Abstract
Background and Objectives: The cytokine IL-6, methyltransferase NSD2, pro-protein convertase Furin, and growth factor receptor IGF-1R are essential factors in the proliferation of cancer cells. These proteins are involved in the tumor process by generating several cell-signaling pathways. However, the interactions of these oncogenic biomarkers, Furin, IL-6, and NSD2, and their links with the inhibitor SERPINA-1 remain largely unknown. Materials and Methods: Cell proliferation is measured by colorimetric and enzymatic methods. The genetic expressions of SERPINA-1, Furin, IL-6, and NSD2 are measured by qRT-PCR, while the expression of IGF-1R on the cell surface is measured by flow cytometry. Results: The proliferation of cells overexpressing SERPINA-1 (JP7pSer+) is decreased by more than 90% compared to control cells (JP7pSer-). The kinetics of the gene expression ratios of Furin, IL-6, and NSD2 show an increase for 48 h, followed by a decrease after 72 h for the three biomarkers in JP7pSer+ cells compared to JP7pSer- cells. The expression of IGF-1R on the cell surface in both cell lines is low, with JP7pSer- cells expressing 1.33 times more IGF-1R than JP7pSer+ cells. Conclusions: These results suggest gene correlations of SERPINA-1 overexpression with decreased cell proliferation and modulation of gene expression of Furin, IL-6, and NSD2. This study should be complemented by molecular transcriptomic and proteomic experiments to better understand the interaction of SERPINA-1 with IL-6, Furin, and NSD2, and their effect on tumor progression.
1. Introduction
Cancer is a complex disease characterized by uncontrolled cell proliferation, driven by a combination of genetic, environmental, and epigenetic factors [1]. The cellular signaling pathways are central to these events, which regulate cell growth, survival, and differentiation [2,3]. These pathways are complex networks of proteins involved in genetic and epigenetic regulation, as well as in modulating the cellular environment, influencing fundamental processes such as apoptosis, cell proliferation, and tissue invasion [3,4,5]. Prominent signaling pathways frequently deregulated in cancer development include the PI3K/AKT/mTOR, MAPK/ERK, and JAK/STAT pathways [6,7,8,9].
These pathways are often abnormally activated by genetic mutations or aberrant activation mechanisms, leading to excessive stimulation of proliferation and inhibition of apoptosis mechanisms [10,11,12]. Indeed, the JAK/STAT pathway, when activated by IL-6, a cytokine implicated in several cancers [9], induces the activation of the transcription factor STAT3 in tumor cells. The activation of the NF-κB and STAT3 signaling pathways promotes resistance to apoptosis, angiogenesis, and metastasis of tumor cells [13,14]. Furthermore, IL-6 contributes to the resistance of tumor cells to treatment, such as chemotherapy, by promoting survival and invasion and by inhibiting apoptosis of malignant cells [9,15].
IL-6 plays a role in modulating the tumor microenvironment, promoting inflammation and tumor growth, such as in multiple myeloma [16,17]. Song and colleagues demonstrated that NSD2 can methylate and activate STAT3, creating a potential positive feedback loop that amplifies IL-6 signaling [18]. NSD2 is a methyltransferase that plays a key role in the regulation of gene expression by methylating histones. This enzyme is responsible for the catalysis of mono- and di-methylation of lysine 36 on histone H3 [19]. This histone modification can recruit or exclude other chromatin-associated proteins, leading to either activation or repression of gene expression, depending on the cellular context [20]. NSD2 expression is frequently elevated in various types of cancers [18,19,20,21]. The evolution of cancer involves complex biological events that depend on the activity of various enzymes. Among these, proprotein convertases, particularly Furin, are involved in the maturation of numerous precursor proteins into their active forms, thereby promoting tumor progression [22,23]. Furin, a subtilisin-like protease, cleaves precursor proteins at the consensus site of Arg—X—Lys/Arg—Arg or Arg—X—X—Arg, facilitating their maturation [24,25,26].
Furin is involved in the maturation of precursors such as pro-TNF-α, pro-TGF-β1, and pro-IGF-1R, as well as various growth factors. These molecules play important roles in diverse cellular processes, including inflammation, growth, and angiogenesis, which are frequently deregulated in malignant cells [26,27,28]. In the context of the tumor microenvironment, pro-inflammatory cytokines matured by Furin, such as pro-TNF-α and pro-TGF-β1, can induce the activation of IL-6 gene expression [29], which in turn promotes tumor growth, angiogenesis, and metastasis [13]. Indeed, IL-6 produced by tumor microenvironment cells induces the activation of the JAK/STAT3 pathway in tumor cells, stimulating their proliferation, survival, and invasion [9]. Furin, therefore, contributes to the regulation of several signaling pathways crucial for tumor progression, including the maturation of the pro-IGF-1R precursor into IGF-1R, enabling its expression on the cell surface [30,31]. By interacting with its ligands IGF1 and IGF2, IGF-1R activates cell proliferation and promotes tumor progression [32]. Furin is often overexpressed in various carcinomas, suggesting its potential role in the survival and proliferation of tumor cells [26].
However, the involvement of Furin in different cancers, such as malignant hematological diseases, remains largely unknown, particularly regarding its potential interactions with IL-6 and NSD2. Studies on the links between these markers would help identify common mechanistic pathways and specificities in different cancer pathologies. To date, no study has established a definitive mechanistic link between these three players (Furin, IL-6, and NSD2) in cancer, which limits the understanding of their synergies or common regulatory pathways. In a proliferative hetero-hybrid lymphoblastoid cell model, an in vitro approach involves verifying whether there are significant correlations between the expression of Furin, IL-6, and NSD2 genes under the effect of overexpression of the SERPINA-1 gene, a serine protease inhibitor, notably neutrophil elastase, and a potent anti-inflammatory agent [33,34].
2. Materials and Methods
2.1. Maintenance of the JP7 Cell Line
The JP7 cell line, established in our laboratory according to the protocol described by Tissent and al. [35], originated from the transformation of human lymphocytes by the Epstein-Barr virus. These transformed lymphocytes were then stabilized by fusion with the murine X63Ag8 myeloma cell line derived from BALB/c mice. The resulting hetero-lymphoblastoid cells are maintained in continuous culture through successive passages in RPMI 1640 culture medium supplemented with 10% fetal bovine serum (FBS), 200 mM glutamine, and 100 IU/mL penicillin and streptomycin. Cultures are maintained in sterile containers and incubated at 37 °C in a humidified atmosphere with 5% CO2.
2.2. Establishment, Identification, and Selection of JP7pSer+ and JP7pSer- Cell Lines
JP7 cells were cultured in a 6-well plate at a density of 5 × 105 cells/well in 3 mL of culture medium. After 24 h, the cells were washed and supplemented with 1 mL/well of culture medium containing 1 µg of either the pCMV6-SERPINA1-GFP or pCMV6-GFP plasmid vector, along with 8 µL of transfection reagent (CANFAST, Canvaxbiotech; Valladolid, Spain). Cells transfected with pCMV6-SERPINA1-GFP were designated JP7pSer+, while cells transfected with pCMV6-GFP were designated JP7pSer-. Following a 5-h incubation period, the cells were washed by centrifugation with RPMI medium without FBS and returned to culture as described above, with the addition of the selective antibiotic G418 at 400 µg/mL. After 48 h, the cells were harvested for downstream assays. Cell condition and proliferation were monitored by microscopy (EVOS™ FL Digital Inverted Fluorescence Microscope). Under these selective conditions, only transfected cells that had integrated the G418 resistance gene and expressed green fluorescent protein (GFP) were able to proliferate, as indicated by green fluorescence. Transfection efficiency was assessed by determining the percentage of fluorescent cells using microscopy (EVOS™ FL Digital Inverted Fluorescence Microscope) at 200-x magnification, and SERPINA1 gene expression was confirmed by qRT-PCR.
2.3. Evaluation of the Effect of Serpina-1 Gene Transfection on Cell Lines in Culture
2.3.1. Cell Proliferation
The colorimetric study using Trypan blue exclusion, 20 × 103 cells/well were seeded into six 96-well plates. For the enzymatic study employing MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) (Toronto Research Chemicals; Vaughan, ON, Canada), 5 × 103 cells/well were seeded into three 96-well plates. The JP7pSer+ and JP7pSer- cells were cultured for 6 days under standard conditions as previously described, but without the selective G418. The culture medium was not renewed during this period. Cell viability and proliferation were assessed daily and in parallel using both the Trypan blue exclusion and MTT assays. All tests were performed in triplicate across three independent experiments. The percentage of viable cells, as determined by Trypan blue exclusion, was quantified by microscopy using a Malassez counting chamber. The enzymatic activity of the cells was evaluated by spectrophotometric measurement at 570 nm, which quantifies the optical density of the formazan product resulting from the reduction of MTT by succinate dehydrogenase, a mitochondrial metabolic enzyme.
2.3.2. Gene Expression of SERPINA-1, Furin, IL-6 and NSD2
The total RNA from the cell lines was extracted using the kit (PRImeZOL RNA Isolation, Canvaxbiotech; Valladolid, Spain), in accordance with the manufacturer’s instructions, and quantified by spectrophotometry (MaestroNano, MaestroGen, Hsinchu City, Taiwan). A quantity of 250 ng/μl of RNA was used for each qRT-PCR using the kit one-step (qMAXSen, Canvaxbiotech; Valladolid, Spain). The specific primers used for the genes (Table 1), provided and reconstituted according to the manufacturer’s recommendations (NeoBiotech; Nanterre—France). Amplification in 35 cycles was performed in a thermocycler (BIO-RAD CFX 96, C1000) calibrated for SYBR Green fluorescence quantification. The qRT-PCR experiments were performed in triplicate daily for 5 days. The results obtained were normalized separately to the human beta-actin reference gene according to the Livak method before calculating the means of the observed expression ratios each day [36].

Table 1.
Primers used during one-step qRT-PCR for Furin, IL-6, NSD2, and Beta-actin genes.
2.3.3. Membrane Expression of the IGF-1 Receptor
The membrane expression of IGF-1R was measured by flow cytometry (BD FACS Lyric Flow Cytometry System, BD Biosciences). For this purpose, the JP7pSer+ and JP7pSer- cells, suspended at 30 × 103/100 µL in RPMI-1640 medium without additives, were incubated in 5 mL tubes with 10 µL of the anti-CD221-PE monoclonal antibody (Clone IH7, Origène Technologie; Rockville, MD, USA) or the non-specific anti-IgG1-PE isotype antibody (Clone HP6001, BD Pharmingen; San Diego, CA, USA). After 30 min of incubation in the dark at room temperature, the cells were washed 3 times by centrifugation with RPMI-1640 medium and then suspended for flow cytometer analysis.
2.4. Statistical Analysis
The normality of the distribution of values was verified by the Shapiro-Wilcoxon test. Statistical differences according to the t test, Welch, and Fisher tests were verified using GraphPad Prism8 software (version 8.0.2).
3. Results
3.1. Establishment, Identification, and Selection of JP7pSer+ and JP7pSer- Cell Lines
The selection of transfected cell lines was based on observations from both fluorescence microscopy and the quantification of mRNA for A1AT. Culture wells containing fluorescent cells at greater than 95% and an SERPINA-1 gene mRNA level at least equal to 1 relative to the Beta-actin reference gene were selected.
3.1.1. Microscopy
To evaluate the stability of the JP7pSer- and JP7pSer+ model lines, the cells were observed under a fluorescence microscope and digitized to have more than 95% of the fluorescent cells, in order to perform the genetic expression and proliferation tests.
After 48 h of culture with selective agent G418 to generate stable cell lines, more than 95% of JP7pSer+ and JP7pSer- cells emitted green fluorescence under excitation at 490 nm (blue light). These results show that JP7pSer+ are fluorescent and express GFP; this expression is indicative of A1AT expression in the JP7pSer+ line because the same cytomegalovirus promoter governs both the SERPINA-1 and GFP genes (Figure 1).
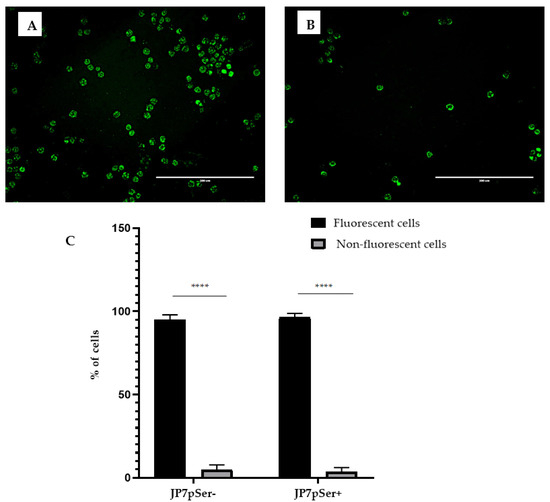
Figure 1.
JP7pSer- (A) and JP7pSer+ (B) cells observed under a microscope at 200× magnification. The majority of the cells under 490 nm excitation emit the green fluorescence of the GFP protein. Scale 200 µm. (C) Graphical representation of the percentages of fluorescent and non-fluorescent cells in the two cell lines JP7pSer- and JP7pSer+ treated by ImageJ. Data are expressed as mean ± standard deviation of at least three independent experiments, and significant differences between the different groups in each cell line are indicated by **** p-value < 0.0001.
3.1.2. qRT-PCR
qRT-PCR analysis was performed on several wells of cells to measure SERPINA-1 gene expression in both JP7pSer+ and JP7pSer- cell clones in order to confirm SERPINA-1 overexpression by JP7pSer+ cells. Beta-actin was used as the housekeeping gene in this experiment.
The average expression of the SERPINA-1 gene in JP7pSer- cells is 1.2, while that of JP7-pSer+ cells is 226, relative to the expression of the Beta-actin housekeeping gene (Figure 2). These results therefore confirm the overexpression of the SERPINA-1 in the JP7pSer+ cell line.
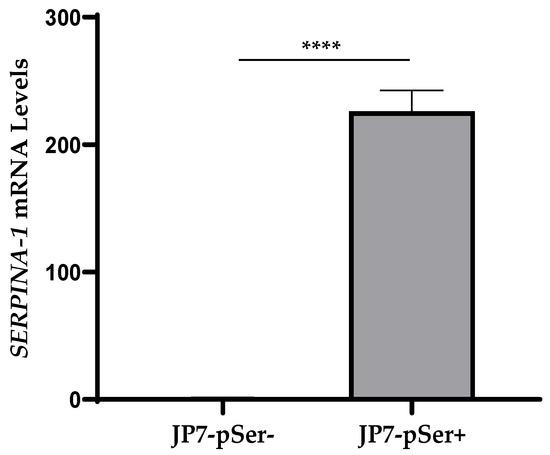
Figure 2.
Relative expression of the SERPINA-1 gene in JP7pSer- and JP7pSer+ cells referred to the expression of Beta-actin housekeeping gene. Data are mean ± SD of at least three independent experiments, and significant differences between the two cell lines were shown as **** p-value < 0.0001.
3.2. Evaluation of the Effect of SERPINA-1 Gene Transfection on Cultured Cell Lines
3.2.1. Cell Proliferation
The proliferation test was designed to determine whether the proliferative capacity of JP7pSer+ cells overexpressing SERPINA-1 is lower than that of JP7pSer- cells.
To confirm this, two proliferation tests were used: a colorimetric method based on counting viable cells with trypan blue (Figure 3A), and an enzymatic method by MTT based on the metabolic activity of the cells (Figure 3B).
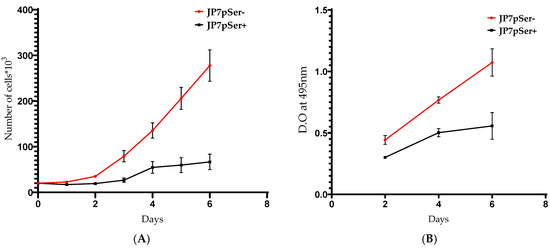
Figure 3.
(A) The proliferation of 20 × 103 cells/well of JP7pSer+ or JP7pSer- evaluated by the colorimetric Trypan blue method. (B) The proliferation of 5 × 103 cells/well of the two cell lines, measured by enzymatic method (MTT). O.D: Optical density. Data are mean ± SD of at least three independent experiments and significant differences between the two cell lines, with p < 0.05 for both experiments.
The colorimetric proliferation assay of JP7pSer+ cells shows the same trends, with a decrease in proliferation of more than 64% in 4 days and more than 91% after 6 days (Figure 3A). The proliferation measured by MTT of JP7pSer+ cells decreases by 38% in 4 days and by more than 85% in terms of 6 days compared to JP7pSer- cells (Figure 3B).
3.2.2. Kinetics of Furin, IL-6, and NSD2 Gene Expression
The kinetic study of the expression by qRT-PCR of Furin, IL-6, and NSD2 was designed to observe the inhibitor effect of the SERPINA-1 gene on JP7pSer+ cells compared to JP7pSer- cells. These gene expressions of Furin, IL-6, and NSD2 were calculated using the Livak and Schmittgen method [36], relative to the expression of the Beta actin gene.
The qRT-PCR analysis revealed that the JP7pSer+/JP7pSer- ratio of Furin gene expression was 4.88 on the 1st day and became 1.01 on the 3rd day, decreasing to 0.37 on the 6th day of culture. However, the same cellular ratio of IL-6 gene expression was 3.07 on the 1st day and decreased to 0.74 on the 3rd day, reaching 0.47 on the 6th day. As for NSD2 expression, the JP7pSer+/JP7pSer- ratio was 1.59 on the 1st day and decreased to 0.7 on the 3rd day, continuing to decrease to 0.51 on the 6th day (Figure 4). Three experiments were conducted to study the effect of the SERPINA-1 gene over-expression on the expression kinetics of the three genes encoding Furin, IL-6, and NSD2. All three experiments yielded reproducible and highly similar results.
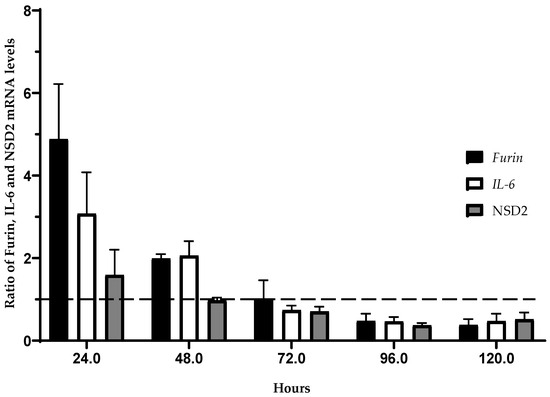
Figure 4.
Kinetics of the Furin, IL-6, and NSD2 gene expression measured by qRT-PCR in the JP7pSer+ line compared to that of the control line JP7pSer-. Data are mean ± SD of at least three independent experiments. Furin: Y = −0.04386X + 4.908; R2 = 0.7251; r = −0.85 ∈ CI95: [−1; −0.6019] p = 0.0001. IL-6: Y = −0.02834X + 3.404; R2 = 0,7463; r = −0.86 ∈ CI95: [−1; −0.7463 ] p = 0.0001. NSD2: Y = −0.01150X + 1.661; R2 = 0.6295; r = −0.79 ∈ CI95: [−1; −0.7071] p = 0.0004.
These results indicate an influence of SERPINA-1 gene over-expression on the kinetics of Furin, IL-6, and NSD2 expression, with an initial increase in expression followed by a progressive decrease.
3.2.3. Membrane Expression of the IGF-1 Receptor (IGF-1R, CD221)
This section will likely examine the inhibitor effect of the SERPINA-1 gene on the surface expression of the Insulin-like Growth Factor 1 Receptor (IGF-1R), also known as CD221.
Cytofluorimetric analysis using anti-CD221-PE showed a near-absence of labelling in JP7 cells. The JP7pSer- and JP7pSer+ cell lines showed low and nearly similar percentages with JP7pSer- (3.9%) vs. JP7pSer+ (3.46%), t-test = 1.2 with a p-value = 0.22 > 0.05, not significant. The average fluorescence intensity over three measurements for each cell line shows that JP7pSer- cells have an average MFI = 278.33 AU, while for JP7pSer+, the average MFI = 209.33 AU, with different confidence intervals CI99: [272.48; 284.17] for JP7pSer- and CI99: [177.31; 241.34] for JP7pSer+. The p-value < 0.01 with a significance threshold of 0.05. These results indicate that CD221 expression is globally very low in both JP7pSer- and JP7pSer+ cell lines, but with a 25% decrease in JP7pSer+ compared to JP7pSer- (Figure 5E).
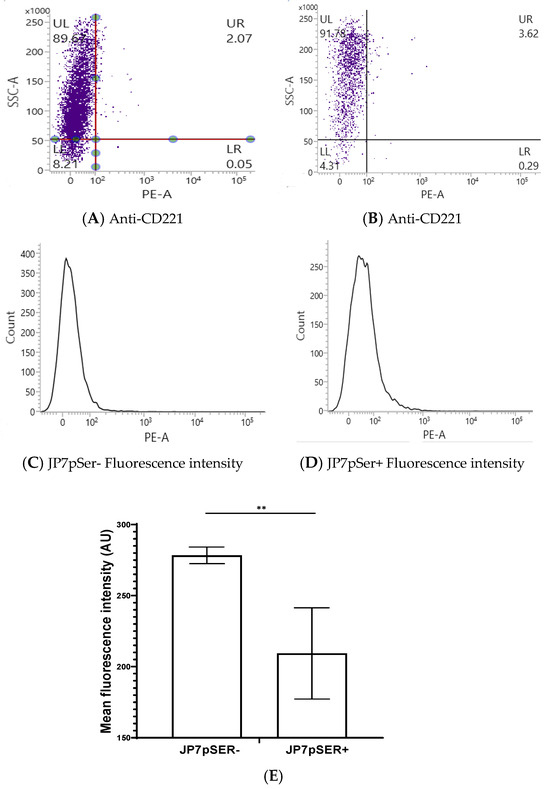
Figure 5.
Graphs of one of the three flow cytometry measurements analysis of membrane expression of IGF-1R by JP7pSer- (A) vs. JP7pSer+ (B) cells, and fluorescence intensity of JP7pSer- (C) vs. JP7pSer+ (D). Mean fluorescence intensity (MFI) was calculated from the three measurements (E), and the average MFI was 278.33 AU for JP7pSer- cells with a CI99: [272.48; 284.17], and an average MFI of 209.33 for JP7pSer+ cells with a CI99: [177.31; 241.34]. The ** p-value < 0.01, with a significance threshold of 0.05.
4. Discussion
The proliferative JP7 cell line, derived from a fusion between a human EBV-transformed lymphocyte and a murine myeloma, has been used as a malignant lymphoblastoid model comparable to Burkitt’s lymphoma [37] to study the effects of SERPINA-1 gene expression on cell proliferation and the expression of Furin, IL-6, and NSD2 biomarkers. JP7pSer+ cells, overexpressing SERPINA-1, and JP7pSer- control cells were generated using the vector under the control of the cytomegalovirus promoter. This approach allowed us to isolate the effect of SERPINA-1 overexpression in JP7pSER+ cells, while maintaining identical experimental conditions for both cell lines. This model helps to study the effect of SERPINA-1 expression on various cellular processes. Alpha-1 antitrypsin (A1AT), a 52 kDa glycoprotein, belongs to the serine protease inhibitor superfamily and is encoded by the SERPINA-1 locus, also known as the Pi locus, located on the long arm of chromosome 14 [38]. However, studies show the involvement of A1AT in tumors such as non-small cell lung cancer [33]. Cancer cells exhibit variable expression of the SERPINA-1 gene, often resulting in limited production of endogenous A1AT protein [39]. This suggests the potential role of A1AT in intracellular signaling pathways or other biological processes within tumor cells [40]. However, in tumor contexts, serine proteases, which can be regulated by A1AT, could modulate the mobilization of hematopoietic progenitor cells into the blood [41]. Current results have revealed that overexpression of the SERPINA-1 gene resulted in slower proliferation of JP7pSer+ cells compared to JP7pSer- cells.
This observation suggests that A1AT may possess anti-proliferative properties in this Burkitt’s lymphoma comparable model, which contradicts previous studies that implicate A1AT in tumor progression in other cancer types. The sustained anti-proteolysis properties of A1AT are important given its physiological role [42]. In this context, our results support a potential role for A1AT in regulating the growth and proliferation of malignant lymphoid cells. Given the well-established role of Furin in cancer progression, we evaluated the impact of SERPINA-1 overexpression on Furin gene expression. Furin, a proprotein convertase, plays a critical role in the activation of various proteins involved in cell growth, angiogenesis, and metastasis [43]. Our results revealed that JP7pSer+ cells exhibited a significant decrease in Furin gene expression compared to JP7pSer- cells after three days of culture. These results suggest that A1AT may exert its anti-proliferative effects by modulating Furin expression, which could affect the activation of downstream proteins that are involved in cancer progression. Therefore, further research is needed to identify the precise molecular mechanisms by which A1AT regulates Furin gene expression and to evaluate the potential therapeutic significance of this process in Burkitt’s lymphoma. However, in the JP7pSer- and JP7pSer+ cell lines, the expression of Furin substrate IGF-1R [27] is present in a minority of cells in both lines, suggesting that this signaling pathway may not play a major role in this cell model. In contrast, in the JP7pSer+ cell line, the mean fluorescence intensity of IGF-1R expression was reduced by 67%, suggesting that SERPINA-1 overexpression even decreases IGF-1R expression on the cell surface. Thus, this reduction in IGF-1R expression on the surface of JP7pSer+ cells provides an indirect argument in favor of the biological activity of A1AT, demonstrating not only its impact on a specific signaling pathway but also on cell proliferation as assessed by enzymatic and colorimetric methods. Other studies show a link between Furin and the IGF-1R axis [31,44]. It is well established that a functional IGF-1R is required for cell growth and plays a crucial role in the survival of various transformed cells in vitro and in vivo [44]. In addition, in the current study, we evaluated the impact of SERPINA-1 overexpression on the expression of IL-6, a cytokine known to promote the growth and survival of Burkitt’s lymphoma cells [45]. Interleukin-6, a pleiotropic cytokine, plays a crucial role in various biological processes, including immune regulation, inflammation, and tumor growth. IL-6 can promote tumor cell proliferation, survival, and angiogenesis, thereby contributing to cancer progression [13]. IL-6 can promote tumor growth via an auto-amplification loop for Th17 cells in the tumor microenvironment. These results suggest that the role of IL-17 in cancer is context- and system-dependent, like many other cytokines such as TNF-α [13]. In contrast, A1AT has demonstrated its ability to interact directly with cytokines such as TNF-α or IL-1 in inflammatory cells, thereby modulating their actions [46]. The latter are pro-inflammatory cytokines produced in response to various stimuli such as infections or tissue damage and stimulate the production of IL-6 [47]. In this study, we observed that SERPINA-1 gene overexpression resulted in a significant decrease in IL-6 gene expression in JP7pSer+ cells compared to JP7pSer- cells. Current observations suggest that A1AT could potentially regulate IL-6 gene expression in Burkitt’s lymphoma comparable model cells, thus offering insight into its potential role in controlling the tumor microenvironment. As for the methyltransferase NSD2/MMSET, it is an enzyme responsible for H3K36 methylation and is involved in various cellular processes, including DNA repair and transcription regulation [48]. Importantly, the study observed a decrease in methyltransferase NSD2/MMSET expression in JP7pSer+ cells compared to JP7pSer- cells. Although the essential role of NSD2 in JAK-STAT signaling via IL-6 activation has been well established, it is also responsible for cancer cell proliferation, which can be modulated with available treatments [49]. These results suggest that A1AT may influence the expression of methyltransferases, potentially affecting epigenetic regulation.
The link between IL-6 and Furin is established via the cytokines TGF-β1, IL-1β, and TNF-α, which are substrates of Furin and major activators of IL-6 expression. Furthermore, peritoneal macrophages from Furin knockout mice have shown reproducible positive regulation of numerous expressed genes, such as Serpinb1a, IL-6, and IL-1β [9,43]. Further studies are needed to further characterize the molecular mechanisms underlying these regulations, providing comprehensive information on the impact of A1AT on various cell-signaling pathways and its potential effects on cancer development and progression.
5. Conclusions
Considering all these results, our study provides evidence that SERPINA-1 overexpression may exert anti-proliferative effects on Burkitt’s lymphoma model cells by modulating the expression of various key proteins involved in cancer progression. More specifically, we observed that SERPINA-1 overexpression resulted in decreased expression of Furin, IL-6, and NSD2, concomitant factors involved in tumor cell proliferation, survival, and epigenetic regulation. Overall, these results suggest that A1AT could potentially serve as a therapeutic target for the treatment of Burkitt’s lymphoma, thus warranting further research, proteomic, epigenetic, and transcriptomic, to fully elucidate its mechanisms of action and evaluate its efficacy in preclinical and clinical studies.
Author Contributions
Conceptualization, N.T., A.T. and N.H.; Methodology, N.T. and N.H.; Validation, A.T. and N.H.; Formal analysis, N.T. and N.H.; Resources, N.T. and H.A.; Data curation, N.T.; Writing—original draft, N.T.; Writing—review & editing, N.T. and N.H.; Visualization, N.T. and A.T.; Supervision, N.H.; Project administration, N.H.; Funding acquisition, H.A. and N.H. All authors have read and agreed to the published version of the manuscript.
Funding
The National Blood Transfusion Center of Morocco and the Laboratory of Cellular and Molecular Inflammatory, Degenerative, and Oncological Pathophysiology (LPCMIDO) funded this research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| IGF-1 | Insulin-like growth factor-1 |
| PI3K | Phosphatidylinositol 3-kinase |
| AKT | Protein kinase B |
| mTOR | Mammalian target of rapamycin |
| MAPK | Mitogen-activated protein kinase |
| ERK | Extracellular signal-regulated kinases |
| JAK | Janus kinases |
| STAT | Signal transducer and activator of transcription proteins |
| IGF-1R | Insulin-like growth factor-1 receptor |
| IL-6 | Interleukin 6 |
| TNF-α | Tumor necrosis factor |
| NSD2 | Probable histone-lysine N-methyltransferase |
| EBV | Epstein-Barr virus |
| STAT3 | Signal transducer and activator of transcription 3 |
| Lys | Lysine |
| Arg | Arginine |
| TGF-β1 | Transforming growth factor beta 1 |
| PC | Proprotein convertase |
| A1AT | Alpha-1 antitrypsin |
| Alpha- 1PDX | Alpha-1 antitrypsin Portland variant |
| PACE4 | Proprotein convertase subtilisin/kexin type 6 |
| PC7 | Proprotein convertase subtilisin/kexin type 7 |
| CMV | Cytomegalovirus |
| GFP | Green fluorescent protein |
| mRNA | Messenger ribonucleic acid |
| qRT-PCR | Quantitative Reverse Transcription—Poly Chain Réaction |
| CI | confidence interval |
References
- Casotti, M.C.; Meira, D.D.; Zetum, A.S.S.; Campanharo, C.V.; da Silva, D.R.C.; Giacinti, G.M.; da Silva, I.M.; Moura, J.A.D.; Barbosa, K.R.M.; Altoé, L.S.C.; et al. Integrating frontiers: A holistic, quantum and evolutionary approach to conquering cancer through systems biology and multidisciplinary synergy. Front. Oncol. 2024, 14, 1419599. [Google Scholar] [CrossRef]
- Caloian, A.D.; Cristian, M.; Calin, E.; Pricop, A.-R.; Mociu, S.-I.; Seicaru, L.; Deacu, S.; Ciufu, N.; Suceveanu, A.-I.; Suceveanu, A.-P.; et al. Epigenetic Symphony in Diffuse Large B-Cell Lymphoma: Orchestrating the Tumor Microenvironment. Biomedicines 2025, 13, 853. [Google Scholar] [CrossRef]
- Dakal, T.C.; Dhabhai, B.; Pant, A.; Moar, K.; Chaudhary, K.; Yadav, V.; Ranga, V.; Sharma, N.K.; Kumar, A.; Maurya, P.K.; et al. Oncogenes and tumor suppressor genes: Functions and roles in cancers. MedComm 2024, 5, e582. [Google Scholar] [CrossRef]
- Wang, S.; Guo, S.; Guo, J.; Du, Q.; Wu, C.; Wu, Y.; Zhang, Y. Cell death pathways: Molecular mechanisms and therapeutic targets for cancer. MedComm 2024, 5, e693. [Google Scholar] [CrossRef]
- Yuan, S.; Zhang, P.; Zhang, F.; Yan, S.; Dong, R.; Wu, C.; Deng, J. Profiling signaling mediators for cell-cell interactions and communications with microfluidics-based single-cell analysis tools. iScience 2024, 28, 111663. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Tan, J. The Relationship between IGF Pathway and Acquired Resistance to Tyrosine Kinase Inhibitors in Cancer Therapy. Front. Biosci. 2023, 28, 163. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Du, R.; Liu, W.; Huang, G.; Dong, Z.; Li, X. PI3K/Akt/mTOR Signaling Pathway: Role in Esophageal Squamous Cell Carcinoma, Regulatory Mechanisms and Opportunities for Targeted Therapy. Front. Oncol. 2022, 12, 852383. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Hu, Z.; Xu, X.; Dai, X.; Liu, Z. Key signal transduction pathways and crosstalk in cancer: Biological and therapeutic opportunities. Transl. Oncol. 2022, 26, 101510. [Google Scholar] [CrossRef]
- Huang, B.; Lang, X.; Li, X. The role of IL-6/JAK2/STAT3 signaling pathway in cancers. Front. Oncol. 2022, 12, 1023177. [Google Scholar] [CrossRef]
- Derakhshani, A.; Rostami, Z.; Taefehshokr, S.; Safarpour, H.; Astamal, R.V.; Taefehshokr, N.; Alizadeh, N.; Argentiero, A.; Silvestris, N.; Baradaran, B. An Overview of the Oncogenic Signaling Pathways in Different Types of Cancers. Preprints 2020. [Google Scholar] [CrossRef]
- Aprile, M.; Cataldi, S.; Perfetto, C.; Federico, A.; Ciccodicola, A.; Costa, V. Targeting metabolism by B-raf inhibitors and diclofenac restrains the viability of BRAF-mutated thyroid carcinomas with Hif-1α-mediated glycolytic phenotype. Br. J. Cancer 2023, 129, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yi, T.; Kortylewski, M.; Pardoll, D.M.; Zeng, D.; Yu, H. IL-17 can promote tumor growth through an IL-6–Stat3 signaling pathway. J. Exp. Med. 2009, 206, 1457–1464. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Belka, M.; Papierska, K. Targeting STAT3 and NF-κB Signaling Pathways in Cancer Prevention and Treatment: The Role of Chalcones. Cancers 2024, 16, 1092. [Google Scholar] [CrossRef]
- Liu, J.; Geng, X.; Hou, J.; Wu, G. New insights into M1/M2 macrophages: Key modulators in cancer progression. Cancer Cell Int. 2021, 21, 389. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. Interleukin (IL-6) Immunotherapy. Cold Spring Harb. Perspect. Biol. 2017, 10, a028456. [Google Scholar] [CrossRef]
- Harmer, D.; Falank, C.; Reagan, M.R. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Front. Endocrinol. 2019, 9, 788. [Google Scholar] [CrossRef]
- Song, D.; Lan, J.; Chen, Y.; Liu, A.; Wu, Q.; Zhao, C.; Feng, Y.; Wang, J.; Luo, X.; Cao, Z.; et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene 2021, 40, 2952–2967. [Google Scholar] [CrossRef]
- Li, W.; Tian, W.; Yuan, G.; Deng, P.; Sengupta, D.; Cheng, Z.; Cao, Y.; Ren, J.; Qin, Y.; Zhou, Y.; et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 2020, 590, 498–503. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, M.W. Histone lysine methyltransferases as anti-cancer targets for drug discovery. Acta Pharmacol. Sin. 2016, 37, 1273–1280. [Google Scholar] [CrossRef]
- Gao, B.; Liu, X.; Li, Z.; Zhao, L.; Pan, Y. Overexpression of EZH2/NSD2 Histone Methyltransferase Axis Predicts Poor Prognosis and Accelerates Tumor Progression in Triple-Negative Breast Cancer. Front. Oncol. 2021, 10, 600514. [Google Scholar] [CrossRef]
- Mehranzadeh, E.; Crende, O.; Badiola, I.; Garcia-Gallastegi, P. What Are the Roles of Proprotein Convertases in the Immune Escape of Tumors? Biomedicines 2022, 10, 3292. [Google Scholar] [CrossRef] [PubMed]
- Poyil, P.K.; Siraj, A.K.; Padmaja, D.; Parvathareddy, S.K.; Diaz, R.; Thangavel, S.; Begum, R.; Haqawi, W.; Al-Mohanna, F.H.; Al-Sobhi, S.S.; et al. Overexpression of the pro-protein convertase furin predicts prognosis and promotes papillary thyroid carcinoma progression and metastasis through RAF/MEK signaling. Mol. Oncol. 2023, 17, 1324. [Google Scholar] [CrossRef] [PubMed]
- Cevenini, A.; Orrù, S.; Mancini, A.; Alfieri, A.; Buono, P.; Imperlini, E. Molecular Signatures of the Insulin-like Growth Factor 1-Mediated Epithelial-Mesenchymal Transition in Breast, Lung and Gastric Cancers. Int. J. Mol. Sci. 2018, 19, 2411. [Google Scholar] [CrossRef] [PubMed]
- Ianza, A.; Sirico, M.; Bernocchi, O.; Generali, D. Role of the IGF-1 Axis in Overcoming Resistance in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 641449. [Google Scholar] [CrossRef]
- Zhou, B.; Gao, S. Pan-Cancer Analysis of FURIN as a Potential Prognostic and Immunological Biomarker. Front. Mol. Biosci. 2021, 8, 648402. [Google Scholar] [CrossRef]
- Fu, J.; Bassi, D.E.; Zhang, J.; Li, T.; Nicolas, É.; Klein-Szanto, A.J. Transgenic Overexpression of the Proprotein Convertase Furin Enhances Skin Tumor Growth. Neoplasia 2012, 14, 271. [Google Scholar] [CrossRef][Green Version]
- Bassi, D.; Lopez De Cicco, R.; Zucker, S.; Thomas, G.; Klein-Szanto, A.J. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc. Natl. Acad. Sci. USA 2001, 98, 10326. [Google Scholar] [CrossRef]
- Seong, G.J.; Hong, S.; Jung, S.-A.; Lee, J.-J.; Lim, E.; Kim, S.-J.; Lee, J.H. TGF-beta-induced interleukin-6 participates in transdifferentiation of human Tenon’s fibroblasts to myofibroblasts. Mol. Vis. 2009, 15, 2123. [Google Scholar]
- Declercq, J.; Brouwers, B.; Pruniau, V.P.E.G.; Stijnen, P.; Tuand, K.; Meulemans, S.; Prat, A.; Seidah, N.G.; Khatib, A.-M.; Creemers, J.W.M. Liver-Specific Inactivation of the Proprotein Convertase FURIN Leads to Increased Hepatocellular Carcinoma Growth. BioMed Res. Int. 2015, 2015, 148651. [Google Scholar] [CrossRef]
- He, Z.; Khatib, A.; Creemers, J.W.M. Loss of Proprotein Convertase Furin in Mammary Gland Impairs proIGF1R and proIR Processing and Suppresses Tumorigenesis in Triple Negative Breast Cancer. Cancers 2020, 12, 2686. [Google Scholar] [CrossRef] [PubMed]
- Werner, H.; Meisel-Sharon, S.; Bruchim, I. Oncogenic fusion proteins adopt the insulin-like growth factor signaling pathway. Mol. Cancer 2018, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Ercetin, E.; Richtmann, S.; Delgado, B.M.; Gomez-Mariano, G.; Wrenger, S.; Korenbaum, E.; Liu, B.; DeLuca, D.; Kühnel, M.P.; Jonigk, D.; et al. Clinical Significance of SERPINA1 Gene and Its Encoded Alpha1-antitrypsin Protein in NSCLC. Cancers 2019, 11, 1306. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, G.; Arciniega, M.; Quezada, A. Specific and Selective Inhibitors of Proprotein Convertases Engineered by Transferring Serpin B8 Reactive-Site and Exosite Determinants of Reactivity to the Serpin α1PDX. Biochemistry 2019, 58, 1679. [Google Scholar] [CrossRef]
- Tissent, A.; Habti, N.; Sadiq, F.; El Amrani, N.; Benchemsi, N. Production d’un réactif monoclonal anti-B humain pour la détection des groupes sanguins ABO. Immuno-Anal. Biol. Spécialisée 2007, 22, 68–71. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kubacz, M.; Kusowska, A.; Winiarska, M.; Bobrowicz, M. In Vitro Diffuse Large B-Cell Lymphoma Cell Line Models as Tools to Investigate Novel Immunotherapeutic Strategies. Cancers 2022, 15, 235. [Google Scholar] [CrossRef]
- Balduyck, M.; Odou, M.-F.; Zerimech, F.; Porchet, N.; Lafitte, J.-J.; Maitre, B. Diagnosis of alpha-1 antitrypsin deficiency: Modalities, indications and diagnosis strategy. Rev. Des Mal. Respir. 2014, 31, 729–745. [Google Scholar] [CrossRef]
- Seixas, S.; Marques, P.I. Known Mutations at the Cause of Alpha-1 Antitrypsin Deficiency an Updated Overview of SERPINA1 Variation Spectrum. Appl. Clin. Genet. 2021, 14, 173–194. [Google Scholar] [CrossRef]
- Maslakova, A.A.; Telkov, M.V.; Orlovsky, I.V.; Sokolova, O.S. Comparative analysis of SERPINA1 gene expression in tumor cell lines. Mosc. Univ. Biol. Sci. Bull. 2015, 70, 127–131. [Google Scholar] [CrossRef]
- Stanke, F.; Janciauskiene, S.; Tamm, S.; Wrenger, S.; Raddatz, E.L.; Jonigk, D.; Braubach, P. Effect of Alpha-1 Antitrypsin on CFTR Levels in Primary Human Airway Epithelial Cells Grown at the Air-Liquid-Interface. Molecules 2021, 26, 2639. [Google Scholar] [CrossRef] [PubMed]
- Ostermann, L.; Maus, R.; Stolper, J.; Schütte, L.; Katsarou, K.; Tumpara, S.; Pich, A.; Mueller, C.; Janciauskiene, S.; Welte, T.; et al. Alpha-1 antitrypsin deficiency impairs lung antibacterial immunity in mice. J. Clin. Investig. 2021, 6, e140816. [Google Scholar] [CrossRef] [PubMed]
- Cordova, Z.M.; Grönholm, A.; Kytölä, V.; Taverniti, V.; Hämäläinen, S.; Aittomäki, S.; Niininen, W.; Junttila, I.; Ylipää, A.; Nykter, M.; et al. Myeloid cell expressed proprotein convertase FURIN attenuates inflammation. Oncotarget 2016, 7, 54392–54404. [Google Scholar] [CrossRef] [PubMed]
- Khatib, A.-M.; Siegfried, G.; Prat, A.; Luis, J.; Chrétien, M.; Metrakos, P.; Seidah, N.G. Inhibition of Proprotein Convertases Is Associated with Loss of Growth and Tumorigenicity of HT-29 Human Colon Carcinoma Cells. J. Biol. Chem. 2001, 276, 30686–30693. [Google Scholar] [CrossRef]
- Ma, Y.-C.; Fan, W.-J.; Rao, S.-M.; Gao, L.; Bei, Z.-Y.; Xu, S.-T. Effect of Furin inhibitor on lung adenocarcinoma cell growth and metastasis. Cancer Cell Int. 2014, 14, 43. [Google Scholar] [CrossRef][Green Version]
- Lockett, A.D.; Kimani, S.; Ddungu, G.; Wrenger, S.; Tuder, R.M.; Janciauskiene, S.M.; Petrache, I. α1-Antitrypsin Modulates Lung Endothelial Cell Inflammatory Responses to TNF-α. Am. J. Respir. Cell Mol. Biol. 2013, 49, 143–150. [Google Scholar] [CrossRef]
- Jain, S.; Gautam, V.; Naseem, S. Acute-phase proteins: As diagnostic tool. J. Pharm. Bioallied Sci. 2011, 3, 118–127. [Google Scholar] [CrossRef]
- Xie, Z.; Chooi, J.Y.; Toh, S.H.M.; Yang, D.; Basri, N.B.; Ho, Y.S.; Chng, W.J. MMSET I acts as an oncoprotein and regulates GLO1 expression in t(4;14) multiple myeloma cells. Leukemia 2018, 33, 739–748. [Google Scholar] [CrossRef]
- Vougiouklakis, T.; Hamamoto, R.; Nakamura, Y.; Saloura, V. The NSD Family of Protein Methyltransferases in Human Cancer. Epigenomics 2015, 7, 863–874. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).