Abstract
The coronavirus disease 2019 (COVID-19), a life-threatening pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has resulted in massive destruction and is still continuously adding to its death toll. The advent of this global outbreak has not yet been confirmed; however, investigation for suitable prophylaxis against this lethal virus is being carried out by experts all around the globe. The SARS-CoV-2 belongs to the Coronaviridae superfamily, like the other previously occurring human coronavirus variants. To better understand a new virus variant, such as the SARS-CoV-2 delta variant, it is vital to investigate previous virus strains, including their genomic composition and functionality. Our study aimed at addressing the basic overview of the virus’ profile that may provide the scientific community with evidence-based insights into COVID-19. Therefore, this study accomplished a comprehensive literature review that includes the virus’ origin, classification, structure, life cycle, genome, mutation, epidemiology, and subsequent essential factors associated with host–virus interaction. Moreover, we summarized the considerable diagnostic measures, treatment options, including multiple therapeutic approaches, and prevention, as well as future directions that may reduce the impact and misery caused by this devastating pandemic. The observations and data provided here have been screened and accumulated through extensive literature study, hence this study will help the scientific community properly understand this new virus and provide further leads for therapeutic interventions.
1. Introduction
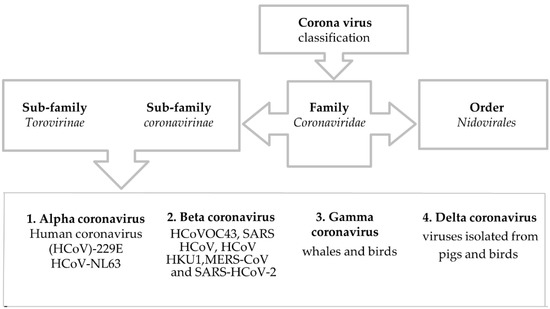
In late December 2019, the novel coronavirus strain, severe acute respiratory syndrome (SARS-CoV-2), was first traced at the Wuhan metropolis of China’s Hubei province [1]. Though the virus was limited in China during the very early phase of the coronavirus disease 2019 (COVID-19) outbreak, the current scenario is quite devastating throughout the globe. The ongoing pandemic has not only created a massive burden on the universal health care system but also impacted global human beings’ social, economic, and health measures [1,2]. The causative agent of this outbreak, a type of ß-coronavirus that emerged from unknown or mysterious sources, is SARS-CoV-2, which belongs to the Coronaviridae family [3]. The world health organization (WHO) declared this public health emergency as a pandemic on 11 March 2020 [4]. As of September 2021, the global community has counted around 234 million cases and near 4.8 million deaths from this infectious disease and its imposed complications. However, at the current time more than 210 million SARS-CoV-2 infected patients have recovered from this infection [5]. As this pathogenic virus has newly emerged, the exact mechanism of pathogenesis and its biochemical pathway are still vague and unclear. The spike protein of SARS-COV-2 might have a strong affinity towards the host cell receptor, enhancing its much higher human-to-human transmission [6]. The primary investigation has revealed that this viral particle infects the respiratory tract and causes respiratory complications ranging from very simple clinical manifestations of hypoxia to acute respiratory distress syndrome (ARDS) [7,8].
Although both the infection and mortality rates were improving, the most virulent variant, the delta variant, has evolved in most countries of the world and worsened the situation with record-breaking infections [9,10]. Furthermore, WHO has officially declared the earliest onset of symptoms as having been on 8 December 2019 and set a Public Health Emergency of International Concern (PHEIC) on 1 February 2020 [11,12]. There are two opinions towards this pathogen. The first is that the origin of this virus is laboratory-based. The second opinion proffers evidence from genomic information suggesting this virus is unique compared to previous coronaviruses [13]. The cause of COVID-19’s severity is well known, and is because it has s1, s2 polybasic cleavage sites in its spike glycoprotein which lead to high binding affinity with ACE-2 receptors [14,15]. According to a comparison between SARS-CoV-2, SARS (Severe Acute Respiratory Syndrome), and MERS (Middle East Respiratory syndrome) concerning their appearance in community transmission across the globe, SARS-COV-2 appeared at the top. The SARS-COV-2 has shown the highest projected basic reproductive number (R0) of (2–3.58) whereas the R0 was (1.7–1.9) and <1 for SARS and MERS, respectively [16,17]. However, the aim of this comprehensive study is to give an overview of the virus and its origin, including its classification, structure, life cycle, genomic constitution, mutation, epidemiology, prognosis, and pathogenesis. We also summarize the aftermath of the infection, treatments, disease associated factors, and the methods of prevention as guided by public health experts in different sections of our literature review.
2. Invasion of SARS-CoV-2
COVID-19 is an infectious disease caused by the infection of SARS severe acute respiratory syndrome coronavirus 2. Coronavirus (CoV) belongs to the RNA virus families that are organized varyingly in animal species. It can invade the respiratory tract, gastrointestinal systems, and the hepatic and nervous systems in humans [18]. Underneath the electron microscope, it appears to be a crown-like structure because of spike glycoproteins [19]. CoVs cause about 5 to 10% of acute respiratory infections. It is reported that 2% of the global population is regarded as healthy carriers of these viruses [18]. In immune-competent individuals, these CoVs result in self-limiting respiratory infections and common colds. In the case of aged people and immune-compromised individuals, CoVs can affect the lower respiratory tracts [19]. Yet, functional deactivation of these viruses can be attained by using ether (75%), ethanol (60%), and chlorine-containing disinfectants [20].
3. Origin of SARS-CoV-2
Like SARS-CoV and MERS, SARS-CoV-2 also displays strong similarities with viruses of bat origin [21]. The whole-genome alignment between the SARS-CoV-2 and the strains from Rhinolophus affinis species (Bat-CoV RaTG13) from Yunnan province matched up to 96%. [22]. Additionally, it is clearly seen that SARS-CoV-2 and previously occurring SARS CoV and MERS harbor bats as a common source (Table 1) [23]. In addition, the study suspects pangolins as the natural stockpile of SARS-CoV-2. This claim was based upon the result inferred by aligning the genome contigs among SARS-CoV-2-like coronaviruses such as Pangolin-CoV, previously sampled from the lung tissue of two dead Malayan pangolins [24]. The overall genome sequence of Pangolin-CoV had a 91.02% similarity with SARS-CoV-2 and a 90.55% similarity with Bat-CoV RaTG13 [23,25].

Table 1.
Synopsis of the natural reservoir, median host, and target host for major coronaviruses.
5. Structure of SARS-CoV-2
SARS-CoV-2 possesses a single and positive-stranded RNA virus enfolded in a lipid bilayer, as shown in (Figure 2) [30,31].
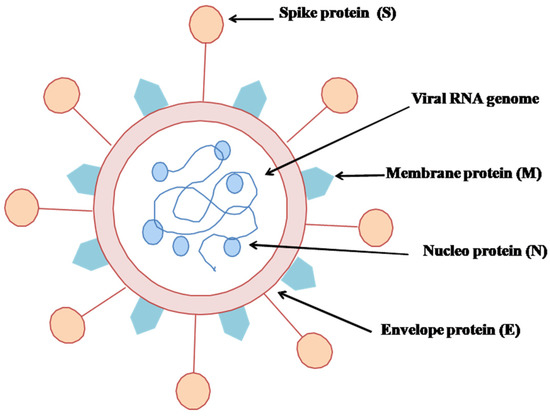
Figure 2.
Structural view of SARS-CoV-2.
The lipid bilayer merges into the host’s cell membrane, delivering a positive RNA strand into the cytoplasm and seeding the translation of different viral proteins. The newly replicated viral proteins and RNA genome reconcile into new viruses, which eventually rush out of the cell [32,33]. SARS-CoV-2 is very weak when susceptible to heat and ultraviolet [19]. The spike protein (S-protein) is a glycoprotein expressed as a homotrimer on the viral counterpart, called the viral envelope [34]. Each S-protein is comprised of S1 and S2 subunits. S1 incorporates a receptor-binding domain that spots receptors on the host cells, and S2 modulates the membrane fusion. This viral S-protein ties up with the human ACE2 (Angiotensin Converting Enzyme 2) receptor protein [35]. Tissues such as lung, heart, kidney, and adipose are rich with ACE2 receptors [36,37].
6. Life Cycle of SARS-CoV-2
The virus particle seems to follow two stages of their complete lifespan: (1) Early stage (entry and initiation, S protein cleavage, membrane fusion) and (2) Advanced stage (translation and RNA replication, virion release) (Figure 3) [22,38,39].
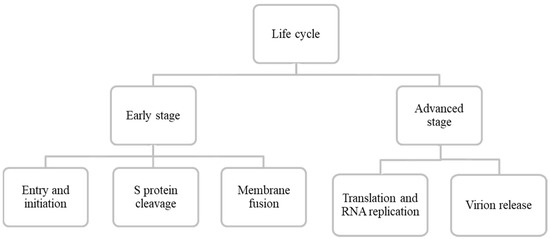
Figure 3.
Schematic representation of SARS-CoV-2 entire life cycle.
6.1. Early Stage
6.1.1. Entry and Initiation
All SARS CoVs, MERS, and SARS-CoV-2 possess an S protein that goes through a cleavage event by fusion proteins (Figure 4). An RBD (receptor-binding domain) at the spike protein enables ACE2 (Angiotensin-converting enzyme 2), the host receptor protein, to initiate membrane fusion activity. SARS-CoV-2 shows a higher binding affinity with the ACE2 receptor than any other coronavirus variant, even 20-fold greater than SARS-CoV [40]. ACE2 receptors are located in different parts of our body, including oral mucosa, nasal epithelium, vascular endothelium, lungs, kidney, small intestine, testis, colon, heart, and brain [41,42]. A study conducted by Devaux et al. [43] reported 32 different ACE2 receptors, including seven hotspot variants specified in the different populations. In addition, it is experimentally found that proteases such as cathepsins, human airway trypsin-like proteases, trypsin, furin, and transmembrane protease serine protease (2/4) (TMPRSS) assist in proteolytic cleaving of the S protein into S1/S2 subunits [44,45,46,47]. The furin protease entry is called endocytosis, whereas TMPRSS (2/4) adopts a direct fusion mechanism [48].
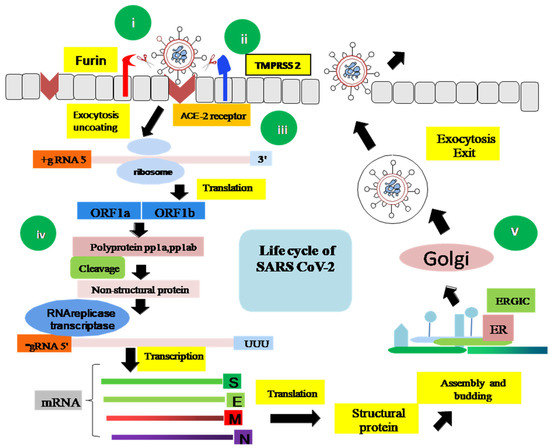
Figure 4.
A schematic representation of the entire life cycle of SARS-CoV-2. (i) The entry of the virus particle is initiated by binding of Spike (S) glycoprotein to the ACE2 receptor of the host cell. (ii) The S glycoprotein is cleaved into S1/S2 either by TMPRSS2 or by the furin protease, in association with some cofactors. (iii) S2 stalk domain undergoes a conformational change and facilitates the fusion between the host cell membrane and the virus envelope protein. (iv) In this advanced stage, host cell machinery is used by the virus to translate viral polyprotein pp1a and pp1ab into nsps. The RNA genome and N protein are made in the host cell cytoplasm and the S, E, and M structural proteins are packaged in the endoplasmic reticulum (ER). (v) The mature virion is transferred to the Golgi body followed by exocytosis, budding, or cell death.
6.1.2. S Protein Cleavage
There are two pathways to S protein cleavage. The first is by protease TMPRSS (2/4) in the host cell, and the second is by furin and furin-like proteases [49]. TMPRSS (2/4) belongs to type II transmembrane serine protease found extensively in gastrointestinal, urogenital, and respiratory tracts [50]. In contrast, the furin and furin-like proteases belong to a group called proprotein convertases, a type I transmembrane protein. It converts the S protein into globular S1 and bio-membrane anchored stalk S2 domain [51]. Interestingly there is no compensation activity between TMPRSS (2/4) and furin and furin-like proteases; therefore, halting either of them can be a promising target zone for drug discovery [52].
6.1.3. Membrane Fusion
There are three mechanisms of membrane fusion. The first is endocytosis, the second is direct fusion, and the third is the proteolytic activity of cathepsins, TMPRSS (2/4), the furin proteases, or other trypsin-like proteases that penetrate through into the endosomes or lysosomes of the cell [49].
6.2. Advanced Stage
6.2.1. Translation and RNA Replication
The step of replication and transcription occurs after the entry of viral RNA (positive single-stranded) [53]. It involves activities like (1) RNA synthesis, (2) template proofreading, and (3) mRNA capping. The NSPs (non-structural protein) domain initially comprising two polypeptides further generates non-structural proteins like papain-like protease, PLpro (nsp3), main protease, Mpro (nsp5), and RNA-dependent RNA polymerase, RdRp (nsp12). Host proteases are utilized during the processing of the initial two polypeptides. The further processing activity is undergone by the PLpro and Mpro of the virus [31,54,55]. RdRp plays the focal role in viral replication by the catalysis activity of nsp12, along with nsp7 and nsp8 as cofactors [56]. The RNA proofreading and capping of the mRNA are processed by the nsp14. Apart from nsp14, the capping machinery comprises nsp13, nsp16, and nsp10 as cofactors. mRNA capping is inevitable for viral stability and protection against host immunity [57,58,59,60,61]. In the host cell cytoplasm, two of the most vital proteins, including N structural protein and the virus RNA, are biosynthesized, and the rest of the structural proteins(S, E, M) are produced in the host’s endoplasmic reticulum (ER) [49].
6.2.2. Virion Packing and Release
The genome is affected by the infection if it occurs within the middle of the ER–Golgi compartment [48,62,63,64]. The virus may leave the host cell either via exocytosis, budding, or cell death. Most often, undeveloped virion adopts the budding process. At the same time, a mature virus is ejected from the host cell through exocytosis. In some cases, virus particles crack the host cell lysosomes and, in the long run, prompt cell demise. The new virus particles either can contaminate new cells or be delivered into the climate, focusing on another host [65,66].
7. Genome Organization of SARS-CoV-2
Human coronaviruses have initially been traced back to the late 1960s, expressing some mild symptoms in the population, similar to the common cold [67]. The date of the first published genomic sequence of SARS-CoV-2 was 24 January 2020, which was an open-access publication. It was found by Wei et al. as a recombinant sequence after the codon usage analysis. However, this notion was disproved by Paraskevis’ full-genome evolutionary analysis and Chen’s Simplot investigation [68,69]. According to the present study, it is found that SARS-CoV-2 is a new type of positive-sense, single-stranded RNA virus in the Betacoronavirus genus from the Coronaviridae family [31,69,70]. This novel virus’ non-segmented whole genome length is 29,891 to 29,903 nucleotides, making it one of the largest viruses among RNA viruses [19]. Akin to SARS-CoV and MERS-CoV, the novel SARS-CoV-2 genome carries two untranslated regions (UTRs), 5′-methylated cap and 3′poly-A tail structure, and a single open reading frame (ORF) that codes for a single polyprotein [29,70]. The 5’-end of the SARS-CoV-2 genome organization is composed of viral replicase (ORF1a and ORF1b)-4 structural proteins (nucleocapsid (N), envelope protein (E), spike protein (S), and membrane protein (M)) and the 3′-end encodes some accessory protein genes, such as ORF 3a, 7, and 8, which are arranged alongside the structural proteins (Figure 5) [29,30,31,69,70,71].
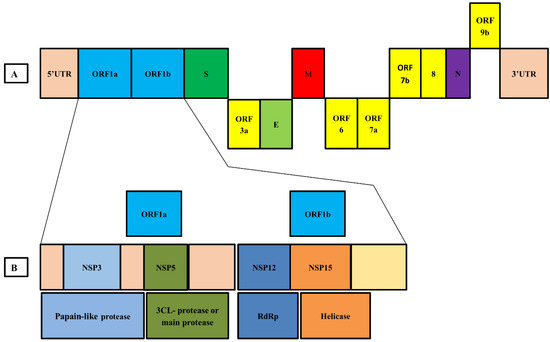
Figure 5.
Genome organization of SARS-CoV-2. (A) The whole genome is arranged in the following order; 5′ UTR-ORF1a—ORF1b-Spike protein (S)—ORF 3a—Envelope protein (E)—Membrane protein (M)-ORF 6-ORF 7a—ORF 8—Nucleocapsid (N)—ORF 9b—3′ UTR. (B) Viral replicase proteins (ORF1a and ORF1b) include nsp3 (Papain-like protease), nsp5 (Main proteas), nsp12 (RNA dependent RNA polymerase), and nsp13 (Helicase).
In the SARS-CoV-2 genome, the ORF1a and ORF1b cover about 66% of the entire genome, encoding 16 non-primary proteins (nsps), whereas the remaining 33% encodes adornment proteins and underlying proteins [10,70]. The source of tests was gathered from bronchoalveolar lavage liquid or throat swabs, the Huanan fish market, and lung injuries of patients. A phylogenetic report showed about a 96.2% arrangement comparability among Bat-CoV and SARS-CoV-2 genomes. Genomic grouping character information likewise showed 79.0% and 50.0% succession similarity with SARS-CoV and MERS-CoV, respectively [72]. Surprisingly, genomic comparison from a dead Malayan Pangolin (Manis javanica) with SARS-CoV-2 matched up to 91.02%, indicating that Pangolin could have contributed as an intermediary host in virus transmission [24]. In recent times, sequences from diverse regions have been recorded in various public databases, making virus tracking a lot more feasible than previously [73].
8. Mutation in SARS-CoV-2
SARS-CoV-2 displays very little sequence diversity due to having a proofreading mechanism, yet there are still chances for natural selection to occur in its favor [74,75,76]. The more SARS-CoV-2 pandemic remains in the population, the greater its possibility to gather immunologically associated mutations, even after the vaccine is available [77,78,79,80,81]. Scientists are solely focused on understanding the evolutionary mechanisms of SARS-CoV-2, including mutation, recombination, and tracing for indels in the genome as they were found in previously discovered coronaviruses [82]. Experiments with the evidence of antigenic drift were seen in previous strains, such as OC43, 229E, and SARS CoV-1, but have not yet been found in SARS-CoV-2 [77,78,79,80,81,83,84]. In 2003/2004 viruses, the point mutation D480A/G on the receptor binding domain (RBD) spread with an impactful prevalence in humans and civet cats. In vitro, recapitulated elevation of D480A/G was induced due to immune pressure by neutralizing antibody 80R [85]. Likewise, point mutations were detected to be worthwhile as an inhibitor for neutralizing antibodies in SARS-CoV-1 and MERS-CoV [85,86]. An evolutionary analysis on 351 sequences found two mutations in nsp6 and near the ORF10 region in earlier COVID-19 strains may confer less stability to protein structures. Notably, the nsp6 creates autophagosomes and is available in the endoplasmic reticulum (ER) of both alpha and beta coronaviruses. Accordingly, the mutation in nsp6 may confer a noteworthy change in the expression of SARS-CoV to its host, especially in the autophagic lysosomal system [87]. In SARS-CoV-2, another significant amino acid change, D614G, has been traced to appear recurrently in the spike protein worldwide. Before the G614 variant, the original D614 form was firmly retained. An A-to-G mutation gave rise to D614G substitution in the Wuhan reference strain at positions 23,403 [88]. In early March of 2019, G614 was infrequent worldwide, but gradually thriving in Europe. Eventually, variants carrying the D614G substitution were designated as “G clade”. Three additional mutations are also found to be present concurrently with the D614G variant, namely: (1) a C-to-T mutation at position 241, according to Wuhan reference sequence in the 5′ UTR, (2) a C-to-T silent mutation at position 3037, and (3) a C-to-T mutation at position 14,408 (RdRp P323L). Currently, the haplotype containing these four interlinked mutations is starting to be found frequently in Europe, continuing to North America, Oceania, and finally Asia. A study found evidence of higher titers (elevated 2.6–9.3 times) for the G614 variant in infected patients’ samples with lower Ct (cycle threshold) value in vivo, indicating higher viral load; yet, this does not confirm disease severity [16]. However, G614 may induce spike stability and membrane fusion [89].
9. Epidemiology of COVID-19
A few measures are regularly used to decide the death pace of a pestilence flare-up [90]. Regular epidemiological terms incorporate the case casualty rate (CFR), which focuses on the extent of people with a specific case who passed on from that condition. The case-casualty proportion is an assessment of the seriousness of that condition. Although a massive loss of life has occurred, still, overall, the CFR of SARS-CoV-2 is lower than earlier variations, specifically SARS CoV-1 and MERS [61,91]. The SARS-CoV-2 is zoonotic, and human-to-human transmission transformed this disease into a pandemic. The medical clinics’ flood with patients having clinical manifestations of SARS-CoV-2 demonstrates the raised degree of human-to-human transmission rate [92]. Since the day the principal contamination was affirmed, the number of tainted people proceeded consistently (Table 2) [61]. More than 220 countries have already reported confirmed cases all around the globe [71].

Table 2.
Top 10 countries with the count of SARS-CoV-2 related cases, along with the total case count worldwide as of 27 July 2021.
Evidence revealed that intra-family and local area transmission is the potential triggering factor for the mass spread of the SARS-CoV-2 virus. Contamination might occur when people unconsciously come into contact with an infected individual who is sniffling, coughing, or releasing respiratory vapor through their nose or mouth [93,94,95]. Even these droplets remain for a long period in the environment, and may cause infection to a person staying at a distance of more than a meter [95]. Still, more investigation is required to get a clear insight into the community transmission process of SARS-CoV-2 [96].
10. Fatality Rate of the Pandemic
Fatality rates can be an indicator of the disease severity and healthcare quality of an outbreak like COVID-19. By now, two distinguished criteria are used to facilitate the understanding of the death proportion of an infected population, namely infection fatality ratio (IFR) and case fatality ratio (CFR). The IFR is calculated by considering all the deaths against the total number of infected individuals. In comparison, CRF counts the death rate by dividing total deaths by individuals with confirmed cases. However, use of the term “rate” here is theoretically incorrect, as the “rate” assigns a time component, and the time factor lacks in CRF. Therefore, curators have proposed calling this the “case fatality proportion’’ or “case fatality ratio’’. The accurate estimation of the IFR and CFR depends upon the definite counting of infected people, and the number of deaths resulting from the disease. In the case of an ongoing pandemic like COVID-19, most of the fatality ratio records being conducted depend upon surveillance and crude tracking. Consequently, the CFR varies between countries, ranging from below 0.1% to over 25% [97,98]. The IFR and CFR equations recommended by WHO [97] are as follows:
In the case of an ongoing pandemic, using the above-mentioned formula provides conditional CFR estimation due to reporting lag dates of the deaths and cases. As a result, the curated version [97] is as follows:
A study on laboratory-confirmed cases of COVID-19 declared the global CFR for Singapore to be from ~0.1% to ~6.9%, and from ~0.1% to ~16.3% in Belgium [99]. Furthermore, CFR has been found to be low in people below 19 (0–0.1%) and from 20 to 54 (0.1–0.8%) years of age [100,101]. There is a significant rise in the CFR along with an increase in age, such as between 55 and 74 (1.4–4.9%) years, 75 and 84 (4.3–10.5%) years, and ≥85(10.4–27.3%) years [102,103]. The situation was, surprisingly, more dreadful in patients with comorbidity, including diabetes, cardiovascular problems, threatening tumors, liver, and kidney-related intricacies, or bargained invulnerable systems [104]. Additionally, weight and hypertension are reasons for higher CFR. Conversely, high COVID-19 testing facilities, healthcare facilities, and people living in rural areas are significantly less prone to CFR [98]. Surprisingly, SARS-CoV-2 has comparatively lower CFR value than previous SARS CoV-1 (CFR 9.5%) and MERS (CFR 34.4%) [105]. Furthermore, SARS-CoV-2 has a higher transmission ability with a Ro (reproduction rate) of about 2.5%, by WHO analysis [106]. Elevated values of Ro and CFR create more vulnerable and fragile situations for the healthcare system [107,108].
11. Pathogenesis Process
11.1. Transmission
At first, the cases were identified with direct exposure to contaminated creatures (transmission from animal to human) at a fish market in Wuhan, China. After that, clinical cases with community transmission came to light. This supported the idea regarding a doable human-to-human transmission of the infection. In this way, human-to-human transmission is currently viewed as the principal type of transmission. The virus can also be transmitted from an asymptomatic person; yet, symptomatic individuals are the predominant transmission source. The transmission process begins with the passing of respiratory droplets via sneezing or coughing [19]. The study also reports that transmission can occur due to nearby interaction between individuals [109]. Current statistics suggest three to seven days as the incubation period for the virus, whereas some studies depicted it to be closer to five days and the incubation time between 2 and 14 days [110,111,112,113]. These outcomes have been recorded from earlier cases. Therefore, advanced studies are required to track down transmission dynamics and incubation times [20]. Transmission can be traced down to the patient’s natural liquids, for instance, respiratory beads, spit, excrement, and pee [9]. The most appropriate temperature for virion endurance demonstrates that 22 °C has a less noteworthy endurance rate than 4 °C [114,115]. As SARS-CoV-2 virions are held all throughout the clinical course, patients with asymptomatic symptoms of COVID-19 can spread the disease as well, during the indicative course, and at the hour of the clinical recuperation time frame. Furthermore, additional alerts ought to be kept up during the endurance time of the SARS-CoV-2 virions on a superficial level. SARS-CoV-2 infection half-life on copper, vaporizers, cardboard, hardened steel, and plastic surfaces are 1, 1.5, 3.4, 5.6, and 6.8 h individually, as per the information given by specific tests [116].
11.2. Pathological Process
Although β coronavirus shows a higher species conservancy than the other coronavirus families, the minor mutation in its genome can suddenly change to its pathogenicity, tissue tropism, and host range. The introduction of this zoonotic disease (SARS CoVs and MERS) into the human chronicle provides an indisputable sign of adaptability of these virus particles [117,118]. There is a censorious role played by the median hosts that completes the cross-species transmission process, and this is important at the same time for building the connection between the virus and the new hosts to facilitate the viral replication process in a new environment [119].
In order to limit the quick pervasiveness of SARS-CoV-2, immediate surveillance is required to monitor its further host adaptation, transmissibility, infectivity, viral evolution, and pathogenicity. The receptor and multiple molecular interactions are some of the modulating factors for the viral host range. Regardless of the difference between the SARS CoV and SARS-CoV-2, structural similarity was found in their spike protein’s receptor-binding domain (RBD) [31]. Moreover, critical residues such as Asn501 and Gln493 modulate the interaction between the ACE2 from host receptors and RBD of SARS-CoV-2, which ultimately facilitates the person-to-person transmission process [120].
11.3. Host Response
SARS-CoV-2 adopts a dodgy mechanism to escape the host’s innate immune response and successfully infect healthy individuals. Since SARS-CoV-2 and SARS-CoV show close clinical manifestations, SARS-CoV-2 has unique pathogenesis and biochemical processes comparable to SARS CoV [121]. Following SARS-CoV infections, the release of IFN-stimulated genes (ISGs) induced by the type I interferon (IFN) occurs against the viral replication process. In response to this defense activity by host cells, SARS-CoV-2 secretes a minimum of eight antagonizing factors that neutralize the ISG’s activity [122]. However, the immune response mediated by the host’s defense system poses a critical role in limiting viral dissemination. Nevertheless, excess promptness of the immune cells, coupled with lytic stress by viral invasion, results in severe disease outcomes. The most common symptoms manifested at the early stage include patients with acute body pain, pneumonia, dry cough, and fever [121,123]. Further progress rapidly results in acute respiratory stress syndrome (ARDS), septic shock, and organ failure, followed by death [123]. After COVID-19 infection, the development of ARDS and the subsequent damages occurring at the alveolar type II pulmonary cells as well as in the ciliated epithelium cells of the airways suggest that ACE2 is abundant in lung cells, acting as a gateway for virus entry [124]. A similar mode of the inflammatory cascade has been reported in both SARS-CoV-2 and SARS-CoV infected patients. A bulk load of pro-inflammatory cytokines (e.g., interleukin (IFN-γ, IFN-γ-induced protein 10 (IP10), IL-1, IL-6, IL-12)), monocyte chemoattractant protein-1 (MCP1), and macrophage inflammatory proteins1A (MIP1A) were found in serum collected from previously infected patients with SARS-CoV. These immune cells are also related to severe lung damage and pulmonary inflammation [125]. In addition, a high level of pro-inflammatory cytokines such as TNF-α, IL1β, IL-2, IL7, and MCP1were found in samples of SARS-CoV-2 infected patients. On the other hand, (ICU) intensive care unit patients of COVID-19 contained a greater load of cytokines such as TNF-α, GSCF, MCP1, and IP10 compared to the non-ICU patients. This suggests that cytokine storms can be the underlying cause of severe disease manifestation [31]. The study reports that a high cytokine concentration has a reversible correlation with the total immune cell (T, CD4+, and CD8+) counts in SARS-CoV-2 infection. Hence, lower T cell counts can be a potential biomarker for risk stratification of patients infected with COVID-19 [126]. Besides, other findings reported that adults (males) are more vulnerable to SARS-CoV-2 infection than young children [121,123]. A similar finding was observed in the experiment on a primate model called Cynomolgus macaque (crab-eating monkey) [127]. Further follow-up studies are warranted from the scientific community to get a better understanding of the virulence factors of SARS-CoV-2 [128].
12. Post-Infection Outcomes
12.1. Clinical Features
Clinical signs of SARS-CoV-2 infection have close likenesses with SARS-CoV, where the most well-known indications incorporate fever, dry hack, dyspnea, chest torment, exhaustion, and myalgia [31,129]. Less continuous signs include migraine, unsteadiness, stomach torment, loose bowels, queasiness, and regurgitating [31,130]. Given the data of the first 425 affirmed cases from Wuhan, the most well-known signs incorporate fever, dry hack, myalgia, and weariness. Then again, more uncommon side effects are sputum creation, migraine, hemoptysis, stomach torment, and looseness of the bowels (Table 3) [131]. Around 75% of patients have bilateral pneumonia [132]. Apart from SARS-CoV and MERS viral infections, COVID-19 patients display noteworthy upper respiratory tract symptoms, including sore throat, rhinorrhea, and sneezing [31]. Extreme inconveniences and intricacies, such as hypoxemia, intense ARDS, arrhythmia, stun, severe cardiovascular damage, and serious renal damage, have been noted in COVID-19 patients [31,132]. A study of 99 patients revealed that approximately 17% of patients displayed ARDS, and 11% of patients died of several organ failures. [132]. The median time from onset of symptoms to ARDS is eight days [133].

Table 3.
Clinical manifestations and laboratory-based findings of a patient with SARS-CoV-2 viral infection.
12.2. Disease Prognosis
Reports from the study based on a large number of positive cases of COVID-19, especially the severely ill patients, found complications such as ARDS, acute cardiac injury, acute renal injury, shock, and secondary infection. The fatality rate due to COVID-19 infection fluctuated from 0 to 14.6% [121,123,134,135,136]. A comparative data analysis between the ICU patients and the non-ICU patients showed that most aged adults with comorbidities occupied the ICU and had more frequent abdominal pain, anorexia, and dyspnea [134]. Concurrently, it is suggested that patients in ICU deposited a bulk amount of chemokine and plasma cytokines that include TNFα, GSCF, MCP1, MIP1A, IL2, IL7, IL10, and IP10 [121]. Conversely, the non-surviving patients develop more acute lymphopenia and enriched levels of neutrophil, d-dimer, and fibrin derogation products than the surviving individuals [134,137]. As of 22 January 2020, an analysis conducted on 17 death cases of COVID-19 by Wang et al. found that the median days from the onset of early symptoms until death were 14.0 (range 6–41) days, and consistently appeared to be shorter for older individuals [138]. Symptoms such as abdominal pain and dyspnea, comorbidities, age, and significant clinical anomalies (elevated d-dimer, lymphopenia, etc.) may all lead to the risk of poor outcomes (Table 4) [134,138,139,140]. However, SARS-CoV-2 has a comparatively lower mortality rate than SARS-CoV and MERS [72,141].

Table 4.
Risk factors associated with COVID-19 patients.
13. Diagnostic Strategies for COVID-19
To track down the disease severity, complicated detection methods are inevitable. There are two types of diagnosis methods—(1) molecular or viral test and (2) serological or antibody test [142]. The molecular tests involve the testing of a nasopharyngeal (NP) swab or oropharyngeal (OP) swab, stool, sputum, or bronchoalveolar lavage for the existence of viral RNA. However, the FDA suggests patients’ sputum for testing in the laboratory as an “Emergency Use Authorization” [142]. WHO prescribed that samples be collected from both lower and upper respiratory tracts [20]. The molecular test consists of two types, the (1) PCR test and (2) antigen test. Additionally, a serology test involves tracing antibodies in blood samples to ensure prior viral infection of a patient [143]. Molecular tests provide the utmost accuracy relative to other detection methods [144]. Combining both of the methods gradually increases the detection sensitivity (p < 0.001) [145]. The FDA has authorized two hundred manufacturers for molecular tests and 50 manufacturers for serological tests [146]. Serology tests can be executed as an elective identification approach on the off chance that the sub-atomic test is inaccurate; however, both of the discovery techniques have a few downsides. The sub-atomic perceptibility may be diminished due to inappropriate sample handling or issues incurred when blood tests are performed before the principal seven-day stretch of the disease. Serology test results will be flawed when not performed at the appropriate time, since IgM does not appear until seven days after infection and IgG until 15 days [145]. RT-PCR is a foremost testing technique that utilizes warm cycling to create about 35 billion DNA duplicates from RNA records. Fluorescent color is utilized to recognize the presence of target tests. The outcome is positive after it arrives at a specific cycle limit (Ct-esteem). For the same RT-PCR, another atomic procedure RT-LAMP (Reverse Transcriptional Loop-Mediated Isothermal Amplification) uses a similar method. However, rather than applying a progression of various temperature cycles, like RT-PCR, it prompts a steady temperature size of around (60–65) °C. The uniqueness of this technique is that it very well may be estimated with unaided eyes, since a compound called “magnesium pyrophosphate” makes an overcast shading. Simultaneously, serological tests including ELISA and lateral flow measures distinguish antigens and neutralizers in blood tests [147]. Besides, computerized tomography (CT) can be a compensative choice for the deficit of RT-PCR technology. An experimental report from Wuhan depicts that CT images referred to 98% detection sensitivity compared to RT-PCR (78%). CT images have also been used in Turkey because of test kit shortage [148]. However, cell culture is not recommended due to its numerous drawbacks. Additionally, CRISPR-Cas mediated technology can become another potential diagnostic method over time [144].
14. Prevention against COVID-19
14.1. Preventive Measures
The shock of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in 2002 and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012 were viewed as the most effective variation of COVIDs until the emergence of COVID-19, the third novel Covid to cause a wide-scale scourge in the 21st century [149]. COVID-19 has far stronger transmissibility compared to prior variants, causing emergent outbreaks throughout the world. Though COVID-19 has been considered a category “B” infectious disease by law, it is being treated and handled as a category “A” contagious disease due to its rapid transmissibility. Transmission root impeding, source controlling, and protecting susceptive people can effectively impede the illness’ severity [150]. The spread of COVID-19 can be limited by agreeing with the accompanying rules given by WHO to the worldwide populace, which include the following [151].
- Optimizing infection control protocols, self-isolation, and isolated accommodation for patients.
- Immunocompromised, aged persons must take special care; health care personnel must use PPE such as N95, FFP3 mask, gowns, face shield, etc.
- Frequent handwashing with soap and water, with hand sanitizer as an alternative disinfection process; alcohol can also be used.
- Avoid public gatherings and maintain social distancing of at least 1 m. Must wear face masks and cover coughs and sneezes to help prevent aerosol transmission.
Public health measures are daily precautionary activities that include:
- All of us should put on face masks and maintain respiratory and hand hygiene.
- Staying home during sickness.
- Covering mouth and nose with bent elbow or tissue at the time of sneezing or coughing. Used tissue should be disposed of instantly.
- Washing hands regularly with soap and water. Hand hygiene refers to the use of alcohol-based hand sanitizers before and after each patient’s contact as well as after touching any surface, transport, money, etc. Washing hands with soapy water for 20 s is also very effective and essential for everyone. It is also advised to maintain hand hygiene before and after cooking, before eating, before and after removing gloves and face masks, and after using the toilet. Distinct reusable utensils can be used by the patient after proper cleaning.
- Avoiding handshaking is also advised for the time being for your protection.
- Cleaning regularly touched surfaces and objects. Disinfecting the surrounding environment of the patient, including toilet and furniture [152]. Clean and disinfect bathroom and toilet surfaces at least once every day. Regular household soap or detergent should be used first for cleaning and rinsing purposes; regular household disinfectant and germ killer containing 0.5% sodium hypochlorite (NaOCl) should be applied [153].
- Provisionally, prevention is the best way to stop the spread of infection. WHO has boldly recommended hand and respiratory hygiene for cases, contacts, and health care workers [154].
- The medical musk is not needed for the general public, excluding doctors and people associated with the health care system [155]. Patients and caregivers are recommended to remove the mask from behind and practice hand hygiene after removing it [153].
- The doctors and health workers are particularly advised to wear personal protective equipment (PPE), including an N95 respirator mask and eye shield, while collecting respiratory samples. Avoiding contact with body fluids is mandatory [156].
- The duration of quarantine is 14 days from the last contact with PCR confirmed COVID-19 case. They should be checked daily for fever and other symptoms associated with COVID-19 [157].
- Children’s health monitoring is necessary during this time. Children with a history of close contact with infected patients need to be routinely observed for body temperature and clinical features. When presenting with doubtful symptoms, children should be taken to a delegated hospital for screening. Newborns delivered by COVID-19 infected mothers must complete a pathogen test and be isolated in a single cabin room or at home according to their medical conditions [150].
14.2. Treatment
As new immunizations are intermittently gaining endorsement, this doesn’t guarantee accessibility, nor the adequacy of the antibodies. Experts assume that around 90% of individuals of lower-income nations will be denied immunization until 2023 or later. At the same time, immunization aversion in wealthier countries like the USA may transform this into an even more mind-boggling circumstance. Subsequently, even though immunizations are continuously being provided locally, this is no time to overlook security measures (individual cleanliness, face masks, social separating) [158]. However, researchers are trying to globally defend against this virus attack with every possible means. After a comprehensive literature review, we categorized the whole treatment approach into two major groups: (1) physiological treatment and (2) mediated treatment (Figure 6). The physiological treatment mainly involves monitoring viral symptoms and providing equipment support (oxygen saturation, balanced water-electrolyte level, balanced diet, etc.) [159].
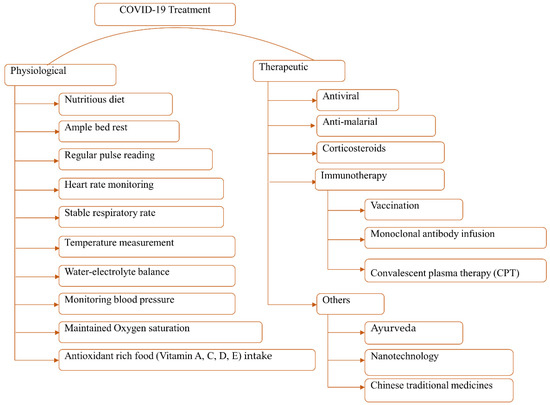
Figure 6.
Overview of the possible COVID-19 treatment measures.
Coronavirus patient management depends upon steady estimates that incorporate ventilation, liquid administration, oxygenation, low–portion antiviral complex, and interferon inward breath [160,161]. The significant commitment before intensive treatment is to confine any presumed patients in a different room, as per the specialist’s proposal. Basic cases are promptly take to the ICU. The resulting treatment course includes sufficient bed rest, consuming sufficient water and calories, and guaranteeing sufficient oxygen immersion [150,162]. A study suggests that mildly symptomatic patients can be treated at home by providing standard care, including an adequate supply of proper nutrition, hydration, fever, and cold management. For patients with fatigue and a temperature above 38.5 °C, it is recommended to provide a lukewarm water bath and antipyretic drug therapy. However, routine medication with antibiotics and anti-viral drugs is highly prohibited [162]. Along with supportive treatment, some potential COVID treatments have been tested in trials, listed in Table 5.

Table 5.
Different approaches of prospective therapeutic trials against nCoV-19.
Moreover, repurposing of existing antiviral medications, including lopinavir, ritonavir, chloroquine, interferon beta, and remdesivir, has been launched as an international clinical trial by the WHO SOLIDARITY trial project [163]. However, these medication trials could not provide any reliable remedy, and two of the reports rejected the use of these antiviral drugs [164]. Surprisingly, remdesivir has been proved to be efficient for severe patients with increased ventilation and proved to be less risky [165].
Moreover, another multidimensional approach for COVID-19 treatment involves 5000-year-old traditional Chinese medicine (TCM), including Huoxiang Zhengqi, Lianhua Qingwen, Shufeng Jiedu, and Xue Bijing used as pills, powder, tea, or tincture, which was proven to be effective in 2002 against the SARS-CoV infection at an early stage by increasing patients’ oxyhemoglobin arterial saturation [168,173]. Furthermore, the intake of antioxidant-rich foods containing Vitamin A, C, D, and E can mitigate the damage via oxidation as well as the risk of cardiovascular disease [159]. Additionally, curcumin, honey, omega-3 fatty acids, and potato starch have some natural anti-inflammatory activities [159,174]. On March 16 of 2020, Regeneron Pharmaceuticals Inc., US, and Sanofi collaboratively announced the phase 2 and 3 clinical trials of a human monoclonal antibody called Sarilumab (Kevzara) against the interleukin-6 (IL-6) receptor on about 400 hospitalized corona patients [167]. On the other hand, a study showed the prophylaxis potential of Tocilizumab (humanized monoclonal antibody) and Melatonin (N-acetyl-5-methoxytryptamine) as two candidate molecules for nCoV-19 treatment [166,175]. Other potential COVID-19 treatments may include nanotechnology, nitric oxide (NO), CRISPR/Cas13d System, Yoga, and Ayurveda [149,176,177,178].
Vaccination
As of now, vaccination is considered a definitive encapsulation comparative with any remaining existing solutions for COVID-19 outbreak. Researchers universally are working next to each other, utilizing accessible innovation and labor to beat this pandemic [159,179]. This overall process may need around 12 to 18 months for achieving promising outcomes and high throughput manufacturing [180]. By now, mainly the mRNA, epitopes, and spike protein RBD (receptor binding domain) structure-based vaccine initiatives are being proposed (Table 6) [181]. At present, 98 vaccines are being evaluated on humans clinically, 32 are in their last phase of the trial, and a minimum of 77 vaccines are in their active preclinical animal trial [182]. Several leading vaccine candidates were listed in Table 6 according to the data of 27 July 2021.

Table 6.
Top 10 leading vaccine candidates with their efficacy level worldwide as of 27 July 2021 [182].
The vaccine pillar of the ACT, COVAX, is co-led by Gavi (the Coalition for Epidemic Preparedness Innovations) and WHO. It is formed to maintain the pace of vaccine development and production and ensure the equal distribution of the vaccine, so that no low-income country lags in vaccination. COVAX set a target of vaccine supply that can compensate a minimum of 20% of the country’s population [183]. At this point, AstraZeneca has agreed to provide vaccines to COVAX, whereas Moderna and Pfizer have not yet confirmed anything. On the other hand, there are also some vital issues relating to the mass supply and conservation of the vaccines in remote places, including cold-chain requirements, logistics, and delays, which has made the situation more challenging [184].
14.3. Boosting Immunity
Boosting resistance includes measures like a reasonable eating regimen, standard exercise, maintaining a strategic distance from additional pressure and weariness, oral wellbeing, and sufficient rest, that are significant not just in forestalling different contaminations, like viral assault, but also in maintaining mental and psychological wellness. Organic citrus products containing Vitamin C (ascorbic corrosive) and different nutrients can stimulate the insusceptible framework, as well. Overall, inoculation is the most essential step to protect against the resistant triggers of a viral assault like SARS-CoV-2 [150].
15. Infection Associated Factors
15.1. Gender Biases
The reports from different case study data indicate a clear contrast between men and women, especially in the case of mortality and other disease outcomes [185]. Studies governed in China and Italy showed that the mortality rate in men is much higher than in women [6,123,186]. The higher ACE2 receptor expression in the lungs of men compared to women can be a causative factor for this [187]. Moreover, the risk appears higher in men in their 50s and starts to mitigate in the 90s. In China, the mortality rate for men and women was 2.8% and 1.7%, respectively [186]. The exact reasons behind the gendered impact of COVID-19 remain unsolved, although behavioral and genetic factors could be plausible indicators for this [185]. Reasons why men may possess greater susceptibility to COVID-19 relative to the female population may include a higher smoking rate in men, sex-based immunological variance, and a higher portion of comorbidity (hypertension, heart disease) in men [186].
15.2. Ethnic Differences
The COVID-19 effects on various ethnic groups are affected fundamentally by the aberrations between medical care availability and financial status. Lower financial conditions and a raised pace of comorbidity impact the COVID-19 withdrawal [188]. In March 2020, The CDC and COVID-19–Associated Hospitalization Surveillance Network conducted a study on 580 hospitalized COVID-19 patients with ethnicity data from 14 states in the US. This study found that, among the patients, about 45.0% were Caucasian (~76% of the US population), 33% were African-American (~13% of the US population), 8% Hispanic (~18% of the US population), and the remainder consisted of 5% Asian, less than 1% American Indian/Alaskan Native, and 7.9% from unknown or other races. However, the racial distribution varied by state depending on the target population size. It has been noted that smaller populations like African Americans have greater COVID-19 infection risk as they are much more prone to have heart disease, asthma, obesity, hypertension, and diabetes [189]. Moreover, minority populations also possess a higher number of people with low socio-economic conditions, contributing to the risk of COVID-19 contraction. A prior investigation on CytoMegalo Virus (CMV) found a significant relationship between cell-mediated immunity and low socio-economic status among different socio-economic populations in the USA, suggesting that people with poor socio-economic condition have access to fewer healthcare facilities, greater life stressors, and a poorer diet, which results in a weak immune system [190,191,192]. On the other hand, higher ACE2 receptor expression in Asian people than in the African American and Caucasian populations may cause dispersed outcomes across different countries [187].
15.3. Environmental Effect
Viruses do not replicate outside of a living cell, yet the infectious viruses may continue on infected natural surfaces, and the span of an active infection is obviously controlled by certain ecological boundaries, for example, temperature and moistness [193]. Dirty surfaces are significant vectors in the infection transmission measure in medical clinic conditions, just as in the local area. A recent study reported that surfaces and suspensions could be recognized as potential transmission sources that might lead to hospital-acquired infections with human coronavirus [193]. The environmental sustainability of SARS-CoV-2 was previously unknown. However, various studies have demonstrated that SARS CoV can withstand at least two weeks in dry conditions. The dried-out virus on smooth surfaces maintains its viability for more than five days at a temperature of 22–25 °C and relative humidity of 40–50%. The virus remains constant for three weeks at room temperature in a fluid environment, but it is easily killed by heat treatment at 56 °C for 15 min [194]. However, virus viability was quickly lost when temperature and humidity were relatively higher (38 °C, and relative humidity of >95%). This indicates that SARS-CoV-2 is a stable virus that may be transmitted through indirect contact or fomites [195]. Such consequences may allude to the significant role of contaminated surfaces in pathogen transmission in the hospital and the community. SARS-CoV-2 is relatively firmer and steadier than the human coronaviruses 229E or OC43, and some other viral respiratory pathogens, such as a respiratory syncytial virus. Generally, direct droplet transmission is an important route of transmission [195]. The robust stability of SARS coronavirus at low temperature and in a low humidity environment may allow for its rapid community transmission in subtropical areas (such as Hong Kong) during the spring season. These findings may also explain the reason why some Asian nations in equatorial areas (for example, Malaysia, Indonesia, or Thailand) with high temperatures and relatively humid environments did not have strong community outbreaks of the SARS virus compared to European regions [196].
15.4. Blood Group and Coronavirus
A study indicates that people having blood group A is related to greater danger of disease by COVID-19 contrasted with non-A blood groups, while blood group O was connected with a similarly lower risk of contamination than non-O blood groups [197]. Ensuing investigation showed that A positive individuals are at a 45%increased respiratory danger, though individuals with blood group O are at a 35% diminished likelihood of having respiratory complications. Late, an Italian–Spanish genome-wide affiliation examination-based investigation uncovered the linkage between blood groups and coronavirus [197]. The effect of age and gender on the ABO blood group distribution in patients with novel coronavirus from two distinct hospitals was analyzed. The results concluded that age and gender do not impact the blood group [198]. However, interestingly, the consequence is significant for A and O blood group type only when the Rh factor is positive in blood samples [199].
16. Conclusions and Future Directions
The current COVID-19 pandemic situation has indeed affected the whole world, socially and economically. The world is now on the move to compensate for the damage done by COVID-19, and to fight back by developing vaccines, many of which are already being delivered globally. However, it should be noted that vaccines are effective only when vaccination is governed properly. Equity distribution to the lower-income countries and willing participation towards vaccination should be maintained to eradicate this crisis. Despite the vaccines’ release, the global population should regularly practice personal hygiene, face masking, and social distancing until the vaccines are proved fully functional and available globally. However, the minimization of the huge load on the healthcare system and the growing mortality rate are two major problems currently receiving global efforts. The lesson taught by COVID-19 should be remembered for a long time, and advanced preparation is needed for any further global hazards that can put global health at stake. Furthermore, paying considerable attention to the plausible cause of this virus’ transmission, there should be a total restriction on trading wild creatures and birds as a food source across the globe, and further investigations on SARS-CoV-2 genomic makeup must be carried out to understand its pathogenicity, epidemiology, and transmission pattern. Disclosing the genomic construction can provide insight into effective drug design and vaccine construction for subsequent variants. Artificial intelligence (AI) could also be an excellent tool to track down the transmission pattern of a deadly virus before it turns into an outbreak. Therefore, uninterrupted surveillance and proper coordination between national and international health stakeholders can balance the situation. Since the advancement in technology gave us the liberty to utilize our utmost potential to contribute to the betterment of human civilization, so this should be the time to make the best use of it.
Author Contributions
Conceptualization, M.T.S., A.Q.F.H. and M.O.R.; validation, M.T.S., M.J.H., A.Q.F.H., M.O.R., H.R.A.E.-M., R.M.E., R.C. and T.B.E.; formal analysis, M.T.S., M.J.H., A.Q.F.H., M.O.R. and T.B.E.; investigation, M.T.S., A.Q.F.H. and M.O.R.; resources, M.T.S., A.Q.F.H., M.O.R., M.J.H. and T.B.E.; data curation, M.T.S., M.J.H., A.Q.F.H. and M.O.R.; writing—original draft preparation, M.T.S., A.Q.F.H. and M.O.R.; writing—review and editing, M.T.S., M.J.H., A.Q.F.H., M.O.R., H.R.A.E.-M., R.M.E., R.C. and T.B.E.; visualization, M M.T.S., M.J.H., A.Q.F.H., M.O.R., H.R.A.E.-M., R.M.E., R.C. and T.B.E.; supervision, M.O.R., R.C. and T.B.E.; project administration M.O.R., R.C. and T.B.E.; funding acquisition, R.C. and T.B.E. All authors have read and agreed to the published version of the manuscript.
Funding
This work was conducted with the individual funding of all authors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Available data are presented in the manuscript and reference list.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Hossain, J.; Ahmmed, F.; Rahman, S.A.; Sanam, S.; Bin Emran, T.; Mitra, S. Impact of online education on fear of academic delay and psychological distress among university students following one year of COVID-19 outbreak in Bangladesh. Heliyon 2021, 7, e07388. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. Mystery deepens over animal source of coronavirus. Nat. Cell Biol. 2020, 579, 18–19. [Google Scholar] [CrossRef] [PubMed]
- Hossain, J.; Kuddus, R.; Rahman, S.M.A. Knowledge, Attitudes, and Behavioral Responses Toward COVID-19 During Early Phase in Bangladesh: A Questionnaire-Based Study. Asia Pac. J. Public Heal. 2021, 33, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Update (Live): 96,095,456 Cases and 2,051,742 Deaths from COVID-19 Virus Pandemic—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 19 January 2021).
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.L.; Chu, D.K.W.; Shiu, E.Y.C.; Chan, K.-H.; McDevitt, J.J.; Hau, B.J.P.; Yen, H.-L.; Li, Y.; Ip, D.K.M.; Peiris, J.S.M.; et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020, 26, 676–680. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Bari, S.; Hossain, J.; Akhter, S.; Bin Emran, T. Delta variant and black fungal invasion: A bidirectional assault might worsen the massive second/third stream of COVID-19 outbreak in South-Asia. Ethic- Med. Public Health 2021, 19, 100722. [Google Scholar] [CrossRef]
- Hossain, M.D.J.; Soma, M.A.; Islam, M.D.R.; Bin Emran, T. Urgent call for actionable measures to fight the current co-epidemic of dengue burden during the SARS-CoV-2 delta variant era in South-Asia. Ethic- Med. Public Health 2021, 19, 100726. [Google Scholar] [CrossRef]
- World Health Organization. Novel Coronavirus—China. Available online: https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ (accessed on 15 January 2021).
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726. [Google Scholar] [CrossRef]
- Dyer, O. Covid-19: WHO says laboratory escape theory is “extremely unlikely” after mission to China. BMJ 2021, 372, n428. [Google Scholar] [CrossRef]
- Samadizadeh, S.; Masoudi, M.; Rastegar, M.; Salimi, V.; Shahbaz, M.B.; Tahamtan, A. COVID-19: Why does disease severity vary among individuals? Respir. Med. 2021, 180, 106356. [Google Scholar] [CrossRef]
- Nao, N.; Yamagishi, J.; Miyamoto, H.; Igarashi, M.; Manzoor, R.; Ohnuma, A.; Tsuda, Y.; Furuyama, W.; Shigeno, A.; Kajihara, M.; et al. Genetic Predisposition To Acquire a Polybasic Cleavage Site for Highly Pathogenic Avian Influenza Virus Hemagglutinin. mBio 2017, 8, e02298-16. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.T.; Leung, K.; Leung, G. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef]
- Wu, P.; Hao, X.; Lau, E.; Wong, J.Y.; Leung, K.; Wu, J.T.K.; Cowling, B.J.; Leung, G.M. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Eurosurveillance 2020, 25, 2000044. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Hassan, S.A.; Sheikh, F.N.; Jamal, S.; Ezeh, J.K.; Akhtar, A. Coronavirus (COVID-19): A Review of Clinical Features, Diagnosis, and Treatment. Cureus 2020, 12, e7355. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Kakodkar, P.; Kaka, N.; Baig, M.N. A Comprehensive Literature Review on the Clinical Presentation, and Management of the Pandemic Coronavirus Disease 2019 (COVID-19). Cureus 2020, 12, e7560. [Google Scholar] [CrossRef]
- Liu, P.; Chen, W.; Chen, J.-P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351. [Google Scholar] [CrossRef]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Genet. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed]
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 May 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-may-2020 (accessed on 15 January 2021).
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Qinfen, Z.; Jinming, C.; Xiaojun, H.; Huanying, Z.; Jicheng, H.; Ling, F.; Kunpeng, L.; Jingqiang, Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med Virol. 2004, 73, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Du, E.-Z.; Luo, W.-T.; Yang, Y.-L.; Zhang, Y.-Q.; Wang, B.; Huang, Y.-W. Characteristics of the Life Cycle of Porcine Deltacoronavirus (PDCoV) In Vitro: Replication Kinetics, Cellular Ultrastructure and Virion Morphology, and Evidence of Inducing Autophagy. Viruses 2019, 11, 455. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Nainu, F.; Abidin, R.S.; Bahar, M.A.; Frediansyah, A.; Emran, T.B.; Rabaan, A.A.; Dhama, K.; Harapan, H. SARS-CoV-2 reinfection and implications for vaccine development. Hum. Vaccines Immunotherap. 2020, 16, 3061–3073. [Google Scholar] [CrossRef]
- Turner, A.J.; Hiscox, J.; Hooper, N. ACE2: From vasopeptidase to SARS virus receptor. Trends Pharmacol. Sci. 2004, 25, 291–294. [Google Scholar] [CrossRef]
- Gupte, M.; Boustany-Kari, C.M.; Bharadwaj, K.; Police, S.; Thatcher, S.; Gong, M.C.; English, V.L.; Cassis, L.A. ACE2 is expressed in mouse adipocytes and regulated by a high-fat diet. Am. J. Physiol. Integr. Comp. Physiol. 2008, 295, R781–R788. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Poduri, R.; Joshi, G.; Jagadeesh, G. Drugs targeting various stages of the SARS-CoV-2 life cycle: Exploring promising drugs for the treatment of Covid-19. Cell. Signal. 2020, 74, 109721. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9. [Google Scholar] [CrossRef]
- Hikmet, F.; Méar, L.; Edvinsson, Å; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.; Puranik, A.; Anand, A.; Zemmour, D.; Yao, X.; Wu, X.; Chilaka, R.; Murakowski, D.K.; Standish, K.; Raghunathan, B.; et al. Knowledge synthesis of 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. eLife 2020, 9. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.-M.; Raoult, D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425–435. [Google Scholar] [CrossRef]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Krämer-Kühl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The Spike Protein of the Emerging Betacoronavirus EMC Uses a Novel Coronavirus Receptor for Entry, Can Be Activated by TMPRSS2, and Is Targeted by Neutralizing Antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef]
- Park, J.-E.; Li, K.; Barlan, A.; Fehr, A.R.; Perlman, S.; McCray, P.B.; Gallagher, T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. USA 2016, 113, 12262–12267. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Kar, S.; Aliter, K.F. Potential Anti-COVID-19 Therapeutics that Block the Early Stage of the Viral Life Cycle: Structures, Mechanisms, and Clinical Trials. Int. J. Mol. Sci. 2020, 21, 5224. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Kar, S. Potential Anti-SARS-CoV-2 Therapeutics That Target the Post-Entry Stages of the Viral Life Cycle: A Comprehensive Review. Viruses 2020, 12, 1092. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Jabeen, N.; Amanullah, A.; Baig, A.A.; Aziz, B.; Shabbir, S.; Raza, F. Structural Basis of SARS-CoV-2 Spike Protein Priming by TMPRSS2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Brielle, E.; Schneidman-Duhovny, D.; Linial, M. The SARS-CoV-2 Exerts a Distinctive Strategy for Interacting with the ACE2 Human Receptor. Viruses 2020, 12, 497. [Google Scholar] [CrossRef]
- Khadim Sheikh, H.; Arshad, T.; Mohammad, Z.S.; Arshad, I.; Hassan, M. Repurposed Single Inhibitor for Serine Protease and Spike Glycoproteins of SAR-CoV-2. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of RNA-Dependent RNA Polymerase from 2019-NCoV, a Major Antiviral Drug Target. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shannon, A.; Le, N.T.-T.; Selisko, B.; Eydoux, C.; Alvarez, K.; Guillemot, J.-C.; Decroly, E.; Peersen, O.; Ferron, F.; Canard, B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020, 178, 104793. [Google Scholar] [CrossRef]
- Shu, T.; Huang, M.; Wu, D.; Ren, Y.; Zhang, X.; Han, Y.; Mu, J.; Wang, R.; Qiu, Y.; Zhang, D.-Y.; et al. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol. Sin. 2020, 35, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Emergence, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef]
- Atri, D.; Siddiqi, H.K.; Lang, J.P.; Nauffal, V.; Morrow, D.A.; Bohula, E.A. COVID-19 for the Cardiologist: Basic Virology, Epidemiology, Cardiac Manifestations, and Potential Therapeutic Strategies. JACC: Basic Transl. Sci. 2020, 5, 518–536. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays J. Pathol. 2020, 42, 3–11. [Google Scholar]
- Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Chiaradia, E.; Urbanelli, L.; Emiliani, C. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int. J. Mol. Sci. 2020, 21, 2576. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.S.; McIntosh, K. History and Recent Advances in Coronavirus Discovery. Pediatr. Infect. Dis. J. 2005, 24, S223–S227. [Google Scholar] [CrossRef]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef]
- Paraskevis, D.; Kostaki, E.; Magiorkinis, G.; Panayiotakopoulos, G.; Sourvinos, G.; Tsiodras, S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020, 79, 104212. [Google Scholar] [CrossRef]
- Chen, L.; Liu, W.; Zhang, Q.; Xu, K.; Ye, G.; Wu, W.; Sun, Z.; Liu, F.; Wu, K.; Zhong, B.; et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020, 9, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Ge, H.; Wang, X.; Yuan, X.; Xiao, G.; Wang, C.; Deng, T.; Yuan, Q.; Xiao, X. The epidemiology and clinical information about COVID-19. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1011–1019. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Sevajol, M.; Subissi, L.; Decroly, E.; Canard, B.; Imbert, I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014, 194, 90–99. [Google Scholar] [CrossRef]
- Smith, E.C.; Blanc, H.; Vignuzzi, M.; Denison, M.R. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathog. 2013, 9, e1003565. [Google Scholar] [CrossRef]
- Fauver, J.R.; Petrone, M.E.; Hodcroft, E.B.; Shioda, K.; Ehrlich, H.Y.; Watts, A.G.; Vogels, C.B.; Brito, A.F.; Alpert, T.; Muyombwe, A.; et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell 2020, 181, 990–996. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhang, Y.; Li, J.; Xiao, Y.; Zhang, J.; Wang, Y.; Chen, L.; Paranhos-Baccalà, G.; Wang, J. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015, 5, 11451. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Moës, E.; Li, S.; Vandamme, A.-M.; Van Ranst, M. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology 2005, 337, 85–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chibo, D.; Birch, C. Analysis of human coronavirus 229E spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. J. Gen. Virol. 2006, 87, 1203–1208. [Google Scholar] [CrossRef]
- Guan, Y.J.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Butt, K.M.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-D.; Tu, C.-C.; Zhang, G.-W.; Wang, S.-Y.; Zheng, K.; Lei, L.-C.; Chen, Q.-X.; Gao, Y.-W.; Zhou, H.-Q.; Xiang, H.; et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- Benvenuto, D.; Giovanetti, M.; Salemi, M.; Prosperi, M.; De Flora, C.; Junior Alcantara, L.C.; Angeletti, S.; Ciccozzi, M. The global spread of 2019-nCoV: A molecular evolutionary analysis. Pathog. Glob. Health 2020, 114, 64–67. [Google Scholar] [CrossRef]
- Cohen, J. COVID-19 shot protects monkeys. Science 2020, 368, 456–457. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; Mcmahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Sui, J.; Aird, D.R.; Tamin, A.; Murakami, A.; Yan, M.; Yammanuru, A.; Jing, H.; Kan, B.; Liu, X.; Zhu, Q.; et al. Broadening of Neutralization Activity to Directly Block a Dominant Antibody-Driven SARS-Coronavirus Evolution Pathway. PLoS Pathog. 2008, 4, e1000197. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-C.; Agnihothram, S.S.; Jiao, Y.; Stanhope, J.; Graham, R.L.; Peterson, E.C.; Avnir, Y.; Tallarico, A.S.C.; Sheehan, J.; Zhu, Q.; et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA 2014, 111, E2018–E2026. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, D.; Angeletti, S.; Giovanetti, M.; Bianchi, M.; Pascarella, S.; Cauda, R.; Ciccozzi, M.; Cassone, A. Evolutionary analysis of SARS-CoV-2: How mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020, 81, e24–e27. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Rangarajan, E.S.; Izard, T.; Farzan, M.; Choe, H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Principles of Epidemiology in Public Health Practice, Third Edition an Introduction to Applied Epidemiology and Biostatistics| Lesson 3|Overview. Available online: https://www.cdc.gov/csels/dsepd/ss1978/lesson3/index.html (accessed on 17 January 2021).
- Hawks, L.; Woolhandler, S.; McCormick, D. COVID-19 in Prisons and Jails in the United States. JAMA Intern. Med. 2020, 180, 1041. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, S.; Teng, T.; Abdalla, A.; Zhu, W.; Xie, L.; Wang, Y.; Guo, X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020, 12, 244. [Google Scholar] [CrossRef]
- Coronavirus Update (Live): 78,475,152 Cases and 1,726,535 Deaths from COVID-19 Virus Pandemic—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 23 December 2020).
- Hossain, J.; Ahmmed, F.; Kuddus, R.; Alam, S.; Rahman, S.A. Exploring public awareness and spreading pattern analysis of COVID-19 outbreak in Bangladesh. Bangladesh J. Med. Sci. 2021, 20, 108–117. [Google Scholar] [CrossRef]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alsuliman, M.; Ahmed, G.Y.; Rabaan, A.A.; Al-Tawfiq, J.A.; Al-Omari, A. Knowledge of infection prevention and control among healthcare workers and factors influencing compliance: A systematic review. Antimicrob. Resist. Infect. Control. 2021, 10, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323, 1610. [Google Scholar] [CrossRef] [PubMed]
- Estimating Mortality from COVID-19. Available online: https://www.who.int/news-room/commentaries/detail/estimating-mortality-from-covid-19 (accessed on 13 February 2021).
- Ergönül, Ö; Akyol, M.; Tanrıöver, C.; Tiemeier, H.; Petersen, E.; Petrosillo, N.; Gönen, M. National case fatality rates of the COVID-19 pandemic. Clin. Microbiol. Infect. 2021, 27, 118–124. [Google Scholar] [CrossRef]
- COVID-19 Map—Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/map.html (accessed on 13 December 2020).
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- COVID-19 National Emergency Response Center, Epidemiology and Case Management Team, Korea Centers for Disease Control and Prevention Early Epidemiological and Clinical Characteristics of 28 Cases of Coronavirus Disease in South Korea. Osong Public Health Res. Perspect. 2020, 11, 8–14. [CrossRef]
- Bialek, S.; Boundy, E.; Bowen, V.; Chow, N.; Cohn, A.; Dowling, N.; Ellington, S.; Gierke, R.; Hall, A.; MacNeil, J.; et al. Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19)–United States, February 12–March 16, 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 343–346. [Google Scholar] [CrossRef]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef]
- Rajgor, D.D.; Lee, M.H.; Archuleta, S.; Bagdasarian, N.; Quek, S.C. The many estimates of the COVID-19 case fatality rate. Lancet Infect. Dis. 2020, 20, 776–777. [Google Scholar] [CrossRef]
- Hossain, J. Is Bangladesh Moving toward Herd Immunity? Current COVID-19 Perspective. Bangladesh J. Infect. Dis. 2020, 7, S63–S66. [Google Scholar] [CrossRef]
- Gharizadeh, B.; Yue, J.; Yu, M.; Liu, Y.; Zhou, M.; Lu, D.; Zhang, J. Navigating the Pandemic Response Life Cycle: Molecular Diagnostics and Immunoassays in the Context of COVID-19 Management. IEEE Rev. Biomed. Eng. 2021, 14, 30–47. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Ghinai, I.; McPherson, T.D.; Hunter, J.C.; Kirking, H.L.; Christiansen, D.; Joshi, K.; Rubin, R.; Morales-Estrada, S.; Black, S.R.; Pacilli, M.; et al. First Known Person-to-Person Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the USA. Lancet 2020, 395, 1137–1144. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Uras, U. Coronavirus: Comparing COVID-19, SARS and MERS. Available online: https://www.aljazeera.com/news/2020/4/8/coronavirus-comparing-covid-19-sars-and-mers (accessed on 16 January 2021).
- Rabaan, A.A.; Al-Ahmed, S.H.; Muhammad, J.; Khan, A.; Sule, A.A.; Tirupathi, R.; Mutair, A.; Alhumaid, S.; Al-Omari, A.; Dhawan, M.; et al. Role of inflammatory cytokines in COVID-19 patients: A review on molecular mechanisms, immune functions, immunopathology and immunomodulatory drugs to counter cytokine storm. Vaccines 2021, 9, 436. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef]
- Otter, J.A.; Donskey, C.; Yezli, S.; Douthwaite, S.; Goldenberg, S.; Weber, D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Guan, Y.; Yuen, K.-Y. Severe acute respiratory syndrome. Nat. Med. 2004, 10, S88–S97. [Google Scholar] [CrossRef] [PubMed]
- Brennan, G.; Kitzman, J.O.; Rothenburg, S.; Shendure, J.; Geballe, A.P. Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range. PLoS Pathog. 2014, 10, e1004002. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Totura, A.; Baric, R.S. SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012, 2, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Rakib, A.; Nain, Z.; Islam, M.A.; Sami, S.A.; Mahmud, S.; Islam, A.; Ahmed, S.; Siddiqui, A.B.F.; Babu, S.M.O.F.; Hossain, P.; et al. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: An in silico investigation. Brief. Bioinform. 2021, 22, 1476–1498. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Translator Wong, C.K.; Lam, C.W.K.; Wu, A.K.L.; Ip, W.K.; Lee, N.L.S.; Chan, I.H.S.; Lit, L.C.W.; Hui, D.S.C.; Chan, M.H.M.; Chung, S.S.C.; et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004, 136, 95–103. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Smits, S.L.; De Lang, A.; Brand, J.M.A.V.D.; Leijten, L.M.; van Ijcken, W.; Eijkemans, M.J.C.; Van Amerongen, G.; Kuiken, T.; Andeweg, A.C.; Osterhaus, A.; et al. Exacerbated Innate Host Response to SARS-CoV in Aged Non-Human Primates. PLoS Pathog. 2010, 6, e1000756. [Google Scholar] [CrossRef]
- Harapan, H.; Itoh, N.; Yufika, A.; Winardi, W.; Keam, S.; Te, H.; Megawati, D.; Hayati, Z.; Wagner, A.L.; Mudatsir, M. Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Health 2020, 13, 667–673. [Google Scholar] [CrossRef]
- Chen, J. Pathogenicity and transmissibility of 2019-nCoV—A quick overview and comparison with other emerging viruses. Microbes Infect. 2020, 22, 69–71. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus Infections—More Than Just the Common Cold. JAMA 2020, 323, 707. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; Van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Genet. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Rakib, A.; Sami, S.A.; Islam, M.A.; Ahmed, S.; Faiz, F.B.; Khanam, B.H.; Marma, K.K.S.; Rahman, M.; Uddin, M.M.N.; Nainu, F.; et al. Epitope-Based Immunoinformatics Approach on Nucleocapsid Protein of Severe Acute Respiratory Syndrome-Coronavirus-2. Molecules 2020, 25, 5088. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Xu, X.-W.; Wu, X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Al-Ahmed, S.H.; Garout, M.A.; Al-Qaaneh, A.M.; Sule, A.A.; Tirupathi, R.; Mutair, A.; Alhumaid, S.; Hasan, A.; Dhawan, M.; et al. Diverse immunological factors influencing pathogenesis in patients with COVID-19: A review on viral dissemination, immunotherapeutic options to counter cytokine storm and inflammatory responses. Pathogens 2021, 10, 565. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Wang, W.; Tang, J.; Wei, F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med Virol. 2020, 92, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical Course and Outcomes of Critically Ill Patients with SARS-CoV-2 Pneumonia in Wuhan, China: A Single-Centered, Retrospective, Observational Study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Zhang, J.-J.; Dong, X.; Cao, Y.-Y.; Yuan, Y.-D.; Yang, Y.-B.; Yan, Y.-Q.; Akdis, C.A.; Gao, Y.-D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East Respiratory Syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Testing FAQ—Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/testing/testing-faq/overview#how-is-covid-19-diagnosed. (accessed on 16 January 2021).
- West, R.; Kobokovich, A.; Connell, N.; Gronvall, G.K. COVID-19 Antibody Tests: A Valuable Public Health Tool with Limited Relevance to Individuals. Trends Microbiol. 2021, 29, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.N.; Chaudhury, A.; Akanda, A.M.; Hossain, M.A.; Islam, T. Genomic diversity and evolution, diagnosis, prevention, and therapeutics of the pandemic COVID-19 disease. PeerJ 2020, 8, e9689. [Google Scholar] [CrossRef]
- Shormin, M.; Yusuf, A. Laboratory Detection of Covid19 Cases: A Systematic Review. Bangladesh J. Infect. Dis. 2020, S11–S17. [Google Scholar] [CrossRef]
- Alyssa Billingsley, PharmD The Latest on Coronavirus Testing: New Methods, Accuracy, and Availability—GoodRx. Available online: https://www.goodrx.com/blog/coronavirus-covid-19-testing-updates-methods-cost-availability/ (accessed on 16 January 2021).
- Green, K.; Winter, A.; Dickinson, R.; Graziadio, S.; Wolff, R.; Mallett, S.; Allen, A.J. What Tests Could Potentially Be Used for the Screening, Diagnosis and Monitoring of COVID-19 and What Are Their Advantages and Disadvantages? Available online: https://www.cebm.net/covid-19/what-tests-could-potentially-be-used-for-the-screening-diagnosis-and-monitoring-of-covid-19-and-what-are-their-advantages-and-disadvantages/ (accessed on 16 January 2021).
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Alanagreh, L.; Alzoughool, F.; Atoum, M. The Human Coronavirus Disease COVID-19: Its Origin, Characteristics, and Insights into Potential Drugs and Its Mechanisms. Pathogens 2020, 9, 331. [Google Scholar] [CrossRef]
- Shen, K.; China National Clinical Research Center for Respiratory Diseases; Yang, Y.; Wang, T.; Zhao, D.; Jiang, Y.; Jin, R.; Zheng, Y.; Xu, B.; Xie, Z.; et al. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: Experts’ consensus statement. World J. Pediatr. 2020, 16, 223–231. [Google Scholar] [CrossRef]
- World Health Organization. Advice for the Public on COVID-19—World Health Organization. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public (accessed on 16 February 2021).
- Key Messages and Actions for COVID-19 Prevention and Control in Schools. Available online: https://www.who.int/publications/m/item/key-messages-and-actions-for-covid-19-prevention-and-control-in-schools (accessed on 16 February 2021).
- World Health Organization. Home Care for Patients with Suspected Novel Coronavirus (nCoV) Infection Presenting with Mild Symptoms and Management of Contacts: Interim Guidance, 20 January 2020; World Health Organization: New York, NY, USA, 2020; ISBN 978-92-4-000083-4. [Google Scholar]
- WHO. Infection Prevention and Control during Health Care When Novel Coronavirus (nCoV) Infection Is Suspected: Interim Guidance; (Ref. WHO/2019-nCoV/IPC/2020.3); World Health Organization: New York, NY, USA, 25 January 2020. [Google Scholar]
- Advice on the Use of Masks in the Community, during Home Care and in Healthcare Settings in the Context of the Novel Coronavirus (COVID-19) Outbreak. Available online: https://www.who.int/publications-detail-redirect/advice-on-the-use-of-masks-in-the-community-during-home-care-and-in-healthcare-settings-in-the-context-of-the-novel-coronavirus-(2019-ncov)-outbreak (accessed on 16 February 2021).
- WHO. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-NCoV) Infection Is Suspected: Interim Guidance; (Ref. WHO/nCoV/Clinical/2020.3); World Health Organization: New York, NY, USA, 28 January 2020. [Google Scholar]
- Considerations for Quarantine of Contacts of COVID-19 Cases. Available online: https://www.who.int/publications-detail-redirect/considerations-for-quarantine-of-individuals-in-the-context-of-containment-for-coronavirus-disease-(covid-19) (accessed on 16 February 2021).
- Su, Z.; Wen, J.; McDonnell, D.; Goh, E.; Li, X.; Šegalo, S.; Ahmad, J.; Cheshmehzangi, A.; Xiang, Y.-T. Vaccines are not yet a silver bullet: The imperative of continued communication about the importance of COVID-19 safety measures. Brain Behav. Immun. Health 2021, 12, 100204. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.; Dalvi, S.; Sălăgean, T.; Pop, I.D.; Bordea, I.R.; Benedicenti, S. Understanding COVID-19 Pandemic: Molecular Mechanisms and Potential Therapeutic Strategies. An Evidence-Based Review. J. Inflamm. Res. 2021, 14, 13–56. [Google Scholar] [CrossRef]
- Cunningham, A.C.; Goh, H.P.; Koh, D. Treatment of COVID-19: Old tricks for new challenges. Crit. Care 2020, 24, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.C. Covid-19 treatment update: Follow the scientific evidence. J. Thromb. Thrombolysis 2020, 50, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.-G.; Shin, H.-J.; Kim, M.-H.; Lee, S.; Kim, H.-S.; Myoung, J.; Kim, B.-T.; Kim, S.-J. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.; Mustafa, F.; Rizvi, T.A.; Loney, T.; Al Suwaidi, H.; Al-Marzouqi, A.H.H.; Eldin, A.K.; Alsabeeha, N.; Adrian, T.E.; Stefanini, C.; et al. SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses 2020, 12, 526. [Google Scholar] [CrossRef]
- Mahmud, S.; Paul, G.K.; Afroze, M.; Islam, S.; Gupt, S.B.R.; Razu, M.H.; Biswas, S.; Zaman, S.; Uddin, M.S.; Khan, M.; et al. Efficacy of Phytochemicals Derived from Avicennia officinalis for the Management of COVID-19: A Combined In Silico and Biochemical Study. Molecules 2021, 28, 2210. [Google Scholar] [CrossRef]
- Young, B.; Tan, T.T.; Leo, Y.S. The place for remdesivir in COVID-19 treatment. Lancet Infect. Dis. 2021, 21, 20–21. [Google Scholar] [CrossRef]
- McCreary, E.K.; Pogue, J.M. Coronavirus Disease 2019 Treatment: A Review of Early and Emerging Options. Open Forum Infect. Dis. 2020, 7, ofaa105. [Google Scholar] [CrossRef]
- Hossain, J.; Jannat, T.; Brishty, S.R.; Roy, U.; Mitra, S.; Rafi, O.; Islam, R.; Nesa, M.L.; Islam, A.; Bin Emran, T. Clinical Efficacy and Safety of Antiviral Drugs in the Extended Use against COVID-19: What We Know So Far. Biologics 2021, 1, 252–284. [Google Scholar] [CrossRef]
- Rosa, S.G.V.; Santos, W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Pub. 2020, 44, e40. [Google Scholar] [CrossRef]
- Toniati, P.; Piva, S.; Cattalini, M.; Garrafa, E.; Regola, F.; Castelli, F.; Franceschini, F.; Airò, P.; Bazzani, C.; Beindorf, E.-A.; et al. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: A single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020, 19, 102568. [Google Scholar] [CrossRef]
- Guaraldi, G.; Meschiari, M.; Cozzi-Lepri, A.; Milic, J.; Tonelli, R.; Menozzi, M.; Franceschini, E.; Cuomo, G.; Orlando, G.; Borghi, V.; et al. Tocilizumab in patients with severe COVID-19: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e474–e484. [Google Scholar] [CrossRef]
- Lan, S.-H.; Lai, C.-C.; Huang, H.-T.; Chang, S.-P.; Lu, L.-C.; Hsueh, P.-R. Tocilizumab for severe COVID-19: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2020, 56, 106103. [Google Scholar] [CrossRef] [PubMed]
- Hossain, J.; Rahman, S.M.A. Repurposing therapeutic agents against SARS-CoV-2 infection: Most promising and neoteric progress. Expert Rev. Anti-infective Ther. 2021, 19, 1009–1027. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Wu, Y.-Q.; Ni, W.-J.; Shen, A.-Z.; Liu, S. The potential insights of Traditional Chinese Medicine on treatment of COVID-19. Chin. Med. 2020, 15, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zahedipour, F.; Hosseini, S.A.; Sathyapalan, T.; Majeed, M.; Jamialahmadi, T.; Al-Rasadi, K.; Banach, M.; Sahebkar, A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother. Res. 2020, 34, 2911–2920. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X.; Ni, L.; Di, X.; Ma, B.; Niu, S.; Liu, C.; Reiter, R.J. COVID-19: Melatonin as a potential adjuvant treatment. Life Sci. 2020, 250, 117583. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, X.; Shu, Y.; Guo, M.; Zhang, H.; Tao, W. Insights from nanotechnology in COVID-19 treatment. Nano Today 2021, 36, 101019. [Google Scholar] [CrossRef]
- Mir, J.M.; Maurya, R.C. Nitric oxide as a therapeutic option for COVID-19 treatment: A concise perspective. New J. Chem. 2021, 45, 1774–1784. [Google Scholar] [CrossRef]
- Tillu, G.; Chaturvedi, S.; Chopra, A.; Patwardhan, B. Public Health Approach of Ayurveda and Yoga for COVID-19 Prophylaxis. J. Altern. Complement. Med. 2020, 26, 360–364. [Google Scholar] [CrossRef]
- Hossain, J.; Kuddus, R.; A Rashid, M.; Sultan, Z. Understanding and Dealing the SARS-CoV-2 Infection: An Updated Concise Review. Bangladesh Pharm. J. 2021, 24, 61–75. [Google Scholar] [CrossRef]
- Stockman, L.J.; Bellamy, R.; Garner, P. SARS: Systematic Review of Treatment Effects. PLoS Med. 2006, 3, e343. [Google Scholar] [CrossRef]
- Scagnolari, C.; Vicenzi, E.; Bellomi, F.; Stillitano, M.G.; Pinna, D.; Poli, G.; Clementi, M.; Dianzani, F.; Antonelli, G. Increased sensitivity of SARS-coronavirus to a combination of human type I and type II interferons. Antivir. Ther. 2004, 9, 1003–1011. [Google Scholar]
- The New York Times. Coronavirus Vaccine Tracker. Available online: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html (accessed on 26 November 2020).
- COVAX. Available online: https://www.who.int/initiatives/act-accelerator/covax (accessed on 19 February 2021).
- Kim, J.H.; Marks, F.; Clemens, J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021, 27, 205–211. [Google Scholar] [CrossRef]
- Wenham, C.; Smith, J.; Morgan, R. Gender and COVID-19 Working Group COVID-19: The Gendered Impacts of the Outbreak. Lancet 2020, 395, 846–848. [Google Scholar] [CrossRef]
- Kocher, K.; Delot-Vilain, A.; Spencer, D.; LoTempio, J.; Délot, E.C. Paucity and Disparity of Publicly Available Sex-Disaggregated Data for the COVID-19 Epidemic Hamper Evidence-Based Decision-Making. Arch. Sex. Behav. 2021, 50, 407–426. [Google Scholar] [CrossRef]
- Sun, P.; Lu, X.; Xu, C.; Sun, W.; Pan, B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020, 92, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Kopel, J.; Perisetti, A.; Roghani, A.; Aziz, M.; Gajendran, M.; Goyal, H. Racial and Gender-Based Differences in COVID-19. Front. Public Health 2020, 8, 418. [Google Scholar] [CrossRef]
- Garg, S.; Kim, L.; Whitaker, M.; O’Halloran, A.; Cummings, C.; Holstein, R.; Prill, M.; Chai, S.J.; Kirley, P.D.; Alden, N.B.; et al. Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019—COVID-NET, 14 States, March 1–30, 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 458–464. [Google Scholar] [CrossRef]
- Dowd, J.; Aiello, A.E. Socioeconomic Differentials in Immune Response. Epidemiology 2009, 20, 902–908. [Google Scholar] [CrossRef]
- Adler, N.E.; Boyce, W.T.; Chesney, M.A.; Folkman, S.; Syme, S.L. Socioeconomic inequalities in health. No easy solution. JAMA 1993, 269, 3140–3145. [Google Scholar] [CrossRef] [PubMed]
- Steptoe, A.; Shamaei-Tousi, A.; Gylfe, A.; Henderson, B.; Bergstrom, S.; Marmot, M. Socioeconomic status, pathogen burden and cardiovascular disease risk. Hear 2006, 93, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, K.H.; Chowdhury, M.R.; Mahmud, S.; Tareq, A.M.; Hanif, N.B.; Banu, N.; Reza, A.S.M.A.; Emran, T.B.; Simal-Gandara, J. Drug Repurposing Approach against Novel Coronavirus Disease (COVID-19) through Virtual Screening Targeting SARS-CoV-2 Main Protease. Biology 2020, 10, 2. [Google Scholar] [CrossRef]
- World Health Organization. Severe Acute Respiratory Syndrome (SARS). Available online: https://www.who.int/westernpacific/health-topics/severe-acute-respiratory-syndrome (accessed on 20 February 2021).
- Seto, W.; Tsang, D.; Yung, R.; Ching, T.; Ng, T.; Ho, M.; Ho, L.; Peiris, J.S.M. Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet 2003, 361, 1519–1520. [Google Scholar] [CrossRef]
- Tumilaar, S.G.; Siampa, J.P.; Fatimawali, F.; Kepel, B.J.; Niode, N.J.; Idroes, R.; Rakib, A.; Emran, T.B.; Tallei, T.E. The potential of leaf extract of Pangium edule Reinw as HIV-1 protease inhibitor: A computational biology approach. J. Appl. Pharm. Sci. 2021, 11, 101–110. [Google Scholar] [CrossRef]
- Ellinghaus, K. The ABO blood group locus and a chromosome 3 gene cluster associate with SARS-CoV-2 respiratory failure in an Italian-Spanish genome-wide association analysis. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship Between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Zietz, M.; Zucker, J.; Tatonetti, N.P. Testing the Association between Blood Type and COVID-19 Infection, Intubation, and Death. medRxiv 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).