Immunotherapy Potential of Animal-Sourced Probiotic Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolatation and Sequence Retrieval
2.2. Bacterial Toxin Investigation
2.3. Selection of Protein-Coding Hypothetical Proteins
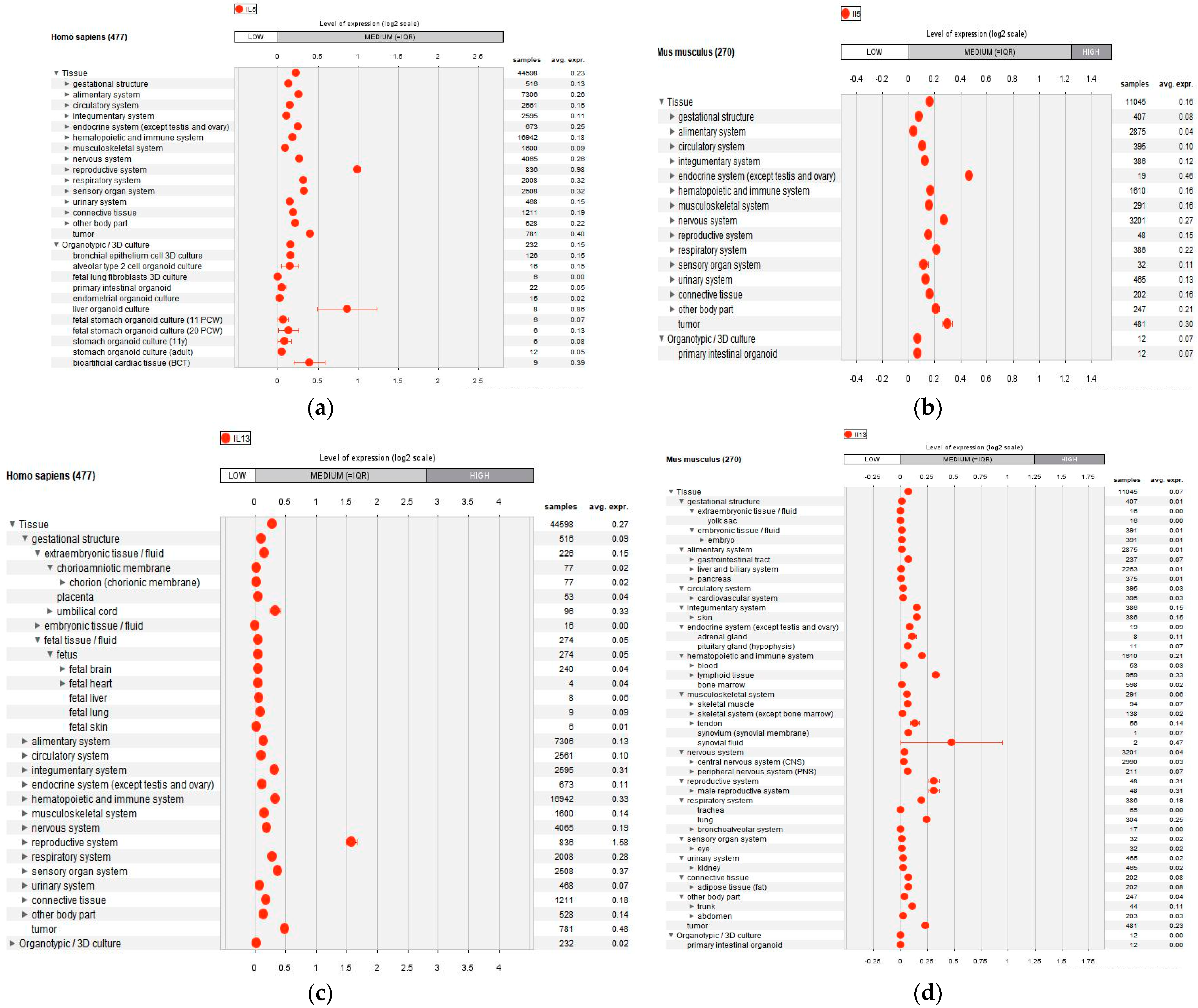
2.4. IL-5 and IL-13 Anaysis
2.5. Statistical Analysis and Gene Expression
3. Results
3.1. Probiotic Characteristics of the Genomes
3.2. Prediction of Bacterial Toxins
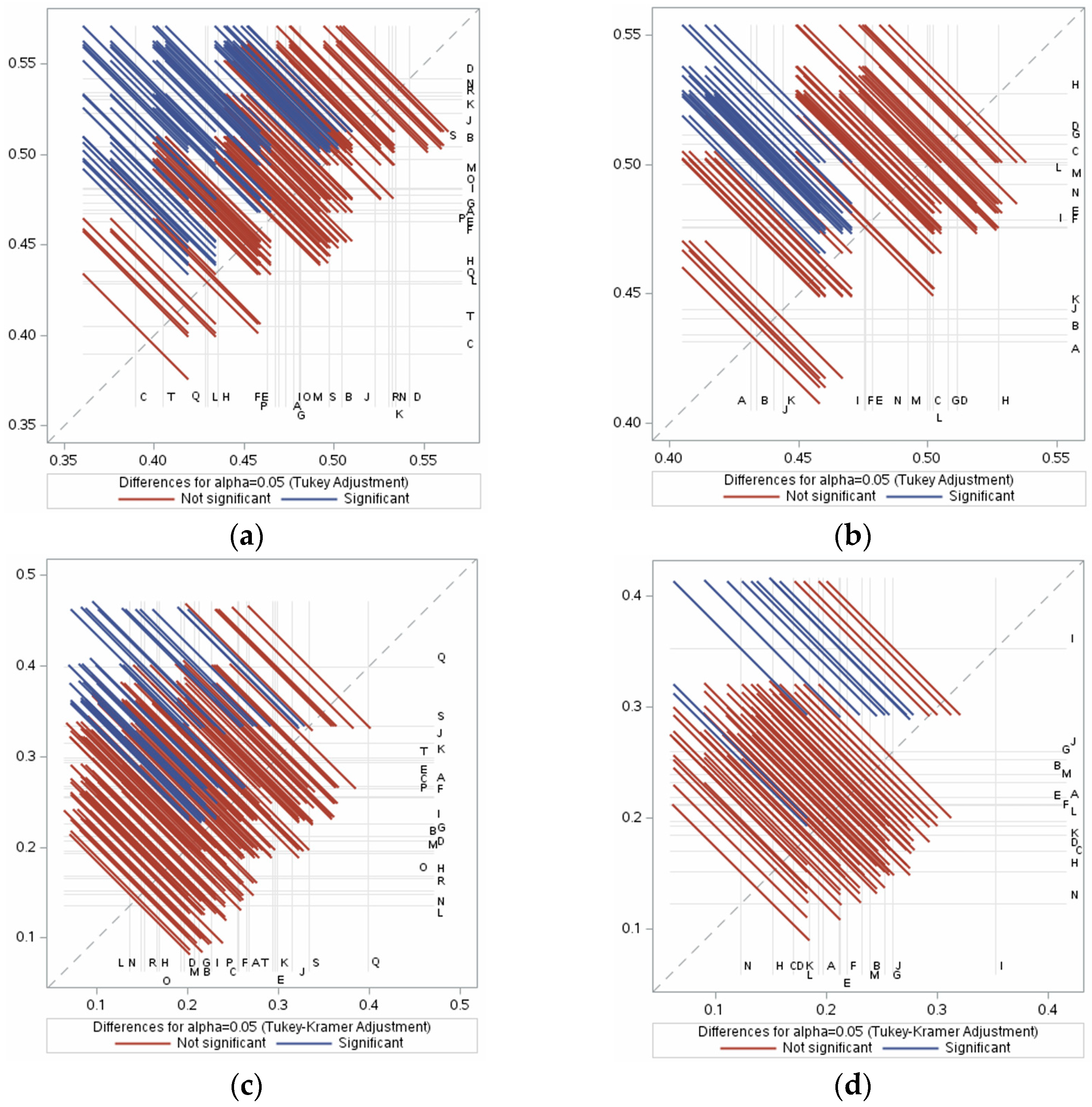
3.3. IL-5- and IL-13-Inducing Capacity
3.4. Immunogenic IL-5- and IL-13-Inducing Peptides
3.5. Physicochemical Properties
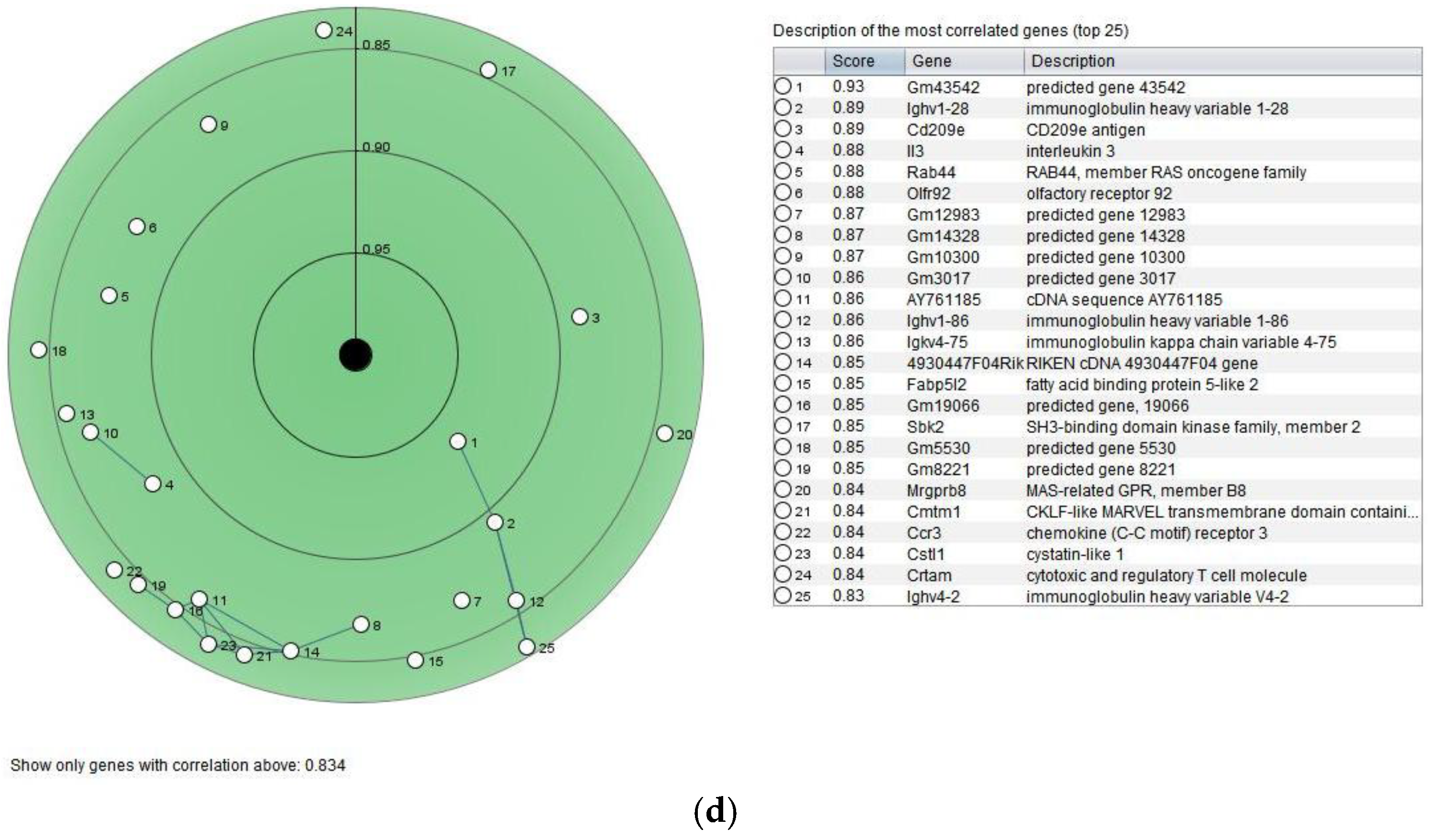
3.6. Gene Expression
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EC | extinction coefficient |
| II | instability index |
| AI | aliphatic index |
| IFN-γ | interferon-gamma |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| IL-3 | interleukin-3 |
| IL-4 | interleukin-4 |
| IL-5 | interleukin-5 |
| IL-13 | interleukin-13 |
| MW | molecular weight |
| NK | natural killer |
| pI | isoelectric point |
| Th2 | T helper 2 |
| NPL | non-probiotic lactobacillus |
| PL | probiotic lactobacillus |
| LAB | lactic acid bacteria |
References
- Avery, S.; Rothwell, L.; Degen, W.D.; Schijns, V.E.; Young, J.; Kaufman, J.; Kaiser, P. Characterization of the first nonmammalian T2 cytokine gene cluster: The cluster contains functional single-copy genes for IL-3, IL-4, IL-13, and GM-CSF, a gene for IL-5 that appears to be a pseudogene, and a gene encoding another cytokinelike transcript, KK34. J. Interferon Cytokine Res. 2004, 24, 600–610. [Google Scholar]
- Watanabe, T.; Terada, T.; Ezaki, R.; Matsuzaki, M.; Furusawa, S.; Horiuchi, H. Chicken Interleukin-5 is expressed in splenic lymphocytes and affects antigen-specific antibody production. J. Poult. Sci. 2024, 61, jpsa.2024002. [Google Scholar] [CrossRef] [PubMed]
- Ologhobo, A.D.; Akangbe, E.I.; Adejumo, I.O.; Adeleye, O. Effect of Moringa oleifera leaf meal as replacement for oxytetracycline on carcass characteristics of the diets of broiler chickens. Annu. Res. Rev. Biol. 2014, 4, 423–431. [Google Scholar] [CrossRef]
- Ologhobo, A.D.; Adejumo, I.O.; Akangbe, E.I. Comparison effect of Moringa oleifera leaf meal and oxytetracycline on haematology and serum biochemical profile of broiler finishers. Int. Blood Res. Rev. 2014, 2, 29–36. [Google Scholar] [CrossRef]
- Adejumo, I.O.; Adetunji, C.O.; Olopade, C.O.; George, K.O. Response of broilers to dietary Moringa oleifera leaf, raw and cooked seed meal and synthetic antibiotics. J. Exp. Agric. Int. 2016, 12, 1–7. [Google Scholar] [CrossRef]
- Odukoya, A.A.; Adejumo, I.O.; Ologhobo, A.D. Chemical constituents of crude extracts of Moringa oleifera (Lam) leaf and biochemical response of weanling Wistar rats administered crude extract. Niger. Agric. J. 2022, 53, 351–356. [Google Scholar]
- Adejumo, I.O. Hypothetical proteins of chicken-isolated Limosilactobacillus reuteri subjected to in silico analyses induce IL-2 and IL-10. Genes Nutr. 2024, 19, 24. [Google Scholar] [CrossRef]
- Adejumo, I.O.; Adebiyi, O.A. Peptides from hypothetical proteins of Lactobacillus acidophilus induce IL-4 and IL-10. Biomed. Res. Ther. 2025, 12, 1–16. [Google Scholar] [CrossRef]
- Adejumo, I.O.; Adebiyi, O.A. IL-4-inducing hypothetical proteins of chicken-isolated Limosilactobacillus reuteri. Biology 2025, in press.
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A machine learning platform for rapid identification of probiotic properties from whole-genome primary sequences. Brief. Bioinform. 2022, 23, 1–12. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. BTXpred: Prediction of Bacterial Toxins. Silico Biol. 2007, 7, 405–412. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P.S. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Dhall, A.; Patiyal, S.; Raghava, G.P. IL13Pred: A method for predicting immunoregulatory cytokine IL-13 inducing peptides. Comput. Biol. Med. 2022, 143, 105297. [Google Scholar] [CrossRef]
- Jain, S.; Dhall, A.; Patiyal, S.; Raghava, G.P.S. In Silico tool for identification, designing, and searching of IL13-inducing peptides in antigens. Methods Mol. Biol. 2023, 2673, 329–338. [Google Scholar]
- Naorem, D.N.; Sharma, N.; Raghava, G.P.S. A web server for predicting and scanning of IL-5 inducing peptides using alignment-free and alignment-based method. Comput. Biol. Med. 2023, 158, 106864. [Google Scholar] [CrossRef]
- Dimitrov, I.; Zaharieva, N.; Doytchinova, I. Bacterial immunogenicity prediction by machine learning methods. Vaccines 2020, 8, 709. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Stedman, A.; van Vliet, A.H.M.; Chambers, M.; Gutierrez-Merino, J. Gut commensal bacteria show beneficial properties as wildlife probiotics. Ann. N. Y. Acad. Sci. 2020, 1467, 112–132. [Google Scholar] [CrossRef]
- Zhai, Q.; Shen, X.; Cen, S.; Zhang, C.; Tian, F.; Zhao, J.; Zhang, H.; Xue, Y.; Chen, W. Screening of Lactobacillus salivarius strains from the feces of Chinese populations and the evaluation of their effects against intestinal inflammation in mice. Food Funct. 2020, 11, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Chaves, B.; Brashears, M.; Nightingale, K. Applications and safety considerations of Lactobacillus salivarius as a probiotic in animal and human health. J. Appl. Microbiol. 2017, 123, 18–28. [Google Scholar] [CrossRef]
- O’Hara, A.M.; O’Regan, P.; Fanning, A.; O’Mahony, C.; Macsharry, J.; Lyons, A.; Bienenstock, J.; O’MAhony, L.; Shanahan, F. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology 2006, 118, 202–215. [Google Scholar] [CrossRef]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prévost, H.; Connil, N.; Chobert, J.M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [CrossRef]
- Dobreva, L.; Atanasova, N.; Donchev, P.; Krumova, E.; Abrashev, R.; Karakirova, Y.; Mladenova, R.; Tolchkov, V.; Ralchev, N.; Dishliyska, V.; et al. Candidate-probiotic lactobacilli and their postbiotics as health-benefit promoters. Microorganisms 2024, 12, 1910. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Chen, H.; Yang, F.; Xu, B.; Wang, K.; Liu, Q.; Liang, G.; Zhang, R.; Jiao, X.; et al. Assessment of beneficial effects and identification of host adaptation-associated genes of Ligilactobacillus salivarius isolated from badgers. BMC Genom. 2023, 24, 530. [Google Scholar] [CrossRef]
- Stedman, A.; Maluquer de Motes, C.; Lesellier, S.; Dalley, D.; Chambers, M.; Gutierrez-Merino, J. Lactic acid bacteria isolated from European badgers (Meles meles) reduce the viability and survival of Bacillus Calmette-Guerin (BCG) vaccine and influence the immune response to BCG in a human macrophage model. BMC Microbiol. 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Li, Y.; Bumann, M.; Raftis, E.J.; Casey, P.G.; Cooney, J.C.; Walsh, M.A.; O’Toole, P.W. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Bacteriol. 2009, 191, 5743–5757. [Google Scholar] [CrossRef] [PubMed]
- Harris, H.M.B.; Bourin, M.J.B.; Claesson, M.J.; O’Toole, P.W. Phylogenomics and comparative genomics of Lactobacillus salivarius, a mammalian gut commensal. Microb. Genom. 2017, 3, e000115. [Google Scholar] [CrossRef]
- Guo, X.H.; Kim, J.M.; Nam, H.M.; Park, S.Y.; Kim, J.M. Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 2010, 16, 321–326. [Google Scholar] [CrossRef]
- Messaoudi, S.; Madi, A.; Prévost, H.; Feuilloley, M.; Manai, M.; Dousset, X.; Connil, N. In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe 2012, 18, 584–589. [Google Scholar] [CrossRef]
- Raftis, E.J.; Forde, B.M.; Claesson, M.J.; O’Toole, P.W. Unusual genome complexity in Lactobacillus salivarius JCM1046. BMC Genom. 2014, 15, 771. [Google Scholar] [CrossRef]
- Kang, C.H.; Han, S.H.; Kim, Y.; Paek, N.S.; So, J.S. In vitro probiotic properties of Lactobacillus salivarius MG242 isolated from human vagina. Probiotics Antimicrob. Proteins 2018, 10, 343–349. [Google Scholar] [CrossRef]
- Kang, M.S.; Lim, H.S.; Oh, J.S.; Lim, Y.J.; Wuertz-Kozak, K.; Harro, J.M.; Shirtliff, M.E.; Achermann, Y. Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog. Dis. 2017, 75, ftx009. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, L.; Zheng, X.; Fu, T.; Guo, H.; Ren, F. Lactobacillus salivarius strain FDB89 induced longevity in Caenorhabditis elegans by dietary restriction. J. Microbiol. 2013, 51, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Gilman, J.; Cashman, K.D. The effect of probiotic bacteria on transepithelial calcium transport and calcium uptake in human intestinal-like Caco-2 cells. Curr. Issues Intest. Microbiol. 2006, 7, 1–5. [Google Scholar]
- Charizani, E.; Dushku, E.; Karathodorou, A.; Kyritsi, M.; Amanetidou, E.; Metallinou, E.T.; Kokkaleniou, M.M.; Passalis, N.; Tefas, A.; Staikou, A.; et al. Predicting the immunomodulatory activity of probiotic lactic acid bacteria using supervised machine learning in a Cornu aspersum snail model. Fish Shellfish Immunol. 2024, 152, 109788. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Mazziotta, C.; Tognon, M.; Martini, F.; Torreggiani, E.; Rotondo, J.C. Probiotics mechanism of action on immune cells and beneficial effects on human health. Cells 2023, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Wells, J.M. Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 2011, 10, S17. [Google Scholar] [CrossRef]
- Clark, A.G.; Glanowski, S.; Nielsen, R.; Thomas, P.D.; Kejariwal, A.; Todd, M.A.; Tanenbaum, D.M.; Civello, D.; Lu, F.; Murphy, B.; et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 2003, 302, 1960–1963. [Google Scholar] [CrossRef]
- Junttila, I.S. Tuning the cytokine responses: An update on interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef]
- McKenzie, A.N.; Culpepper, J.A.; de Waal Malefyt, R.; Brière, F.; Punnonen, J.; Aversa, G.; Sato, A.; Dang, W.; Cocks, B.G.; Menon, S. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc. Natl. Acad. Sci. USA 1993, 90, 3735–3739. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, G.J.; Bancroft, A.; Grencis, R.K.; McKenzie, A.N. A distinct role for interleukin-13 in Th2-cell-mediated immune responses. Curr. Biol. 1998, 8, 339–342. [Google Scholar] [CrossRef]
- Punnonen, J.; Aversa, G.; Cocks, B.G.; McKenzie, A.N. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc. Natl. Acad. Sci. USA 1993, 90, 3730–3734. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xia, Y.; Nguyen, A.; Lai, Y.H.; Feng, L.; Mosmann, T.R.; Lo, D. Effects of Th2 cytokines on chemokine expression in the lung: IL-13 potently induces eotaxin expression by airway epithelial cells. J. Immunol. 1999, 162, 2477–2487. [Google Scholar] [CrossRef]
- Gallo, E.; Katzman, S.; Villarino, A.V. IL-13-producing Th1 and Th17 cells characterize adaptive responses to both self and foreign antigens. Eur. J. Immunol. 2012, 42, 2322–2328. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Periwal, N.; Goyal, Y.; Sood, V.; Kaur, B. iIL13Pred: Improved prediction of IL-13 inducing peptides using popular machine learning classifiers. BMC Bioinform. 2023, 24, 141. [Google Scholar] [CrossRef]
- Mannon, P.; Reinisch, W. Interleukin 13 and its role in gut defence and inflammation. Gut 2012, 61, 1765–1773. [Google Scholar] [CrossRef]
- Wyn, T.A. IL-13 effector functions. Annu. Rev. Immunol. 2003, 21, 425–456. [Google Scholar] [CrossRef]
- Terabe, M.; Park, J.M.; Berzofsky, J.A. Role of IL-13 in regulation of anti-tumor immunity and tumor growth. Cancer Immunol. Immunother. 2004, 53, 79–85. [Google Scholar] [CrossRef]
- Su, T.; Mi, Y.; Zhang, L.; Wang, S.; Lu, H.; Shi, L.; Sun, H.; Wu, X.; Zhang, W.; Zuo, L.; et al. Association between IL13 gene polymorphisms and susceptibility to cancer: A meta-analysis. Gene 2013, 515, 56–61. [Google Scholar] [CrossRef]
- Ya, C.; Kim, J. Association of IL4, IL13, and IL4R polymorphisms with gastrointestinal cancer risk: A meta-analysis. J. Epidemiol. 2017, 27, 215–220. [Google Scholar]
- Liu, H.; Antony, S.; Roy, K.; Juhasz, A.; Wu, Y.; Lu, J.; Meitzler, J.L.; Jiang, G.; Polley, E.; Doroshow, J.H. Interleukin-4 and interleukin-13 increase NADPH oxidase 1-related proliferation of human colon cancer cells. Oncotarget 2017, 8, 38113–38135. [Google Scholar] [CrossRef] [PubMed]
- Traub, B.; Sun, L.; Ma, Y.; Xu, P.; Lemke, J.; Paschke, S.; Henne-Bruns, D.; Knippschild, U.; Kornmann, M. Endogenously expressed IL-4Rα promotes the malignant phenotype of human pancreatic cancer in vitro and in vivo. Int. J. Mol. Sci. 2017, 18, 716. [Google Scholar] [CrossRef]
- Song, X.; Traub, B.; Shi, J.; Kornmann, M. Possible roles of interleukin-4 and -13 and their receptors in gastric and colon cancer. Int. J. Mol. Sci. 2021, 22, 727. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S. Immune surveillance: A balance between protumor and antitumor immunity. Curr. Opin. Genet. Dev. 2008, 18, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Heuvel, F.O.; Rehman, R.; Aousji, O.; Froehlich, A.; Li, Z.; Jark, R.; Zhang, W.; Conquest, A.; Woelfle, S.; et al. Interleukin-13 and its receptor are synaptic proteins involved in plasticity and neuroprotection. Nat. Commun. 2023, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Debinski, W.; Gibo, D.M.; Hulet, S.W.; Connor, J.R.; Gillespie, G.Y. Receptor for interleukin 13 is a marker and therapeutic target for human high-grade gliomas. Clin. Cancer Res. 1999, 5, 985–990. [Google Scholar]
- Pelaia, C.; Paoletti, G.; Puggioni, F.; Racca, F.; Pelaia, G.; Canonica, G.W.; Heffler, E. Interleukin-5 in the pathophysiology of severe asthma. Front. Physiol. 2019, 10, 1514. [Google Scholar] [CrossRef]
- Le Beau, M.M.; Lemons, R.S.; Espinosa, R.; Larson, R.A.; Arai, N.; Rowley, J.D. Interleukin-4 and interleukin-5 map to human chromosome 5 in a region encoding growth factors and receptors and are deleted in myeloid leukemias with a del(5q). Blood 1989, 73, 647–650. [Google Scholar]
- Adachi, T.; Alam, R. The mechanism of IL-5 signal transduction. Am. J. Physiol. 1998, 275, C623–C633. [Google Scholar] [CrossRef]
- Moore, M.L.; Peebles, R.S., Jr. Interleukins |IL-5. Encycl. Respir. Med. 2006, 359–363. [Google Scholar]
- Garcia, G.; Taillé, C.; Laveneziana, P.; Bourdin, A.; Chanez, P.; Humbert, M. Anti-interleukin-5 therapy in severe asthma. Eur. Respir. Rev. 2013, 22, 251–257. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Roufosse, F. Targeting the interleukin-5 pathway for treatment of eosinophilic conditions other than asthma. Front. Med. 2018, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Ikutani, M.; Yanagibashi, T.; Ogasawara, M.; Tsuneyama, K.; Yamamoto, S.; Hattori, Y.; Kouro, T.; Itakura, A.; Nagai, Y.; Takaki, S.; et al. Identification of innate IL-5–producing cells and their role in lung eosinophil regulation and antitumor immunity. J. Immunol. 2012, 188, 703–713. [Google Scholar] [CrossRef]
- Bagnasco, D.; Ferrando, M.; Varricchi, G.; Puggioni, F.; Passalacqua, G.; Canonica, G.W. Anti-Interleukin 5 (IL-5) and IL-5Ra biological drugs: Efficacy, safety, and future perspectives in severe eosinophilic asthma. Front. Med. 2017, 4, 135. [Google Scholar] [CrossRef]
- Harish, A.; Schwartz, S.A. Targeted anti-IL-5 therapies and future therapeutics for hypereosinophilic syndrome and rare eosinophilic conditions. Clin. Rev. Allergy Immunol. 2020, 59, 231–247. [Google Scholar] [CrossRef]
- Nagase, H.; Ueki, S.; Fujieda, S. The roles of IL-5 and anti-IL-5 treatment in eosinophilic diseases: Asthma, eosinophilic granulomatosis with polyangiitis, and eosinophilic chronic rhinosinusitis. Allergol. Int. 2020, 69, 178–186. [Google Scholar] [CrossRef]
- Yoshida, T.; Ikuta, K.; Sugaya, H.; Maki, K.; Takagi, M.; Kanazawa, H.; Sunaga, S.; Kinashi, T.; Yoshimura, K.; Miyazaki, J.-I.; et al. Defective B-1 cell development and impaired immunity against Angiostrongylus cantonensis in IL-5R α-deficient mice. Immunity 1996, 4, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Moon, B.; Takaki, S.; Miyake, K.; Takatsu, K. The role of IL-5 for mature B-1 cells in homeostatic proliferation, cell survival, and Ig production. J. Immunol. 2004, 172, 6020–6029. [Google Scholar] [CrossRef]
- Takatsu, K.; Nakajima, H. IL-5 and eosinophilia. Curr. Opin. Immunol. 2008, 20, 288–294. [Google Scholar] [CrossRef]
- Verma, N.D.; Plain, K.M.; Nomura, M.; Tran, G.T.; Robinson, C.; Boyd, R.; Hodgkinson, S.J.; Hall, B.M. CD4 + CD25 + T cells alloactivated ex vivo by IL-2 or IL-4 become potent alloantigen-specific inhibitors of rejection with different phenotypes, suggesting separate pathways of activation by Th1 and Th2 responses. Blood 2009, 113, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Tran, G.T.; Hodgkinson, S.J.; Carter, N.M.; Verma, N.D.; Plain, K.M.; Boyd, R.; Robinson, C.M.; Nomura, M.; Killingsworth, M.; Hall, B.M. IL-5 promotes induction of antigen-specific CD4 + CD25 + T regulatory cells that suppress autoimmunity. Blood 2012, 119, 4441–4450. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.; Morris, K.; Bruce, M.; O’Neil, T.; Jansen, E.; Coupar, B.; Strom, D. Sustained biological effects of porcine interleukin 5 delivered to pigs as recombinant protein or via a DNA vector. Cytokine 2007, 40, 193–200. [Google Scholar] [CrossRef]
- Wen, T.; Rothenberg, M.E.; Gordon, S. The regulatory function of eosinophils. Microbiol. Spectr. 2016, 4, 257–269. [Google Scholar] [CrossRef]
- Dougan, M.; Dranoff, G.; Dougan, S.K. GM-CSF, IL-3, and IL-5 family of cytokines: Regulators of inflammation. Immunity 2019, 50, 796–811. [Google Scholar] [CrossRef]
- Massey, O.W.; Suphioglu, C. Taking a breather: Advances in interleukin 5 inhibition for asthma relief. Int. J. Mol. Sci. 2022, 23, 11166. [Google Scholar] [CrossRef]
- Gresser, I.; Bourali, C. Antitumor effects of interferon preparations in mice. J. Natl. Cancer Inst. 1970, 45, 365–376. [Google Scholar]
- Spaapen, R.M.; Leung, M.Y.; Fuertes, M.B.; Kline, J.P.; Zhang, L.; Zheng, Y.; Fu, Y.X.; Luo, X.; Cohen, K.S.; Gajewski, T.F. Therapeutic activity of high-dose intratumoral IFN-β requires direct effect on the tumor vasculature. J. Immunol. 2014, 193, 4254–4260. [Google Scholar] [CrossRef] [PubMed]
- Berraondo, P.; Sanmamed, M.F.; Ochoa, M.C.; Etxeberria, I.; Aznar, M.A.; Pérez-Gracia, J.L.; Rodriguez-Ruiz, M.E.; Ponz-Sarvise, M.; Castañón, E.; Melero, I. Cytokines in clinical cancer immunotherapy. Br. J. Cancer 2019, 120, 6–15. [Google Scholar] [CrossRef] [PubMed]
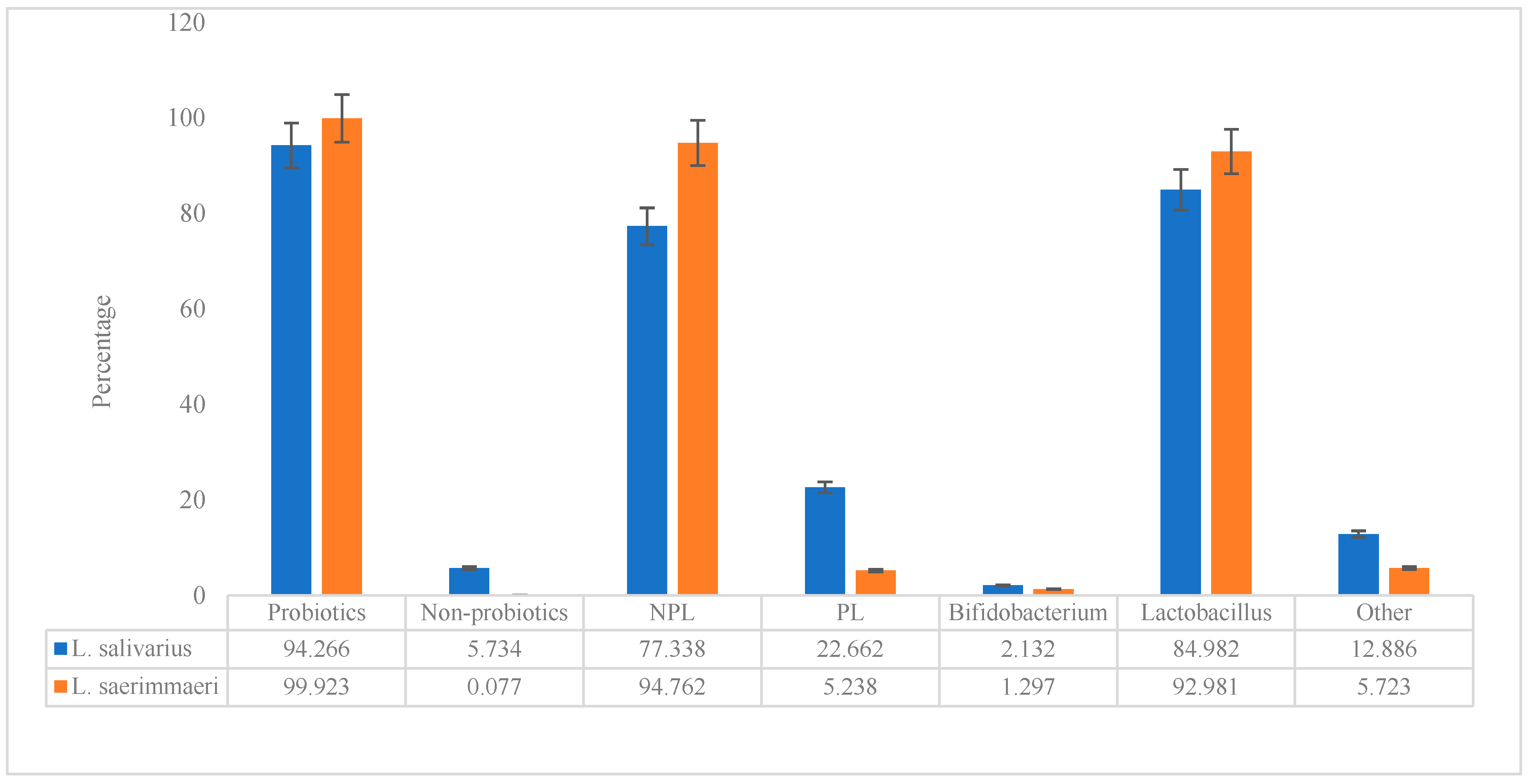
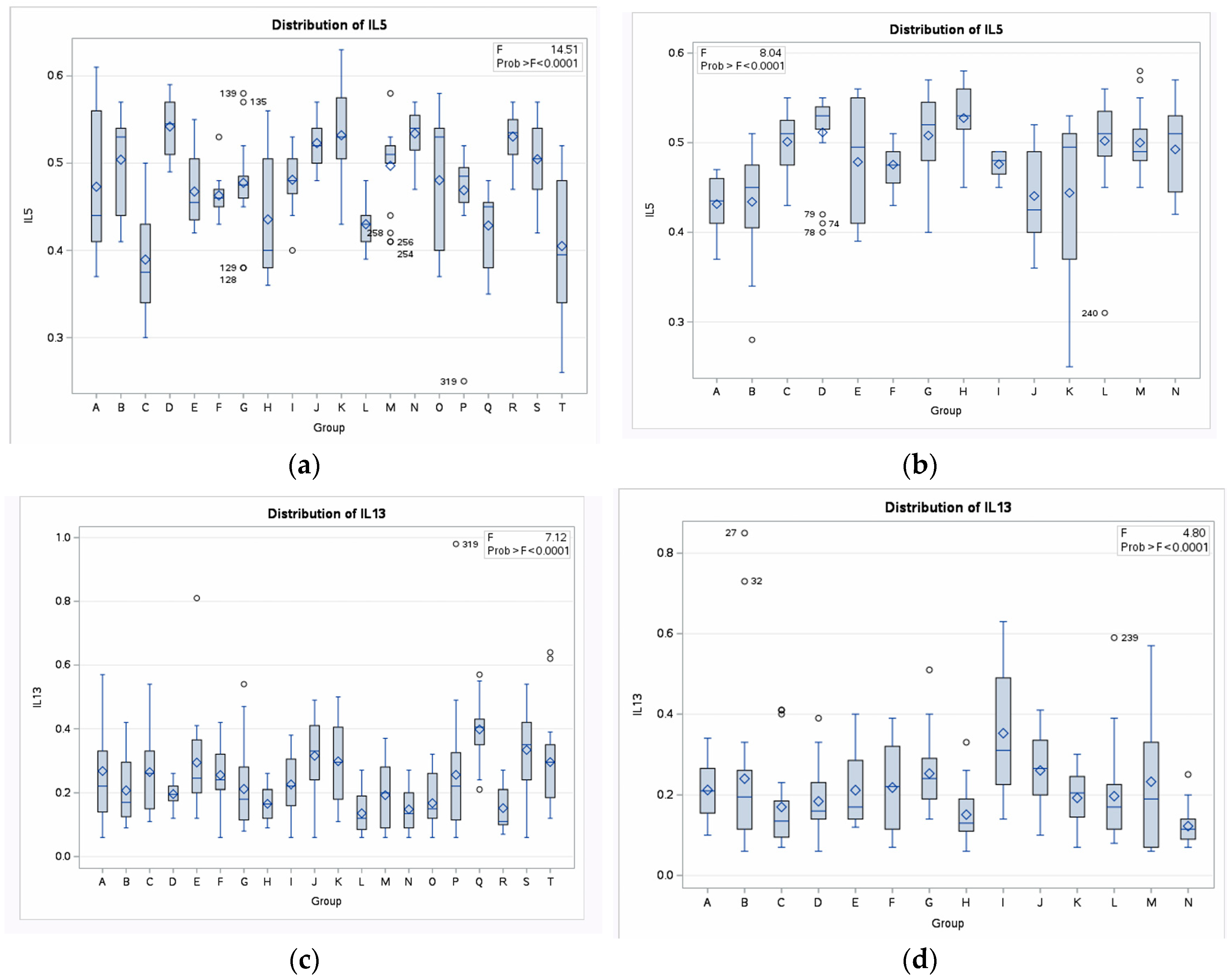
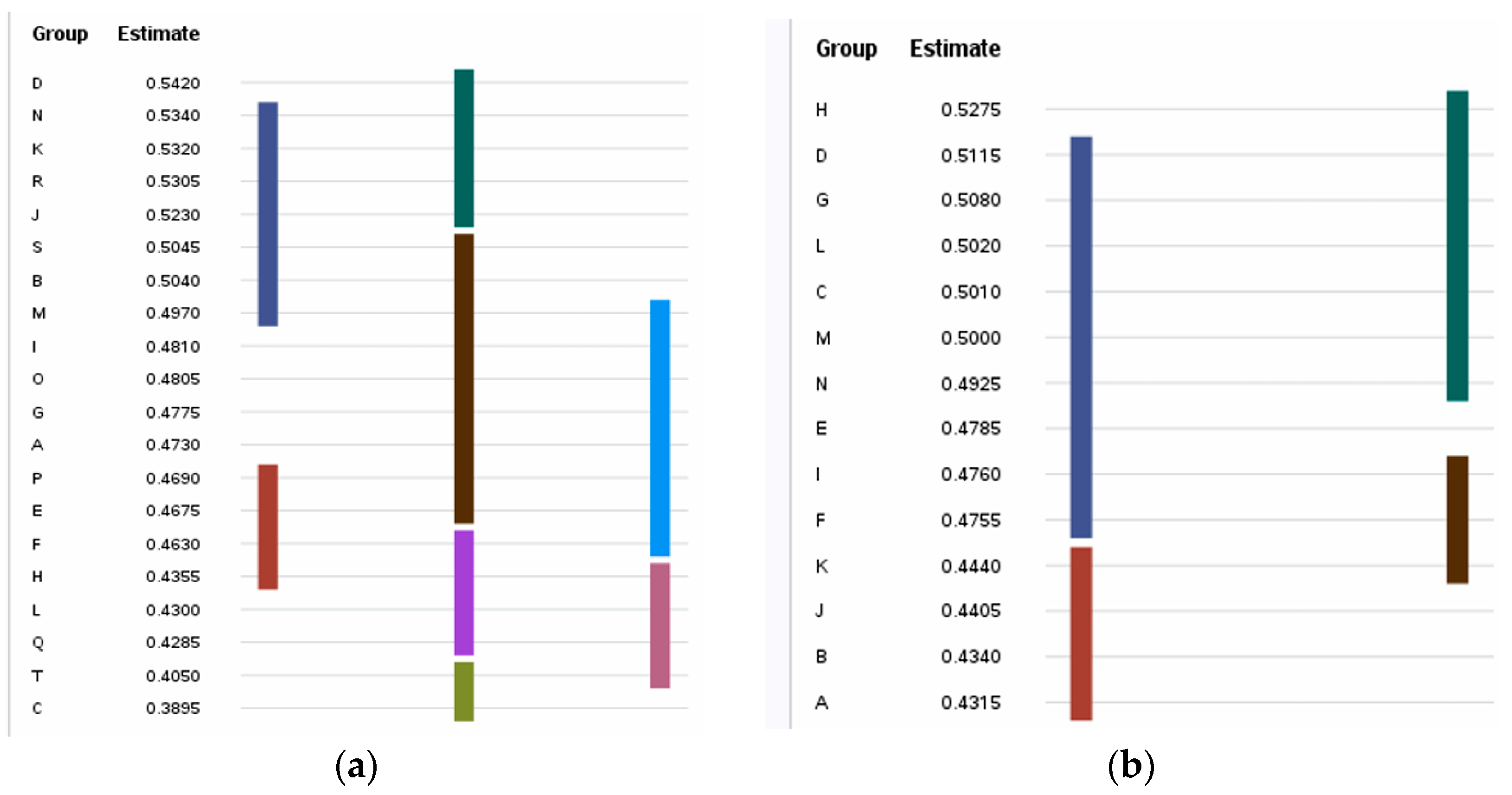
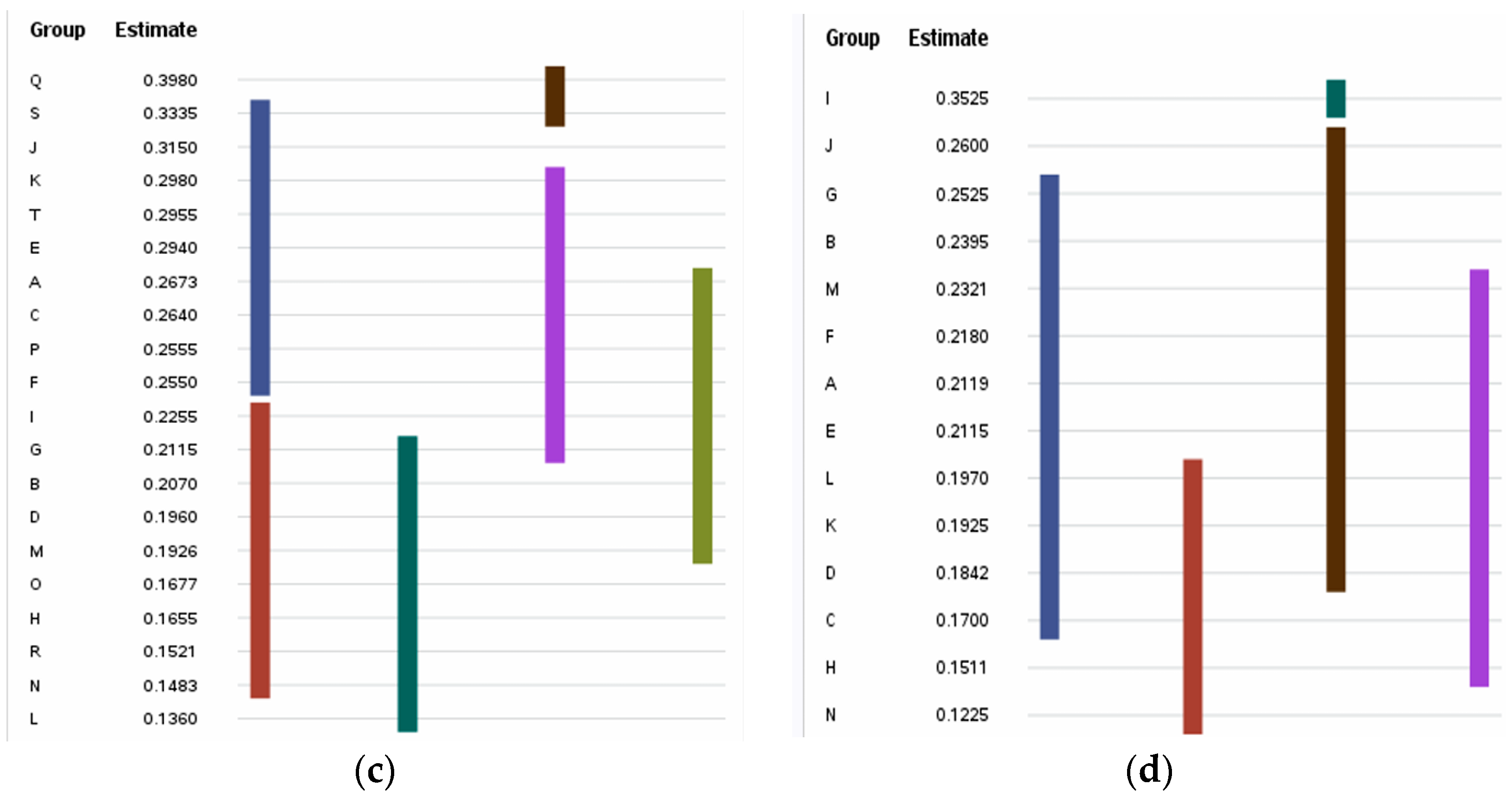


| Sequence Id | ||
|---|---|---|
| Designation | L. saerimneri | L. salivarius |
| A | * 258409048 + | * 003705713 + |
| B | * 155824516 + | * 035162675 + |
| C | * 009553931 + | * 035162669 + |
| D | * 040533884 + | * 192044553 + |
| E | * 009555546 + | * 003707278 + |
| F | * 338434469 + | * 003705087 + |
| G | * 338434518 + | * 003709061 + |
| H | * 338433807 + | * 003699688 + |
| I | * 278838030 + | * 003699582 + |
| J | * 278838849 + | * 003699525 + |
| K | * 338433893 + | * 255320006 + |
| L | * 009553726 + | * 192044603 + |
| M | * 338433902 + | * 255320007 + |
| N | * 338433903 + | * 095758380 + |
| O | * 081537403 + | |
| P | * 162853508 + | |
| Q | * 147759279 + | |
| R | * 003708122 + | |
| S | * 192044672 + | |
| T | * 192044673 + | |
| L. salivarius | L. saerimneri | |||
|---|---|---|---|---|
| Protein Group | ML Score | IL-13 Score | ML Score | IL-13 Score |
| A | 0.473 ± 0.083 cd | 0.267 ± 0.150 bcd | 0.432 ± 0.033 d | 0.212 ± 0.072 bcd |
| B | 0.504 ± 0.054 bc | 0.207 ± 0.099 def | 0.434 ± 0.060 d | 0.240 ± 0.204 bc |
| C | 0.390 ± 0.068 h | 0.264 ± 0.133 bcd | 0.501 ± 0.030 ab | 0.170 ± 0.110 cde |
| D | 0.542 ± 0.033 a | 0.196 ± 0.041 def | 0.512 ± 0.045 ab | 0.184 ± 0.075 bcde |
| E | 0.468 ± 0.040 cde | 0.294 ± 0.149 bc | 0.479 ± 0.067 b | 0.212 ± 0.085 bcd |
| F | 0.463 ± 0.021 def | 0.255 ± 0.098 bcd | 0.476 ± 0.023 bc | 0.218 ± 0.112 bcd |
| G | 0.478 ± 0.048 cd | 0.212 ± 0.127 cdef | 0.508 ± 0.048 ab | 0.253 ± 0.091 bc |
| H | 0.436 ± 0.071 efg | 0.166 ± 0.053 ef | 0.528 ± 0.039 a | 0.151 ± 0.070 de |
| I | 0.481 ± 0.031 cd | 0.226 ± 0.085 cde | 0.476 ± 0.013 bc | 0.353 ± 0.148 a |
| J | 0.523 ± 0.023 ab | 0.315 ± 0.120 b | 0.441 ± 0.053 d | 0.260 ± 0.090 b |
| K | 0.532 ± 0.058 ab | 0.298 ± 0.128 bc | 0.444 ± 0.092 cd | 0.193 ± 0.066 bcde |
| L | 0.430 ± 0.027 fg | 0.136 ± 0.062 f | 0.502 ± 0.054 ab | 0.197 ± 0.119 bcde |
| M | 0.497 ± 0.044 bcd | 0.193 ± 0.104 def | 0.500 ± 0.035 ab | 0.232 ± 0.161 bcd |
| N | 0.534 ± 0.030 ab | 0.148 ± 0.066 ef | 0.493 ± 0.049 ab | 0.123 ± 0.0438 e |
| O | 0.481 ± 0.074 cd | 0.168 ± 0.090 ef | ||
| P | 0.469 ± 0.057 cde | 0.256 ± 0.211 bcd | ||
| Q | 0.429 ± 0.042 fg | 0.398 ± 0.097 a | ||
| R | 0.531 ± 0.029 ab | 0.152 ± 0.071 ef | ||
| S | 0.505 ± 0.040 bc | 0.334 ± 0.129 ab | ||
| T | 0.405 ± 0.084 gh | 0.296 ± 0.142 bc | ||
| IL-5-Inducing Immunogenic Peptides | |||||
|---|---|---|---|---|---|
| L. salivarius | L. saerimneri | ||||
| Group | Start to End | Sequences | Group | Start to End | Sequences |
| D | 15 to 35 | FSKEVADRANVENIEPGLIR | C | 10 to 30 | GVGNDRRPVNAKNIKKRRAQ |
| J | 15 to 35 | TLDKLEINTEEFMDFQKAFM | G | 21 to 41 | GFDTDRYFEENKNEYDWGKP |
| N | 18 to 38 | VRAKNYNAAETQVKVSVIAN | H | 24 to 44 | EKYHLIEAEGIKRVTEEFIW |
| R | 2 to 22 | LYLVEYFINNKLHNMIVRAK | L | 1 to 21 | DNKVPVHVKGVEYAANAEDS |
| IL-13-inducing immunogenic peptides | |||||
| E | 2 to 22 | QRVHITNLYGLSGVAGLAQK | B | 5 to 26 | PLVLGVLFIATGYISYATYR |
| J | 6 to 26 | ALLDELREGTLDKLEINTEE | G | 21 to 41 | GFDTDRYFEENKNEYDWGKP |
| Q | 33 to 53 | RKFIREIANEKQLNELEILI | I | 10 to 30 | DVGSLLINHVLTSTLVMKQA |
| S | 35 to 55 | SEKIKKEILDAAEKGKMNLK | M | 15 to 35 | QDPAITAEEERKIIRDFRKK |
| T | 25 to 45 | MKAYDRKQKENPRPKGWVPW | |||
| IL-5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| L salivarius | |||||||||
| Group | MW | pI | #−ve | #+ve | Formula | EC | II | AI | GRAVY |
| D | 2257.53 | 4.87 | 4 | 3 | C98H161N29O32 | nd | 55.05 | 97.50 | −0.455 |
| J | 2450.80 | 4.18 | 5 | 2 | C110H168N24O35S2 | nd | 28.55 | 63.50 | −0.380 |
| N | 2175.47 | 9.70 | 1 | 3 | C94H159N29O30 | 1490 * | −3.73 | 97.50 | −0.205 |
| R | 2478.98 | 9.53 | 1 | 3 | C116H184N30O28S1 | 2980 * | 24.69 | 131.50 | 0.160 |
| L. saerimneri | |||||||||
| C | 2277.62 | 12.01 | 1 | 7 | C94H169N39O27 | nd | 107.81 | 58.50 | −1.655 |
| G | 2510.61 | 4.30 | 6 | 3 | C113H152N28O38 | 8480 | 27.39 | 0.00 | −2.065 |
| H | 2490.84 | 5.02 | 5 | 3 | C116H176N28O33 | 6990 | 67.14 | 97.50 | −0.510 |
| L | 2142.31 | 4.75 | 4 | 2 | C92H144N26O33 | 1490 * | 61.77 | 73.00 | −0.695 |
| IL-13 | |||||||||
| L salivarius | |||||||||
| E | 2125.46 | 9.99 | 0 | 2 | C94H157N29O27 | 1490 * | 19.71 | 117.00 | 0.090 |
| J | 2301.53 | 4.00 | 7 | 2 | C98H165N25O38 | nd | 37.71 | 122.00 | −0.645 |
| Q | 2469.91 | 6.29 | 4 | 4 | C111H189N31O32 | nd | 50.23 | 141.50 | −0.365 |
| S | 2273.72 | 9.30 | 4 | 6 | C99H177N27O31S1 | nd | 33.47 | 88.00 | −1.000 |
| T | 2514.93 | 10.17 | 2 | 6 | C115H176N34O28S1 | 12,490 | 39.70 | 19.50 | −1.950 |
| L. saerimneri | |||||||||
| B | 2217.64 | 8.90 | 0 | 1 | C108H165N23O27 | 4470 * | 4.39 | 136.50 | 1.110 |
| G | 2510.61 | 4.30 | 6 | 3 | C113H152N28O38 | 8480 | 27.39 | 0.00 | −2.065 |
| I | 2139.54 | 6.74 | 1 | 1 | C94H163N25O29S1 | nd | 17.09 | 146.00 | 0.750 |
| M | 2443.79 | 8.50 | 5 | 6 | C106H179N33O33 | nd | 53.35 | 68.50 | −1.430 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adejumo, I.O. Immunotherapy Potential of Animal-Sourced Probiotic Bacteria. Biologics 2025, 5, 17. https://doi.org/10.3390/biologics5030017
Adejumo IO. Immunotherapy Potential of Animal-Sourced Probiotic Bacteria. Biologics. 2025; 5(3):17. https://doi.org/10.3390/biologics5030017
Chicago/Turabian StyleAdejumo, Isaac Oluseun. 2025. "Immunotherapy Potential of Animal-Sourced Probiotic Bacteria" Biologics 5, no. 3: 17. https://doi.org/10.3390/biologics5030017
APA StyleAdejumo, I. O. (2025). Immunotherapy Potential of Animal-Sourced Probiotic Bacteria. Biologics, 5(3), 17. https://doi.org/10.3390/biologics5030017






