–122, 7–14. [Google Scholar] [CrossRef]
- Kim, Y.B.; Oh, J.H.; McIver, L.J.; Rashkovetsky, E.; Michalak, K.; Garner, H.R.; Kang, L.; Nevo, E.; Korol, A.B.; Michalak, P. Divergence of Drosophila melanogaster Repeatomes in Response to a Sharp Microclimate Contrast in Evolution Canyon, Israel. Proc. Natl. Acad. Sci. USA 2014, 111, 10630–10635. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-Throughput Analysis of the Satellitome Illuminates Satellite DNA Evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ruano, F.J.; Cabrero, J.; López-León, M.D.; Sánchez, A.; Camacho, J.P.M. Quantitative Sequence Characterization for Repetitive DNA Content in the Supernumerary Chromosome of the Migratory Locust. Chromosoma 2018, 127, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Ruiz-Ruano, F.J.; Silva, D.M.Z.A.; Serrano, É.A.; Rosa, I.F.; Scudeler, P.E.S.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. A Glimpse into the Satellite DNA Library in Characidae Fish (Teleostei, Characiformes). Front. Genet. 2017, 8, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios-Gimenez, O.M.; Koelman, J.; Flores, M.P.; Bradford, T.M.; Jones, K.K.; Cooper, S.J.B.; Kawakami, T.; Suh, A. Comparative Analysis of Morabine Grasshopper Genomes Reveals Highly Abundant Transposable Elements and Rapidly Proliferating Satellite DNA Repeats. BMC Biol. 2020, 18, 199. [Google Scholar] [CrossRef]
- Sader, M.; Vaio, M.; Cauz-Santos, L.A.; Dornelas, M.C.; Vieira, M.L.C.; Melo, N.; Pedrosa-Harand, A. Large vs. Small Genomes in Passiflora: The Influence of the Mobilome and the Satellitome. Planta 2021, 253, 86. [Google Scholar] [CrossRef]
- Šatović Vukšić, E.; Plohl, M. Exploring Satellite DNAs: Specificities of Bivalve Mollusks Genomes. In Satellite DNAs in Physiology and Evolution; Ugarković, Đ., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 57–83. [Google Scholar] [CrossRef]
- Bai, C.M.; Xin, L.S.; Rosani, U.; Wu, B.; Wang, Q.C.; Duan, X.K.; Liu, Z.H.; Wang, C.M. Chromosomal-Level Assembly of the Blood Clam, Scapharca (Anadara) broughtonii, Using Long Sequence Reads and Hi-C. Gigascience 2019, 8, giz067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Zeng, Q.; Ren, J.; Yao, H.; Lv, L.; He, L.; Ruan, W.; Xue, Q.; Bao, Z.; Wang, S.; et al. The Chromosome-Level Genome Assembly and Comprehensive Transcriptomes of the Razor Clam (Sinonovacula constricta). Front. Genet. 2020, 11, 664. [Google Scholar] [CrossRef]
- Song, H.; Guo, X.; Sun, L.; Wang, Q.; Han, F.; Wang, H.; Wray, G.A.; Davidson, P.; Wang, Q.; Hu, Z.; et al. The Hard Clam Genome Reveals Massive Expansion and Diversification of Inhibitors of Apoptosis in Bivalvia. BMC Biol. 2021, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.H. A High-Quality Reference Genome for a Parasitic Bivalve with Doubly Uniparental Inheritance (Bivalvia: Unionida). Genome Biol. Evol. 2021, 13, evab029. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Y.; Xu, T.; Zhang, Y.; Mu, H.; Zhang, Y.; Lan, Y.; Fields, C.J.; Hui, J.H.L.; Zhang, W.; et al. Adaptation to Deep-Sea Chemosynthetic Environments as Revealed by Mussel Genomes. Nat. Ecol. Evol. 2017, 1, 0121. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, T.; Kawashima, T.; Koyanagi, R.; Gyoja, F.; Tanaka, M.; Ikuta, T.; Shoguchi, E.; Fujiwara, M.; Shinzato, C.; Hisata, K.; et al. Draft Genome of the Pearl Oyster Pinctada fucata: A Platform for Understanding Bivalve Biology. DNA Res. 2012, 19, 117–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mun, S.; Kim, Y.J.; Markkandan, K.; Shin, W.; Oh, S.; Woo, J.; Yoo, J.; An, H.; Han, K. The Whole-Genome and Transcriptome of the Manila Clam (Ruditapes philippinarum). Genome Biol. Evol. 2017, 9, 1487–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaut, S.; Guerra, D.; Hoeh, W.R.; Stewart, D.T.; Bogan, A.E.; Ghiselli, F.; Milani, L.; Passamonti, M.; Breton, S. Genome Survey of the Freshwater Mussel Venustaconcha ellipsiformis (Bivalvia: Unionida) Using a Hybrid de novo Assembly Approach. Genome Biol. Evol. 2018, 10, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Petitpierre, E.; Gatewood, J.M.; Schmid, C.W. Satellite DNA from the Beetle Tenebrio molitor. Experientia 1988, 44, 498–499. [Google Scholar] [CrossRef]
- Meštrović, N.; Plohl, M.; Mravinac, B.; Ugarković, D. Evolution of Satellite DNAs from the Genus Palorus-Experimental Evidence for the “Library” Hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef]
- Gall, J.G.; Cohen, E.; Polan, M.L. Repetitive DNA Sequences in Drosophila. Chromosoma 1971, 33, 319–344. [Google Scholar] [CrossRef]
- Murgarella, M.; Puiu, D.; Novoa, B.; Figueras, A.; Posada, D.; Canchaya, C. A First Insight into the Genome of the Filter- Feeder Mussel Mytilus galloprovincialis. PLoS ONE 2016, 11, e0151561. [Google Scholar] [CrossRef] [Green Version]
- Uliano-Silva, M.; Dondero, F.; Dan Otto, T.; Costa, I.; Lima, N.C.B.; Americo, J.A.; Mazzoni, C.J.; Prosdocimi, F.; Rebelo, M.d.F. A Hybrid-Hierarchical Genome Assembly Strategy to Sequence the Invasive Golden Mussel, Limnoperna fortunei. Gigascience 2018, 7, gix128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaiero, P.; Vaio, M.; Peters, S.A.; Schranz, M.E.; De Jong, H.; Speranza, P.R. Comparative Analysis of Repetitive Sequences among Species from the Potato and the Tomato Clades. Ann. Bot. 2019, 123, 521–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñaloza, C.; Gutierrez, A.P.; Eory, L.; Wang, S.; Guo, X.; Archibald, A.L.; Bean, T.P.; Houston, R.D. A Chromosome-Level Genome Assembly for the Pacific Oyster (Crassostrea gigas). Gigascience 2021, 10, giab020. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Pritham, E.J. Helitrons, the Eukaryotic Rolling-Circle Transposable Elements. Microbiol. Spectr. 2015, 3, 3–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, W.; Dooner, H.K.; Du, C. Rolling-Circle Amplification of Centromeric Helitrons in Plant Genomes. Plant J. 2016, 88, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Tunjić-Cvitanić, M.; Pasantes, J.J.; García-Souto, D.; Cvitanić, T.; Plohl, M.; Šatović-Vukšić, E. Satellitome Analysis of the Pacific Oyster Crassostrea gigas Reveals New Pattern of Satellite DNA Organization, Highly Scattered across the Genome. Int. J. Mol. Sci. 2021, 22, 6798. [Google Scholar] [CrossRef] [PubMed]
- Kourtidis, A.; Drosopoulou, E.; Pantzartzi, C.N.; Chintiroglou, C.C.; Scouras, Z.G. Three New Satellite Sequences and a Mobile Element Found inside HSP70 Introns of the Mediterranean Mussel (Mytilus galloprovincialis). Genome 2006, 49, 1451–1458. [Google Scholar] [CrossRef] [PubMed]
- Šatović, E.; Plohl, M. Tandem Repeat-Containing MITE Elements in the Clam Donax trunculus. Genome Biol. Evol. 2013, 5, 2549–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vojvoda Zeljko, T.; Pavlek, M.; Meštrović, N.; Plohl, M. Satellite DNA-like Repeats Are Dispersed throughout the Genome of the Pacific Oyster Crassostrea gigas Carried by Helentron Non-Autonomous Mobile Elements. Sci. Rep. 2020, 10, 15107. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-Based Web Server for Genome-Wide Characterization of Eukaryotic Repetitive Elements from Next-Generation Sequence Reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tunjić Cvitanić, M.; Vojvoda Zeljko, T.; Pasantes, J.J.; García-Souto, D.; Gržan, T.; Despot-Slade, E.; Plohl, M.; Šatović, E. Sequence Composition Underlying Centromeric and Heterochromatic Genome Compartments of the Pacific Oyster Crassostrea gigas. Genes 2020, 11, 695. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.S.M.L.; Heringer, P.; Guilherme, B.D.; Svartman, M.; Kuhn, G.C.S. De Novo Identification of Satellite DNAs in the Sequenced Genomes of Drosophila virilis and D. Americana Using the RepeatExplorer and TAREAN Pipeline. PLoS ONE 2019, 14, e0223466. [Google Scholar] [CrossRef] [Green Version]
- Montiel, E.E.; Panzera, F.; Palomeque, T.; Lorite, P.; Pita, S. Satellitome Analysis of Rhodnius Prolixus, One of the Main Chagas Disease Vector Species. Int. J. Mol. Sci. 2021, 22, 6052. [Google Scholar] [CrossRef] [PubMed]
- Bouilly, K.; Chaves, R.; Leitao, A.; Benabdelmouna, A.; Guedes-Pinto, H. Chromosomal Organization of Simple Sequence Repeats in Chromosome Patterns. J. Genet. 2008, 87, 119–125. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šatović-Vukšić, E.; Plohl, M. Classification Problems of Repetitive DNA Sequences. DNA 2021, 1, 84-90. https://doi.org/10.3390/dna1020009
Šatović-Vukšić E, Plohl M. Classification Problems of Repetitive DNA Sequences. DNA. 2021; 1(2):84-90. https://doi.org/10.3390/dna1020009
Chicago/Turabian StyleŠatović-Vukšić, Eva, and Miroslav Plohl. 2021. "Classification Problems of Repetitive DNA Sequences" DNA 1, no. 2: 84-90. https://doi.org/10.3390/dna1020009
APA StyleŠatović-Vukšić, E., & Plohl, M. (2021). Classification Problems of Repetitive DNA Sequences. DNA, 1(2), 84-90. https://doi.org/10.3390/dna1020009