Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway
Abstract
:1. Introduction
2. Results
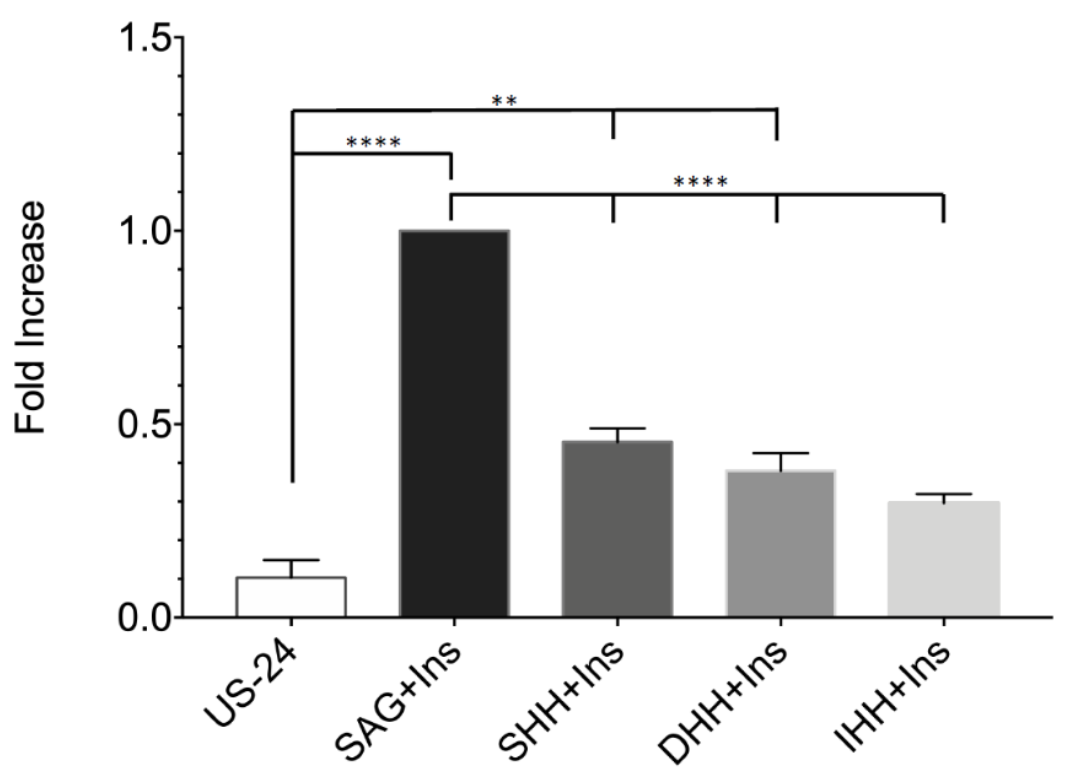
2.1. Generation In Vitro of CD24 Positive Cells from Primary hASCs
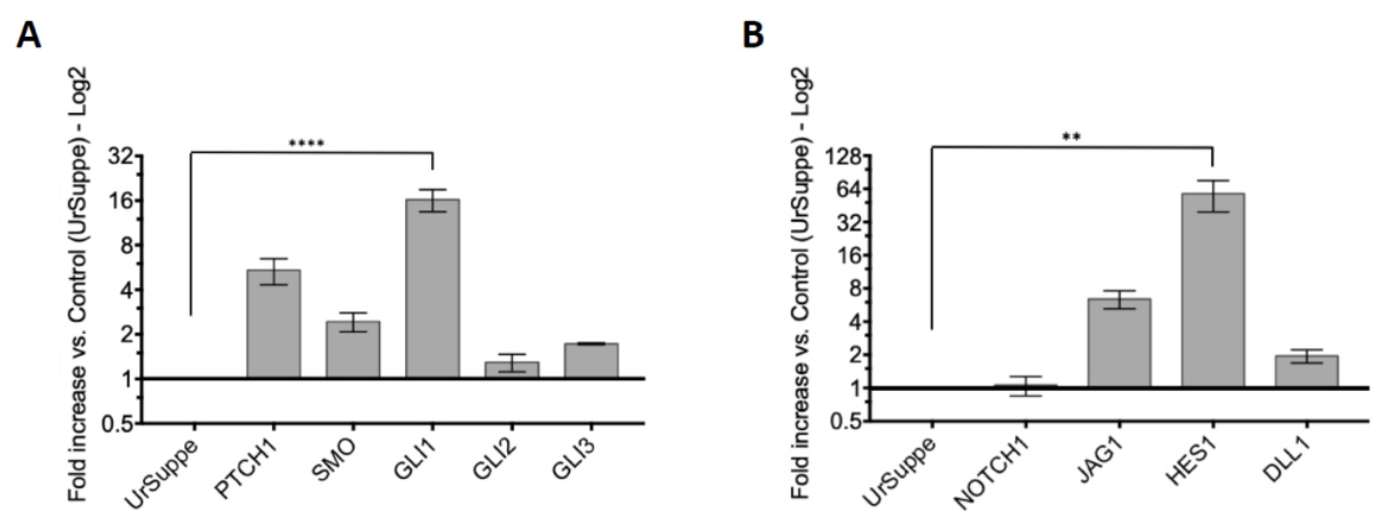
2.2. Induction of the Hedgehog Signaling and Detection by RT-qPCR of Its Core Transducing and Effector Genes
2.3. Monitoring the Expression of Selected Stemness or Cell Differentiation Genes by RT-qPCR
2.4. Transforming Growth Factor Family Members Downregulate the Expression of the CD24 Antigen
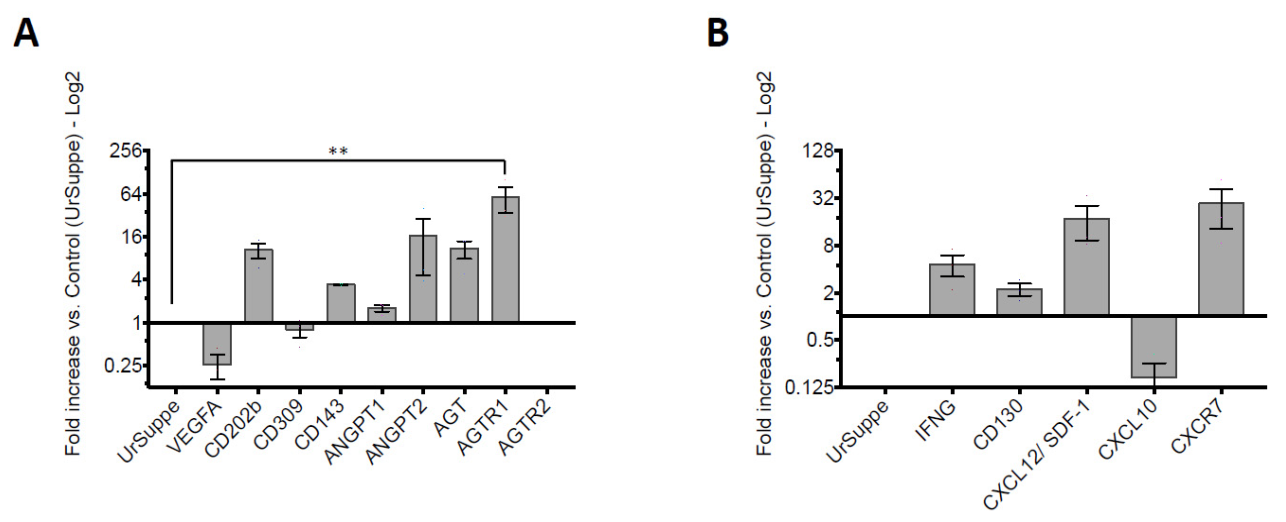
2.5. Monitoring Angiogenic Markers after Triggering the Hh Signaling Pathway in hASCs
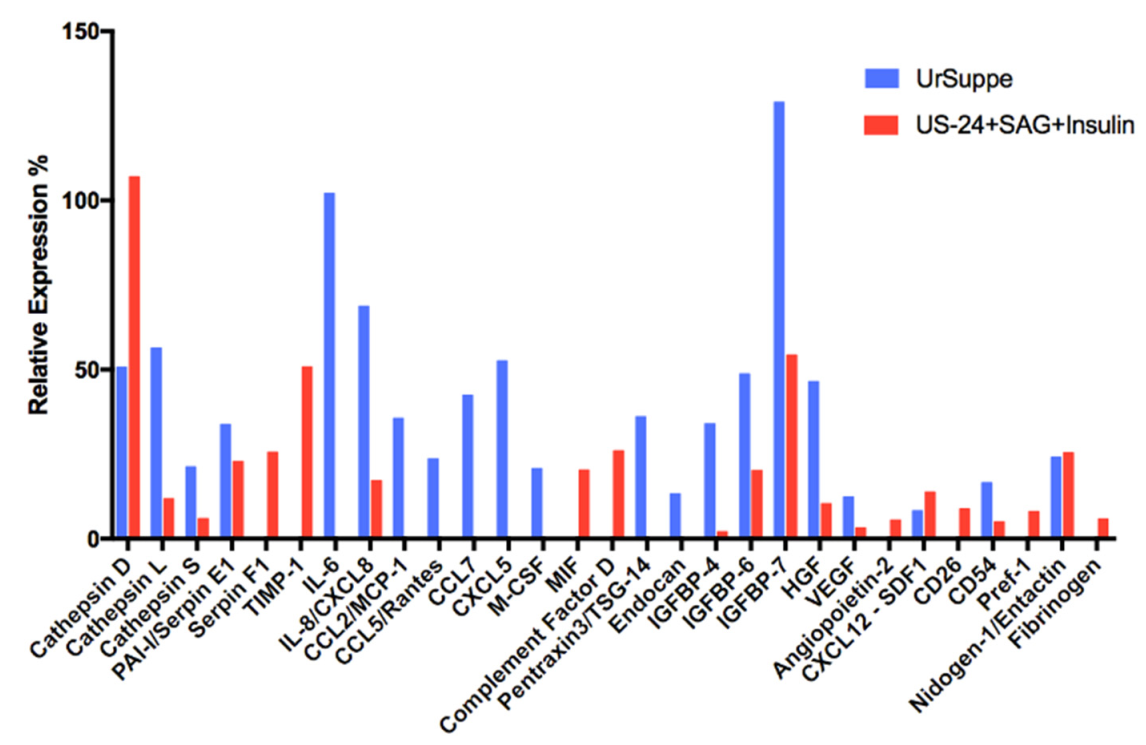
2.6. Secretome Analysis of hASCs Cultured in UrSuppe or Hedgehog Signaling Permissive Conditions
3. Discussion
4. Materials and Methods
4.1. Isolation and Culture of Human Adipose Stem Cells (hASCs)
4.1.1. Extraction of Adipo-Cutaneous Tissue
4.1.2. Isolation of Stromal Vascular Fraction (SVF)
4.1.3. Seeding and Culture of hASCs
4.2. Induction and Characterization of CD24 Positive Cells
4.2.1. Generation of CD24 Positive Cells
4.2.2. Flow Cytometry Analysis
4.2.3. RNA Preparation, cDNA Synthesis and Quantitative Real-Time PCR (RT-qPCR) Analysis
4.2.4. Secretome Profiler
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuk, P.A.; Zhu, M.; Ashjian, P.; De Ugarte, D.A.; Huang, J.I.; Mizuno, H.; Alfonso, Z.C.; Fraser, J.K.; Benhaim, P.; Hedrick, M.H. Human Adipose Tissue Is a Source of Multipotent Stem Cells. Mol. Biol. Cell 2002, 13, 4279–4295. [Google Scholar] [CrossRef]
- Zuk, P.A. The Adipose-derived Stem Cell: Looking Back and Looking Ahead. Mol. Biol. Cell 2010, 21, 1783–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrikoski, M.; Mannerström, B.; Miettinen, S. Perspectives for clinical translation of adipose stromal/stem cells. Stem Cells Int. 2019, 2019, 5858247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argentati, C.; Morena, F.; Bazzucchi, M.; Armentano, I.; Emiliani, C.; Martino, S. Adipose stem cell translational applications: From bench-to-bedside. Int. J. Mol. Sci. 2018, 19, 3475. [Google Scholar] [CrossRef] [Green Version]
- Gottipamula, S.; Chokalingam, K.; Sridhar, K.N. Major clinical application of adipose derived stem cells. J. Stem Cell Regen. Biol. 2018, 4, 4–19. [Google Scholar] [CrossRef]
- Kapur, S.K.; Katz, A.J. Review of the adipose derived stem cell secretome. Biochimie 2013, 95, 2222–2228. [Google Scholar] [CrossRef]
- Ceccarelli, S.; Pontecorvi, P.; Anastasiadou, E.; Napoli, C.; Marchese, C. Immunomodulatory Effect of Adipose-Derived Stem Cells: The Cutting Edge of Clinical Application. Front. Cell Dev. Biol. 2020, 8, 2222–2228. [Google Scholar] [CrossRef]
- Lombardi, F.; Palumbo, P.; Augello, F.R.; Cifone, M.G.; Cinque, B.; Giuliani, M. Secretome of adipose tissue-derived stem cells (ASCs) as a novel trend in chronic non-healing wounds: An overview of experimental in vitro and in vivo studies and methodological variables. Int. J. Mol. Sci. 2019, 20, 3721. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Bourin, P.; Bunnell, B.A.; Casteilla, L.; Dominici, M.; Katz, A.J.; March, K.L.; Redl, H.; Rubin, J.P.; Yoshimura, K.; Gimble, J.M. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: A joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International So. Cytotherapy 2013, 15, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Ménard, C.; Dulong, J.; Roulois, D.; Hébraud, B.; Verdière, L.; Pangault, C.; Sibut, V.; Bezier, I.; Bescher, N.; Monvoisin, C.; et al. Integrated transcriptomic, phenotypic, and functional study reveals tissue-specific immune properties of mesenchymal stromal cells. Stem Cells 2020, 38, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.A.; Park, M.; Kim, Y.H.; Woo, S.Y.; Ryu, K.H. RNA sequencing reveals a transcriptomic portrait of human mesenchymal stem cells from bone marrow, adipose tissue, and palatine tonsils. Sci. Rep. 2017, 7, 17114. [Google Scholar] [CrossRef] [Green Version]
- Ferretti, E.; Hadjantonakis, A.K. Mesoderm specification and diversification: From single cells to emergent tissues. Curr. Opin. Cell Biol. 2019, 61, 110–116. [Google Scholar] [CrossRef]
- Loh, K.M.M.; Chen, A.; Koh, P.W.W.; Deng, T.Z.Z.; Sinha, R.; Tsai, J.M.M.; Barkal, A.A.A.; Shen, K.Y.Y.; Jain, R.; Morganti, R.M.M.; et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 2016, 166, 451–467. [Google Scholar] [CrossRef] [Green Version]
- Mendjan, S.; Mascetti, V.L.; Ortmann, D.; Ortiz, M.; Karjosukarso, D.W.; Ng, Y.; Moreau, T.; Pedersen, R.A. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 2014, 15, 310–325. [Google Scholar] [CrossRef] [Green Version]
- Sheng, G. The developmental basis of mesenchymal stem/stromal cells (MSCs). BMC Dev. Biol. 2015, 15, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebo, Z.L.; Rodeheffer, M.S. Assembling the adipose organ: Adipocyte lineage segregation and adipogenesis in vivo. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodeheffer, M.S.; Birsoy, K.; Friedman, J.M. Identification of White Adipocyte Progenitor Cells In Vivo. Cell 2008, 135, 240–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, R.; Rodeheffer, M.S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 2013, 15, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.; Jeffery, E.; Rodeheffer, M.S. Weighing in on adipocyte precursors. Cell Metab. 2014, 19, 8–20. [Google Scholar] [CrossRef] [Green Version]
- Ayre, D.C.; Pallegar, N.K.; Fairbridge, N.A.; Canuti, M.; Lang, A.S.; Christian, S.L. Analysis of the structure, evolution, and expression of CD24, an important regulator of cell fate. Gene 2016, 590, 324–337. [Google Scholar] [CrossRef]
- Kay, R.; Rosten, P.M.; Humphries, R.K. CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J. Immunol. 1991, 147, 1412–1416. [Google Scholar] [PubMed]
- Eyvazi, S.; Kazemi, B.; Dastmalchi, S.; Bandehpour, M. Involvement of CD24 in Multiple Cancer Related Pathways Makes It an Interesting New Target for Cancer Therapy. Curr. Cancer Drug Targets 2018, 18, 328–336. [Google Scholar] [CrossRef]
- Weichert, W.; Denkert, C.; Burkhardt, M.; Gansukh, T.; Bellach, J.; Altevogt, P.; Dietel, M.; Kristiansen, G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin. Cancer Res. 2005, 11, 6574–6581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, Y.Y.; Jeng, Y.M.; Lee, T.T.; Hu, F.C.; Kao, H.L.; Lin, W.C.; Lai, P.L.; Hu, R.H.; Yuan, R.H. Cytoplasmic CD24 expression is a novel prognostic factor in diffuse-type gastric adenocarcinoma. Ann. Surg. Oncol. 2007, 14, 2748–2758. [Google Scholar] [CrossRef]
- Duex, J.E.; Owens, C.; Chauca-Diaz, A.; Dancik, G.M.; Vanderlinden, L.A.; Ghosh, D.; Leivo, M.Z.; Hansel, D.E.; Theodorescu, D. Nuclear CD24 drives tumor growth and is predictive of poor patient prognosis. Cancer Res. 2017, 77, 4858–4867. [Google Scholar] [CrossRef] [Green Version]
- Kristiansen, G.; Sammar, M.; Altevogt, P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J. Mol. Histol. 2004, 35, 255–262. [Google Scholar] [CrossRef]
- Altevogt, P.; Sammar, M.; Hüser, L.; Kristiansen, G. Novel insights into the function of CD24: A driving force in cancer. Int. J. Cancer 2020, 546–559. [Google Scholar] [CrossRef]
- Fischer, G.F.; Majdic, O.; Gadd, S.; Knapp, W. Signal transduction in lymphocytic and myeloid cells via CD24, a new member of phosphoinositol-anchored membrane molecules. J. Immunol. 1990, 144, 638–641. [Google Scholar]
- Ayre, D.C.; Christian, S.L. CD24: A rheostat that modulates cell surface receptor signaling of diverse receptors. Front. Cell Dev. Biol. 2016, 4, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haynes, B.A.; Huyck, R.W.; James, A.J.; Carter, M.E.; Gaafar, O.U.; Day, M.; Pinto, A.; Dobrian, A.D. Isolation, expansion, and adipogenic induction of CD34+CD31+ endothelial cells from human omental and subcutaneous adipose tissue. J. Vis. Exp. 2018, 2018, 57804. [Google Scholar] [CrossRef]
- Johal, K.S. Adipose-Derived Stem Cells; Selecting for Translational Success. Master’s Thesis, University of Manchester, Manchester, UK, 2014. Available online: https://www.escholar.manchester.ac.uk/uk-ac-man-scw:267164. (accessed on 15 July 2021).
- Hatzmann, F.M.; Ejaz, A.; Wiegers, G.J.; Mandl, M.; Brucker, C.; Lechner, S.; Rauchenwald, T.; Zwierzina, M.; Baumgarten, S.; Wagner, S.; et al. Quiescence, Stemness and Adipogenic Differentiation Capacity in Human DLK1−/CD34+/CD24+ Adipose Stem/Progenitor Cells. Cells 2021, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zheng, P.; Tang, J.; Liu, Y. CD24: From A to Z. Cell. Mol. Immunol. 2010, 7, 100–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Zhao, M.; Xiang, B.; Chang, C.; Lu, Q. CD24: From a Hematopoietic Differentiation Antigen to a Genetic Risk Factor for Multiple Autoimmune Diseases. Clin. Rev. Allergy Immunol. 2016, 50, 70–83. [Google Scholar] [CrossRef]
- Shackleton, M.; Vaillant, F.; Simpson, K.J.; Stingl, J.; Smyth, G.K.; Asselin-Labat, M.L.; Wu, L.; Lindeman, G.J.; Visvader, J.E. Generation of a functional mammary gland from a single stem cell. Nature 2006, 439, 84–88. [Google Scholar] [CrossRef]
- Shakiba, N.; White, C.A.; Lipsitz, Y.Y.; Yachie-Kinoshita, A.; Tonge, P.D.; Hussein, S.M.I.; Puri, M.C.; Elbaz, J.; Morrissey-Scoot, J.; Li, M.; et al. CD24 tracks divergent pluripotent states in mouse and human cells. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Geradts, J.; Dewhirst, M.W.; Lo, H.W. Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene 2012, 31, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Chen, T.; Zhang, Y.; Chen, Q. Hedgehog signaling pathway regulates ovarian cancer invasion and migration via adhesion molecule CD24. J. Cancer 2017, 8, 786–792. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.T.H.; Zhao, Z.; Ingham, P.W. Hedgehog signalling. Development 2016, 143, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Gu, D.; Xie, J. Clinical implications of hedgehog signaling pathway inhibitors. Chin. J. Cancer 2011, 30, 13–26. [Google Scholar] [CrossRef]
- Kopinke, D.; Norris, A.M.; Mukhopadhyay, S. Developmental and regenerative paradigms of cilia regulated hedgehog signaling. Semin. Cell Dev. Biol. 2020, 1–15. [Google Scholar] [CrossRef]
- Ramsbottom, S.A.; Pownall, M.E. Regulation of hedgehog signalling inside and outside the cell. J. Dev. Biol. 2016, 4, 23. [Google Scholar] [CrossRef] [Green Version]
- Suh, J.M.; Gao, X.; McKay, J.; McKay, R.; Salo, Z.; Graff, J.M. Hedgehog signaling plays a conserved role in inhibiting fat formation. Cell Metab. 2006, 3, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cousin, W.; Fontaine, C.; Dani, C.; Peraldi, P. Hedgehog and adipogenesis: Fat and fiction. Biochimie 2007, 89, 1447–1453. [Google Scholar] [CrossRef]
- Fontaine, C.; Cousin, W.; Plaisant, M.; Dani, C.; Peraldi, P. Hedgehog Signaling Alters Adipocyte Maturation of Human Mesenchymal Stem Cells. Stem Cells 2008, 26, 1037–1046. [Google Scholar] [CrossRef]
- Plaisant, M.; Giorgetti-Peraldi, S.; Gabrielson, M.; Loubat, A.; Dani, C.; Peraldi, P. Inhibition of hedgehog signaling decreases proliferation and clonogenicity of human mesenchymal stem cells. PLoS ONE 2011, 6, e16798. [Google Scholar] [CrossRef]
- Pospisilik, J.A.; Schramek, D.; Schnidar, H.; Cronin, S.J.F.; Nehme, N.T.; Zhang, X.; Knauf, C.; Cani, P.D.; Aumayr, K.; Todoric, J.; et al. Drosophila Genome-wide Obesity Screen Reveals Hedgehog as a Determinant of Brown versus White Adipose Cell Fate. Cell 2010, 140, 148–160. [Google Scholar] [CrossRef] [Green Version]
- Varjosalo, M.; Taipale, J. Hedgehog: Functions and mechanisms. Genes Dev. 2008, 22, 2454–2472. [Google Scholar] [CrossRef] [Green Version]
- Stanton, B.Z.; Peng, L.F. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol. Biosyst. 2009, 6, 44–54. [Google Scholar] [CrossRef]
- Panella, S.; Muoio, F.; Jossen, V.; Harder, Y.; Eibl-Schindler, R.; Tallone, T. Chemically Defined Xeno- and Serum-Free Cell Culture Medium to Grow Human Adipose Stem Cells. Cells 2021, 10, 466. [Google Scholar] [CrossRef]
- Tokhunts, R.; Singh, S.; Chu, T.; D’Angelo, G.; Baubet, V.; Goetz, J.A.; Huang, Z.; Yuan, Z.; Ascano, M.; Zavros, Y.; et al. The full-length unprocessed hedgehog protein is an active signaling molecule. J. Biol. Chem. 2010, 285, 2562–2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heretsch, P.; Tzagkaroulaki, L.; Giannis, A. Modulators of the hedgehog signaling pathway. Bioorganic Med. Chem. 2010, 18, 6613–6624. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Lauth, M.; Zwijsen, A.; Huylebroeck, D.; Oswald, F.; Giaimo, B.D. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFβ/BMP and hypoxia pathways. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.T.; Huang, P. Complex crosstalk of Notch and Hedgehog signalling during the development of the central nervous system. Cell. Mol. Life Sci. 2020, 78, 635–644. [Google Scholar] [CrossRef]
- Shan, T.; Liu, J.; Wu, W.; Xu, Z.; Wang, Y. Roles of Notch Signaling in Adipocyte Progenitor Cells and Mature Adipocytes. J. Cell. Physiol. 2017, 232, 1258–1261. [Google Scholar] [CrossRef]
- Bi, P.; Kuang, S. Notch signaling as a novel regulator of metabolism. Trends Endocrinol. Metab. 2015, 26, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, R.; Ohtsuka, T.; Tomita, K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cells 2000, 10, 1–7. [Google Scholar] [CrossRef]
- Liu, Z.H.; Dai, X.M.; Du, B. Hes1: A key role in stemness, metastasis and multidrug resistance. Cancer Biol. Ther. 2015, 16, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Ross, D.A.; Hannenhalli, S.; Tobias, J.W.; Cooch, N.; Shiekhattar, R.; Kadesch, T. Functional analysis of Hes-1 in preadipocytes. Mol. Endocrinol. 2006, 20, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Hudak, C.S.; Gulyaeva, O.; Wang, Y.; Park, S.-M.; Lee, L.; Kang, C.; Sul, H.S. Pref-1 marks very early mesenchymal precursors required for adipose tissue development and expansion. Cell Rep. 2014, 8, 678–687. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sul, H.S. Pref-1 Regulates Mesenchymal Cell Commitment and Differentiation through Sox9. Cell Metab. 2009, 9, 287–302. [Google Scholar] [CrossRef] [Green Version]
- Gulyaeva, O.; Nguyen, H.; Sambeat, A.; Heydari, K.; Sul, H.S. Sox9-Meis1 Inactivation Is Required for Adipogenesis, Advancing Pref-1+ to PDGFRα+ Cells. Cell Rep. 2018, 25, 1002–1017.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammarstedt, A.; Hedjazifar, S.; Jenndahl, L.; Gogg, S.; Grünberg, J.; Gustafson, B.; Klimcakova, E.; Stich, V.; Langin, D.; Laakso, M.; et al. WISP2 regulates preadipocyte commitment and PPARγ activation by BMP4. Proc. Natl. Acad. Sci. USA 2013, 110, 2563–2568. [Google Scholar] [CrossRef] [Green Version]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 2018, 149, 313–323. [Google Scholar] [CrossRef]
- Toshihisa, K. Molecular Mechanism of Runx2-Dependent Bone Development. Mol. Cells 2020, 43, 168–175. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O.A. Adipose tissue stem cells meet preadipocyte commitment: Going back to the future. J. Lipid Res. 2012, 53, 227–246. [Google Scholar] [CrossRef] [Green Version]
- Smith, N.C.; Fairbridge, N.A.; Pallegar, N.K.; Christian, S.L. Dynamic upregulation of CD24 in pre-adipocytes promotes adipogenesis. Adipocyte 2015, 4, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Sarantopoulos, C.N.; Banyard, D.A.; Ziegler, M.E.; Sun, B.; Shaterian, A.; Widgerow, A.D. Elucidating the Preadipocyte and Its Role in Adipocyte Formation: A Comprehensive Review. Stem Cell Rev. Rep. 2018, 14, 27–42. [Google Scholar] [CrossRef]
- Modica, S.; Wolfrum, C. The dual role of BMP4 in adipogenesis and metabolism. Adipocyte 2017, 6, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Bastida, M.F.; Sheth, R.; Ros, M.A. A BMP-Shh negative-feedback loop restricts Shh expression during limb development. Development 2009, 136, 3779–3789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Dréau, G.; Martí, E. Dorsal-ventral patterning of the neural tube: A tale of three signals. Dev. Neurobiol. 2012, 72, 1471–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jessell, T.M. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 2000, 1, 20–29. [Google Scholar] [CrossRef]
- Christiaens, V.; Van Hul, M.; Lijnen, H.R.; Scroyen, I. CD36 promotes adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Volat, F.; Sandhow, L.; Galitzky, J.; Nguyen, T.; Esteve, D.; Åström, G.; Mejhert, N.; Ledoux, S.; Thalamas, C.; et al. CD36 Is a Marker of Human Adipocyte Progenitors with Pronounced Adipogenic and Triglyceride Accumulation Potential. Stem Cells 2017, 35, 1799–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlinson, J.J.; Boudreau, A.; Wu, D.; Atlas, E.; Haché, R.J.G. Modulation of early human preadipocyte differentiation by glucocorticoids. Endocrinology 2006, 147, 5284–5293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapouly, C.; Guimbal, S.; Hollier, P.L.; Renault, M.A. Role of hedgehog signaling in vasculature development, differentiation, and maintenance. Int. J. Mol. Sci. 2019, 20, 3076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausman, G.J.; Richardson, R.L. Adipose tissue angiogenesis. J. Anim. Sci. 2004, 82, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Adipose tissue angiogenesis as a therapeutic target for obesity and metabolic diseases. Nat. Rev. Drug Discov. 2010, 9, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Masson, O.; Prébois, C.; Derocq, D.; Meulle, A.; Dray, C.; Daviaud, D.; Quilliot, D.; Valet, P.; Muller, C.; Liaudet-Coopman, E. Cathepsin-D, a key protease in breast cancer, is up-regulated in obese mouse and human adipose tissue, and controls adipogenesis. PLoS ONE 2011, 6, e16452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Zhang, Y.; Pan, J.; Sun, J.; Liu, J.; Libby, P.; Sukhova, G.K.; Doria, A.; Katunuma, N.; Peroni, O.D.; et al. Cathepsin L activity controls adipogenesis and glucose tolerance. Nat. Cell Biol. 2007, 9, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Vidak, E.; Javoršek, U.; Vizovišek, M.; Turk, B. Cysteine Cathepsins and their Extracellular Roles: Shaping the Microenvironment. Cells 2019, 8, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benes, P.; Vetvicka, V.; Fusek, M. Cathepsin D-Many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008, 68, 12–28. [Google Scholar] [CrossRef] [Green Version]
- Mariman, E.C.M.; Wang, P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010, 67, 1277–1292. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, C.-G.; Jia, Y.-L.; Hu, L. Tissue Inhibitor of Metalloprotease-1 (TIMP-1) Regulates Adipogenesis of Adipose-derived Stem Cells (ASCs) via the Wnt Signaling Pathway in an MMP-independent Manner. Curr. Med. Sci. 2020, 40, 989–996. [Google Scholar] [CrossRef]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.W.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef] [Green Version]
- Isogai, C.; Laug, W.E.; Shimada, H.; Declerck, P.J.; Stins, M.F.; Durden, D.L.; Erdreich-Epstein, A.; DeClerck, Y.A. Plasminogen activator inhibitor-1 promotes angiogenesis by stimulating endothelial cell migration toward fibronectin. Cancer Res. 2001, 61, 5587–5594. [Google Scholar]
- Wang, M.; Wang, J.J.; Li, J.; Park, K.; Qian, X.; Ma, J.X.; Zhang, S.X. Pigment epithelium-derived factor suppresses adipogenesis via inhibition of the MAPK/ERK pathway in 3T3-L1 preadipocytes. Am. J. Physiol. Endocrinol. Metab. 2009, 297, 1378–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.T.; Hsu, L.W.; Chen, K.-D.; Kung, C.P.; Goto, S.; Chen, C.L. Decreased PEDF expression promotes adipogenic differentiation through the up-regulation of CD36. Int. J. Mol. Sci. 2018, 19, 3992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef]
- Asare, Y.; Schmitt, M.; Bernhagen, J. The vascular biology of macrophage migration inhibitory factor (MIF): Expression and effects in inflammation, atherogenesis and angiogenesis. Thromb. Haemost. 2013, 109, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, T.; Cho, Y.; Leng, L.; McDonald, C.; Yu, T.; Danton, C.; Hong, E.; Mitchell, R.A.; Metz, C.; Niwa, H.; et al. The Proinflammatory Cytokine Macrophage Migration Inhibitory Factor Regulates Glucose Metabolism during Systemic Inflammation. J. Immunol. 2007, 179, 5399–5406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calandra, T.; Bernhagen, J.; Metz, C.N.; Spiegel, L.A.; Bacher, M.; Donnelly, T.; Cerami, A.; Bucala, R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 1995, 377, 68–71. [Google Scholar] [CrossRef]
- Pattrick, M.; Luckett, J.; Yue, L.; Stover, C. Dual role of complement in adipose tissue. Mol. Immunol. 2009, 46, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Song, N.J.; Kim, S.; Jang, B.H.; Chang, S.H.; Yun, U.J.; Park, K.M.; Waki, H.; Li, D.Y.; Tontonoz, P.; Park, K.W. Small molecule-induced complement factor D (Adipsin) promotes lipid accumulation and adipocyte differentiation. PLoS ONE 2016, 11, e0162228. [Google Scholar] [CrossRef]
- Haywood, N.J.; Slater, T.A.; Matthews, C.J.; Wheatcroft, S.B. The insulin like growth factor and binding protein family: Novel therapeutic targets in obesity & diabetes. Mol. Metab. 2019, 19, 86–96. [Google Scholar] [CrossRef]
- Blüher, S.; Kratzsch, J.; Kiess, W. Insulin-like growth factor I, growth hormone and insulin in white adipose tissue. Best Pr. Res. Clin. Endocrinol. Metab. 2005, 19, 577–587. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Dang, H.; Liu, P.; Zhang, B.; Xu, F. Insulin-like growth factor 2 regulates the proliferation and differentiation of rat adipose-derived stromal cells via IGF-1R and IR. Cytotherapy 2019, 21, 619–630. [Google Scholar] [CrossRef]
- Bäck, K.; Brännmark, C.; Strålfors, P.; Arnqvist, H.J. Differential effects of IGF-I, IGF-II and insulin in human preadipocytes and adipocytes—Role of insulin and IGF-I receptors. Mol. Cell. Endocrinol. 2011, 339, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Gealekman, O.; Gurav, K.; Chouinard, M.; Straubhaar, J.; Thompson, M.; Malkani, S.; Hartigan, C.; Corvera, S. Control of Adipose Tissue Expandability in Response to High Fat Diet by the Insulin-like Growth Factor-binding Protein-4. J. Biol. Chem. 2014, 289, 18327–18338. [Google Scholar] [CrossRef] [Green Version]
- Headey, S.J.; Leeding, K.S.; Norton, R.S.; Bach, L.A. Contributions of the N- and C-terminal domains of IGF binding protein-6 to IGF binding. J. Mol. Endocrinol. 2004, 33, 377–386. [Google Scholar] [CrossRef]
- Slater, T.; Haywood, N.J.; Matthews, C.; Cheema, H.; Wheatcroft, S.B. Insulin-like growth factor binding proteins and angiogenesis: From cancer to cardiovascular disease. Cytokine Growth Factor Rev. 2019, 46, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Sakai, K.; Nakamura, T.; Matsumoto, K. Hepatocyte growth factor twenty years on: Much more than a growth factor. J. Gastroenterol. Hepatol. 2011, 26, 188–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augustin, H.G.; Young Koh, G.; Thurston, G.; Alitalo, K. Control of vascular morphogenesis and homeostasis through the angiopoietin—Tie system. Nat. Rev. Mol. Cell Biol. 2009, 10, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, A.; Tao, C.; Li, X.; Jin, P. The role of SDF-1-CXCR4/CXCR7 axis in biological behaviors of adipose tissue-derived mesenchymal stem cells in vitro. Biochem. Biophys. Res. Commun. 2013, 441, 675–680. [Google Scholar] [CrossRef]
- Merrick, D.; Sakers, A.; Irgebay, Z.; Okada, C.; Calvert, C.; Morley, M.P.; Percec, I.; Seale, P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 2019, 364. [Google Scholar] [CrossRef]
- Ferrero, R.; Rainer, P.; Deplancke, B. Toward a Consensus View of Mammalian Adipocyte Stem and Progenitor Cell Heterogeneity. Trends Cell Biol. 2020, 30, 937–950. [Google Scholar] [CrossRef]
- Göbel, K.; Eichler, S.; Wiendl, H.; Chavakis, T.; Kleinschnitz, C.; Meuth, S.G. The coagulation factors fibrinogen, thrombin, and factor XII in inflammatory disorders-a systematic review. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Languino, L.R.; Plescia, J.; Duperray, A.; Brian, A.A.; Plow, E.F.; Geltosky, J.E.; Altieri, D.C. Fibrinogen mediates leukocyte adhesion to vascular endothelium through an ICAM-1-dependent pathway. Cell 1993, 73, 1423–1434. [Google Scholar] [CrossRef]
- Tsakadze, N.L.; Zhao, Z.; D’Souza, S.E. Interactions of intercellular adhesion molecule-1 with fibrinogen. Trends Cardiovasc. Med. 2002, 12, 101–108. [Google Scholar] [CrossRef]
- Gardiner, E.E.; D’Souza, S.E. A mitogenic action for fibrinogen mediated through intercellular adhesion molecule-1. J. Biol. Chem. 1997, 272, 15474–15480. [Google Scholar] [CrossRef] [Green Version]
- Pluskota, E.; D’Souza, S.E. Fibrinogen interactions with ICAM-1 (CD54) regulate endothelial cell survival. Eur. J. Biochem. 2000, 267, 4693–4704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellner, M.; Herse, F.; Janke, J.; Gorzelniak, K.; Engeli, S.; Bechart, D.; Lasalle, P.; Luft, F.C.; Sharma, A.M. Endothelial cell specific molecule-1—A newly identified protein in adipocytes. Horm. Metab. Res. 2003, 35, 217–221. [Google Scholar] [CrossRef]
- Béchard, D.; Scherpereel, A.; Hammad, H.; Gentina, T.; Tsicopoulos, A.; Aumercier, M.; Pestel, J.; Dessaint, J.-P.; Tonnel, A.-B.; Lassalle, P. Human Endothelial-Cell Specific Molecule-1 Binds Directly to the Integrin CD11a/CD18 (LFA-1) and Blocks Binding to Intercellular Adhesion Molecule-1. J. Immunol. 2001, 167, 3099–3106. [Google Scholar] [CrossRef] [Green Version]
- Plaisant, M.; Fontaine, C.; Cousin, W.; Rochet, N.; Dani, C.; Peraldi, P. Activation of Hedgehog Signaling Inhibits Osteoblast Differentiation of Human Mesenchymal Stem Cells. Stem Cells 2009, 27, 703–713. [Google Scholar] [CrossRef]
- Nieto-Vazquez, I.; Fernández-Veledo, S.; Krämer, D.K.; Vila-Bedmar, R.; Garcia-Guerra, L.; Lorenzo, M. Insulin resistance associated to obesity: The link TNF-alpha. Arch. Physiol. Biochem. 2008, 114, 183–194. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rehman, K.; Liaqat, A. Tumor Necrosis Factor-Alpha: Role in Development of Insulin Resistance and Pathogenesis of Type 2 Diabetes Mellitus. J. Cell. Biochem. 2018, 119, 105–110. [Google Scholar] [CrossRef]
- Alzamil, H. Elevated Serum TNF- α Is Related to Obesity in Type 2 Diabetes Mellitus and Is Associated with Glycemic Control and Insulin Resistance. J. Obes. 2020, 2020, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Oestreich, A.K.; Collins, K.H.; Harasymowicz, N.S.; Wu, C.L.; Guilak, F. Is Obesity a Disease of Stem Cells? Cell Stem Cell 2020, 27, 15–18. [Google Scholar] [CrossRef]
- Pollard, A.E.; Carling, D. Thermogenic adipocytes: Lineage, function and therapeutic potential. Biochem. J. 2020, 477, 2071–2093. [Google Scholar] [CrossRef]
- Rondini, E.A.; Granneman, J.G. Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochem. J. 2020, 477, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, A.; Feng, D.; Pessin, E. The Impact of Single-Cell Genomics on Adipose Tissue Research. Int. J. Mol. Sci. 2020, 21, 4773. [Google Scholar] [CrossRef]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Invest. 2019, 129, 4022–4031. [Google Scholar] [CrossRef] [PubMed]
- Schwalie, P.C.; Dong, H.; Zachara, M.; Russeil, J.; Alpern, D.; Akchiche, N.; Caprara, C.; Sun, W.; Schlaudraff, K.U.; Soldati, G.; et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 2018, 559, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.P.; Wu, X.; Yu, G.; Dias, I.R.; Gomes, M.E.; Reis, R.L.; Gimble, J.M. The effect of storage time on adipose-derived stem cell recovery from human lipoaspirates. Cells Tissues Organs 2011, 194, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Melkoumian, Z.; Weber, J.L.; Weber, D.M.; Fadeev, A.G.; Zhou, Y.; Dolley-Sonneville, P.; Yang, J.; Qiu, L.; Priest, C.A.; Shogbon, C.; et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotechnol. 2010, 28, 606–610. [Google Scholar] [CrossRef]
- Carvalho, P.P.; Wu, X.; Yu, G.; Dietrich, M.; Dias, I.R.; Gomes, M.E.; Reis, R.L.; Gimble, J.M. Use of animal protein-free products for passaging adherent human adipose-derived stromal/stem cells. Cytotherapy 2011, 13, 594–597. [Google Scholar] [CrossRef]
- Hadden, M.K. Hedgehog pathway agonism: Therapeutic potential and small-molecule development. ChemMedChem 2014, 9, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Tschaikner, P.; Enzler, F.; Torres-Quesada, O.; Aanstad, P.; Stefan, E. Hedgehog and Gpr161: Regulating cAMP Signaling in the Primary Cilium. Cells 2020, 9, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montagnani, V.; Stecca, B. Role of protein kinases in hedgehog pathway control and implications for cancer therapy. Cancers 2019, 11, 449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulloa, F.; Martí, E. Wnt won the war: Antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 2010, 239, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Chahal, K.K.; Parle, M.; Abagyan, R. Dexamethasone and Fludrocortisone Inhibit Hedgehog Signaling in Embryonic Cells. ACS Omega 2018, 3, 12019–12025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Shahawy, M.; Reibring, C.-G.; Neben, C.L.; Hallberg, K.; Marangoni, P.; Harfe, B.D.; Klein, O.D.; Linde, A.; Gritli-Linde, A. Cell fate specification in the lingual epithelium is controlled by antagonistic activities of Sonic hedgehog and retinoic acid. PLoS Genet. 2017, 13, e1006914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todoric, J.; Strobl, B.; Jais, A.; Boucheron, N.; Bayer, M.; Amann, S.; Lindroos, J.; Teperino, R.; Prager, G.; Bilban, M.; et al. Cross-talk between interferon-γ and hedgehog signaling regulates adipogenesis. Diabetes 2011, 60, 1668–1676. [Google Scholar] [CrossRef] [Green Version]
- Price, P.J.; Gregory, E.A. Relationship between in vitro growth promotion and biophysical and biochemical properties of the serum supplement. In Vitro 1982, 18, 576–584. [Google Scholar] [CrossRef]
- Zheng, X.; Baker, H.; Hancock, W.S.; Fawaz, F.; McCaman, M.; Pungor, E. Proteomic analysis for the assessment of different lots of fetal bovine serum as a raw material for cell culture. Part IV. Application of proteomics to the manufacture of biological drugs. Biotechnol. Prog. 2006, 22, 1294–1300. [Google Scholar] [CrossRef]
- Shih, D.T.B.; Burnouf, T. Preparation, quality criteria, and properties of human blood platelet lysate supplements for ex vivo stem cell expansion. Nat. Biotechnol. 2015, 32, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Sul, H.S. Minireview: Pref-1: Role in Adipogenesis and Mesenchymal Cell Fate. Mol. Endocrinol. 2009, 23, 1717–1725. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, C.; Durandt, C.; Kallmeyer, K.; Ambele, M.A.; Pepper, M.S. The Role of Pref-1 during Adipogenic Differentiation: An Overview of Suggested Mechanisms. Int. J. Mol. Sci. 2020, 21, 4104. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef] [Green Version]
- Grünberg, J.R.; Elvin, J.; Paul, A.; Hedjazifar, S.; Hammarstedt, A.; Smith, U. CCN5/WISP2 and metabolic diseases. J. Cell Commun. Signal. 2018, 12, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Asterholm, I.W.; Kusminski, C.M.; Bueno, A.C.; Wang, Z.; Pollard, J.W.; Brekken, R.A.; Scherer, P.E. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proc. Natl. Acad. Sci. USA 2012, 109, 5874–5879. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, M.; Sun, K.; An, Y.; Gu, X.; Scherer, P.E. VEGF-A–Expressing Adipose Tissue Shows Rapid Beiging and Enhanced Survival After Transplantation and Confers IL-4–Independent Metabolic Improvements. Diabetes 2017, 66, 1479–1490. [Google Scholar] [CrossRef] [Green Version]
- Dallabrida, S.M.; Zurakowski, D.; Shih, S.-C.; E Smith, L.; Folkman, J.; Moulton, K.S.; A Rupnick, M. Adipose tissue growth and regression are regulated by angiopoietin-1. Biochem. Biophys. Res. Commun. 2003, 311, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Pahlavani, M.; Kalupahana, N.S.; Ramalingam, L.; Moustaid-Moussa, N. Regulation and Functions of the Renin-Angiotensin System in White and Brown Adipose Tissue. Compr. Physiol. 2017, 7, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Tyurin-Kuzmin, P.A.; Kalinina, N.I.; Kulebyakin, K.Y.; Balatskiy, A.V.; Sysoeva, V.; Tkachuk, V.A. Angiotensin receptor subtypes regulate adipose tissue renewal and remodelling. FEBS J. 2020, 287, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- Burrell, J.A.; Boudreau, A.; Stephens, J.M. Latest advances in STAT signaling and function in adipocytes. Clin. Sci. 2020, 134, 629–639. [Google Scholar] [CrossRef] [Green Version]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waite, K.J.; Floyd, E.; Arbour-Reily, P.; Stephens, J.M. Interferon-γ-induced Regulation of Peroxisome Proliferator-activated Receptor γ and STATs in Adipocytes. J. Biol. Chem. 2001, 276, 7062–7068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGillicuddy, F.C.; Chiquoine, E.H.; Hinkle, C.C.; Kim, R.J.; Shah, R.; Roche, H.; Smyth, E.M.; Reilly, M.P. Interferon γ Attenuates Insulin Signaling, Lipid Storage, and Differentiation in Human Adipocytes via Activation of the JAK/STAT Pathway. J. Biol. Chem. 2009, 284, 31936–31944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.; Heath, J.K. Receptor recognition by gp130 cytokines. EMBO J. 2000, 19, 2399–2411. [Google Scholar] [CrossRef] [Green Version]
- Cron, L.; Allen, T.; Febbraio, M.A. The role of gp130 receptor cytokines in the regulation of metabolic homeostasis. J. Exp. Biol. 2016, 219, 259–265. [Google Scholar] [CrossRef] [Green Version]
- Meijer, K.; De Vries, M.; Al-Lahham, S.; Bruinenberg, M.; Weening, D.; Dijkstra, M.; Kloosterhuis, N.; Van Der Leij, R.J.; Van Der Want, H.; Kroesen, B.-J.; et al. Human Primary Adipocytes Exhibit Immune Cell Function: Adipocytes Prime Inflammation Independent of Macrophages. PLoS ONE 2011, 6, e17154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabir, S.M.; Lee, E.-S.; Son, D.-S. Chemokine network during adipogenesis in 3T3-L1 cells. Adipocyte 2014, 3, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Angiolillo, A.L.; Sgadari, C.; Taub, D.D.; Liao, F.; Farber, J.M.; Maheshwari, S.; Kleinman, H.K.; Reaman, G.H.; Tosato, G. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J. Exp. Med. 1995, 182, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herder, C.; Hauner, H.; Kempf, K.; Kolb, H.; Skurk, T. Constitutive and regulated expression and secretion of interferon-γ-inducible protein 10 (IP-10/CXCL10) in human adipocytes. Int. J. Obes. 2006, 31, 403–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balabanian, K.; Lagane, B.; Infantino, S.; Chow, K.Y.C.; Harriague, J.; Moepps, B.; Arenzana-Seisdedos, F.; Thelen, M.; Bachelerie, F. The Chemokine SDF-1/CXCL12 Binds to and Signals through the Orphan Receptor RDC1 in T Lymphocytes. J. Biol. Chem. 2005, 280, 35760–35766. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Martín, L.; Sánchez-Mateos, P.; Cabañas, C. CXCR7 impact on CXCL12 biology and disease. Trends Mol. Med. 2013, 19, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, W.; Shen, J. CXCR7 Targeting and Its Major Disease Relevance. Front. Pharmacol. 2018, 9, 641. [Google Scholar] [CrossRef] [Green Version]
- Baer, P.C.; Kuci, S.; Krause, M.; Kuçi, Z.; Zielen, S.; Geiger, H.; Bader, P.; Schubert, R. Comprehensive Phenotypic Characterization of Human Adipose-Derived Stromal/Stem Cells and Their Subsets by a High Throughput Technology. Stem Cells Dev. 2013, 22, 330–339. [Google Scholar] [CrossRef]
- Gilliam, D.T.; Menon, V.; Bretz, N.P.; Pruszak, J. The CD24 surface antigen in neural development and disease. Neurobiol. Dis. 2017, 99, 133–144. [Google Scholar] [CrossRef]
- Deng, W.; Gu, L.; Li, X.; Zheng, J.; Zhang, Y.; Duan, B.; Cui, J.; Dong, J.; Du, J. CD24 associates with EGFR and supports EGF/EGFR signaling via RhoA in gastric cancer cells. J. Transl. Med. 2016, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Song, B.-Q.; Chi, Y.; Li, X.; Du, W.; Han, Z.-B.; Tian, J.-J.; Li, J.-J.; Chen, F.; Wu, H.; Han, L.-X.; et al. Inhibition of Notch Signaling Promotes the Adipogenic Differentiation of Mesenchymal Stem Cells through Autophagy Activation and PTEN-PI3K/AKT/mTOR Pathway. Cell. Physiol. Biochem. 2015, 36, 1991–2002. [Google Scholar] [CrossRef]
- Kang, S.; Akerblad, P.; Kiviranta, R.; Gupta, R.K.; Kajimura, S.; Griffin, M.; Min, J.; Baron, R.; Rosen, E.D. Regulation of Early Adipose Commitment by Zfp521. PLoS Biol. 2012, 10, e1001433. [Google Scholar] [CrossRef]
- Mitterberger, M.C.; Lechner, S.; Mattesich, M.; Kaiser, A.; Probst, D.; Wenger, N.; Pierer, G.; Zwerschke, W. DLK1(PREF1) is a negative regulator of adipogenesis in CD105+/CD90+/CD34+/CD31−/FABP4− adipose-derived stromal cells from subcutaneous abdominal fat pats of adult women. Stem Cell Res. 2012, 9, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muoio, F.; Panella, S.; Harder, Y.; Tallone, T. Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway. Biologics 2021, 1, 129-153. https://doi.org/10.3390/biologics1020008
Muoio F, Panella S, Harder Y, Tallone T. Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway. Biologics. 2021; 1(2):129-153. https://doi.org/10.3390/biologics1020008
Chicago/Turabian StyleMuoio, Francesco, Stefano Panella, Yves Harder, and Tiziano Tallone. 2021. "Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway" Biologics 1, no. 2: 129-153. https://doi.org/10.3390/biologics1020008
APA StyleMuoio, F., Panella, S., Harder, Y., & Tallone, T. (2021). Induction of the CD24 Surface Antigen in Primary Undifferentiated Human Adipose Progenitor Cells by the Hedgehog Signaling Pathway. Biologics, 1(2), 129-153. https://doi.org/10.3390/biologics1020008






