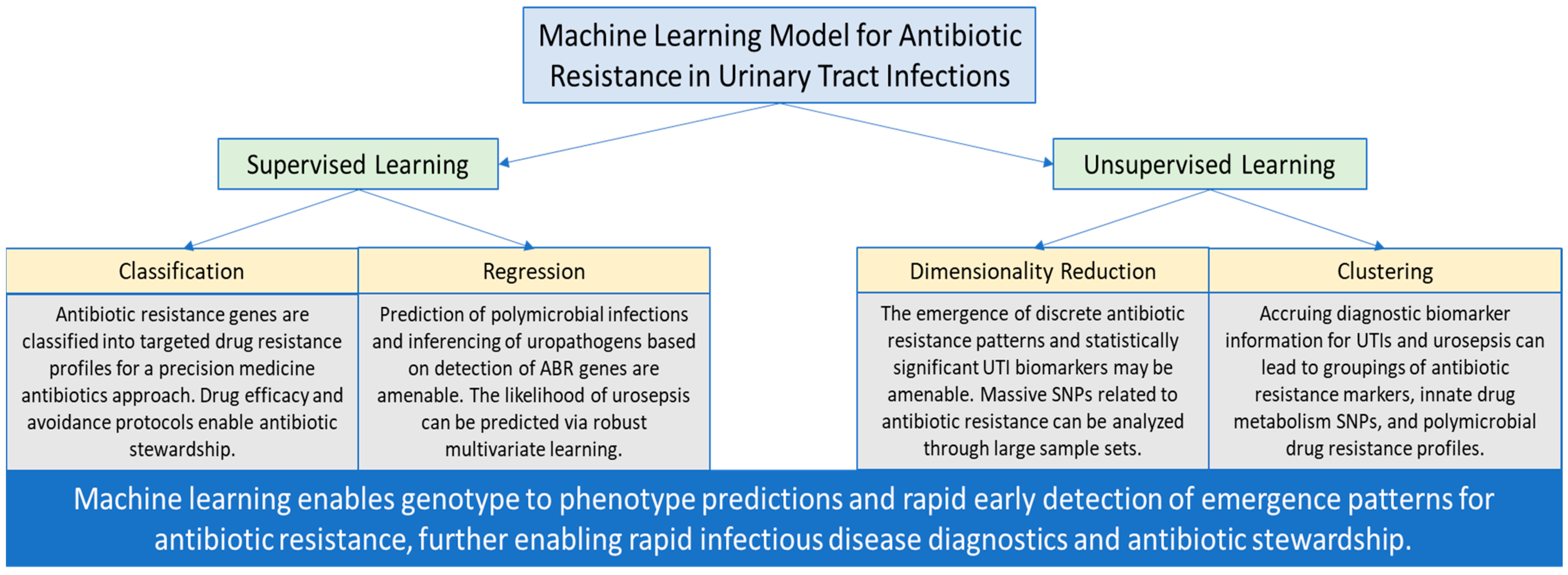
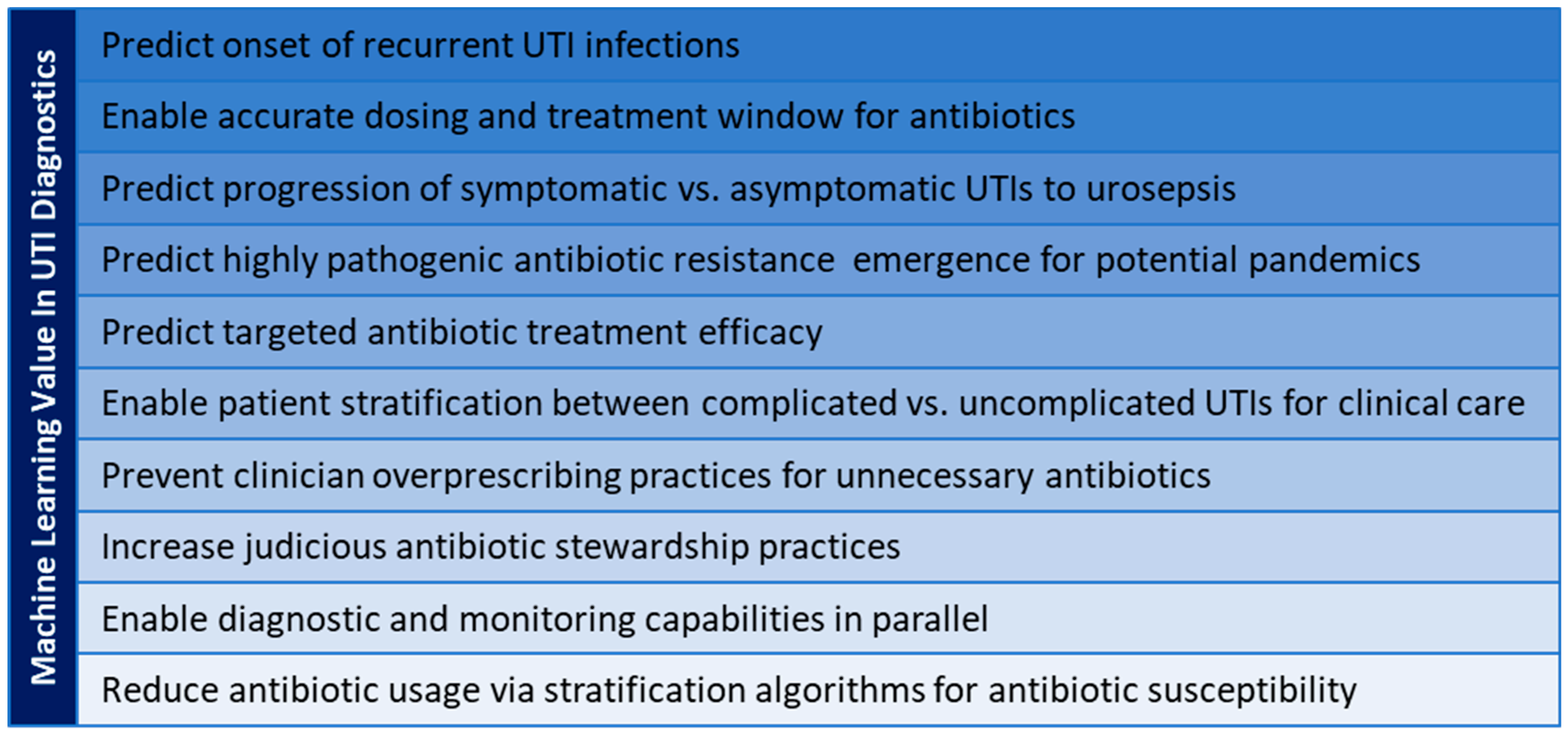
1. Machine Learning: An Introduction
The term “machine learning’ was coined by Arthur Samuel, who described it as “giving computers the ability to learn without being explicitly programmed” [
1]. In machine learning, an automatic computational model is built with available data, and the computer infers patterns from the data and learns from past experiences. Machine learning models rely on the quality and availability of large amounts of data. The process of developing the model is called “training” the model/algorithm, and the data used to build the model are referred to as the “training set”. A developed model/algorithm can be applied to new datasets [
1,
2,
3].
There are two main types of machine learning: supervised and unsupervised learning [
2,
3,
4]. In supervised learning, the goal is to predict an outcome or target. It focuses on classification (choosing among sub-groups to best describe a data point) and prediction. The model is given defined input data or features to learn and outcome measures to be arrived at, and the computer learns relationships and patterns between the datasets. Then, when new inputs are presented, the computer should be able to deliver the outcomes. The performance of such a model is measured by comparing predicted output values to known output values. For example, the model is trained on a training set of patients using input features, such as patient demographics and risk factors, and the algorithm learns to diagnose the presence or absence of disease; the algorithm is then validated on another set of patients [
2,
3,
4].
In unsupervised learning, the computer is presented with unclassified features, and the computer recognizes and determines if there are any relationships or patterns in the data. Unsupervised learning models are useful for clustering or organizing data by patterns. There are no outcomes to predict, but the goal is to determine naturally occurring patterns or groupings within data. The performance of an unsupervised learning model depends on whether the model has captured interesting and useful trends in the data. Such models can often lead to questions and answers not previously thought of by the researchers [
2,
3,
4].
Several models/algorithms are available, such as decision trees, artificial neural networks, and support vector machines, to name a few; however, there is no guide or consensus on the right model to use. Usually, scientists apply different algorithms to their problem and see which works best [
1].
Machine learning offers immense potential to advance medical diagnoses and treatments. Vast amounts of accessible electronic health records may be used to build models that deliver improved and accurate diagnoses, reduce clinical workload by efficiently ruling out negative infections, and predict the risk of infection/disease. With next-generation sequencing generating large amounts of genome sequence data, machine learning models offer new ways to predict and also fight antibiotic resistance and tailor treatments. In the context of antibiotic resistance in UTIs, supervised and unsupervised modalities of machine learning have the potential for early pattern detection and prediction for optimizing clinical decision-making and stewardship (
Figure 1). Machine learning offers accelerated and early prevention capabilities to facilitate guidance for clinicians and patients, translating into value-added capabilities in clinical care and public health (
Figure 2).
2. Machine Learning for Pathogen Identification and Phenotypic AST
Traditional methods for microbial diagnosis of an infection rely on bacterial culture and antimicrobial susceptibility testing (AST) that require a few days. This delay in definitive diagnosis leads to the use of broad-spectrum antibiotics, which in turn leads to increases in antibiotic resistance. Particularly for polymicrobial infections, all the pathogens and their antibiotic susceptibilities need to be identified rapidly for tailored therapy. In this section, we discuss how machine learning has aided pathogen detection and phenotypic or pheno-molecular AST, particularly in urinary tract infections.
A recently developed digital PCR-high-resolution melt platform used a machine learning algorithm to analyze the digital high-resolution melt curves to correctly identify the bacterial species, even in polymicrobial samples [
5]. The microfluidics platform had three independent nanoarray modules, each with ~5000 nanowells. The DNA extracted from the bacterial sample was appropriately diluted such that in each nanowell, the PCR and melt curves were issued from a single DNA molecule. The abundance of each species in a given clinical sample was measured by enumerating the number of species-specific melt curves generated. By simultaneously analyzing samples grown briefly in the presence and absence of antibiotics, antibiotic susceptibility was also tested. This platform could achieve both species identification and AST in just 4 h [
5].
Machine learning models are also aiding pathogen identification with MALDI-TOF. MALDI-TOF is widely used in clinical laboratories for species identification; however, it suffers from the disadvantage that it requires bacteria in pure culture, necessitating additional steps of culturing from clinical samples. Machine learning models that were trained on LC-MS/MS peptidic signatures from bacteria could identify bacteria from urine samples within four hours and without bacterial culture [
6]. Currently, signatures for 15 species that cause 84% of UTIs are available. If the models include peptidic signatures for proteins associated with antimicrobial resistance, MALDI-TOF may be used to predict AST results as well. Machine learning models have already been developed to predict specific antibiotic resistances from MALDI-TOF profiles: colistin-resistant
Acinetobacter baumanii and
Klebsiella pneumoniae [
7], and vancomycin-intermediate Staphylococcus aureus and vancomycin-susceptible
S. aureus [
8].
Machine learning has also benefited microscopic-imaging-based methods of pathogen identification and AST. One method is a low-cost microfluidic dark-field imaging platform where immobilized oligonucleotide probes hybridize to the 16S rRNA of the target pathogen [
9]. A machine learning algorithm examines the images of the agglutinated bacterial clusters and enumerates the bacteria in the clusters. This method delivers pathogen identification results in 30 min and AST results in 3 h [
9]. In another method called MAST, or microscopy-based AST, antibiotics and bacteria dispensed in solid-phase microwells are microscopically imaged to monitor bacterial replication. Machine learning then classified the images as growth or inhibition based on known morphologies of cells. MAST could accurately determine MIC (minimum inhibitory concentration) in 2 h [
10]. In a third method, deep learning algorithms were trained to learn phenotypic features from video microscopy of freely moving single bacteria in urine. The model incorporated multiple features, such as morphology, size, division, and movement. AST was determined in 30 min when tested on
E. coli in urine with five different antibiotics [
11]. Machine learning models have also been used to analyze Fourier-Transform Infrared (FTIR) microscopic images of
E. coli following an initial culture of urine samples from UTI patients and predict AST within a few minutes [
12,
13].
3. Machine Learning for Sequence-Based AST
New diagnostic technologies aim to identify resistance genes and genotypes, rather than resistance phenotypes, and then translate genotype information into treatment. Existing multiplex PCR and microarray methods are used clinically to detect resistance-specific markers, but while these methods succeed in finding enzyme-mediated resistances, they fail at identifying resistances caused by mutations within a gene. Sequence-based diagnostics apply whole-genome sequencing data to predict resistance phenotypes from genotypes. They provide a faster alternative to multiplex PCR screening for phenotypes that are contributed to by multiple loci. Sequence-based diagnostics may use metagenomic sequencing, where DNA is extracted from the sample without culturing, or whole-genome sequencing, where the DNA is extracted from the pure culture of the bacteria obtained from the clinical sample [
14]. Sequencing-based ASTs are currently slower and more expensive than phenotypic ASTs; however, these limitations are likely to decrease in the future with new advances in technology [
15].
Rules-based approaches have been traditionally used to interpret sequence-based diagnostics [
14]. These approaches make predictions based on the presence or absence of resistance genes and are similar to the knowledge-based decisions that a clinician would make. Rules-based models perform well for well-studied pathogens with well-characterized resistance mechanisms, but they require regularly curated databases as new resistance genes and mechanisms are discovered. It is not straightforward to predict phenotypes merely from the presence or absence of resistance genes, as many parameters affect genotype-to-phenotype translations [
15]. Rules-based models do not consider interactions between loci when multiple loci contribute to a phenotype, they do not consider the effect of the strain background on loci activity, and they assume a complete understanding of the genetic background for any resistance phenotype [
14]. Differential expression of genes under different contexts is a confounding factor for the interpretation of genotypes—genes may be expressed differently if they are plasmid-borne or chromosomal, or if they are under the control of regulatory sequences or different promoters [
15].
The large-scale coupling of antimicrobial resistance data and whole-genome sequencing data for thousands of isolates has enabled many researchers to apply machine learning to aid sequence-based diagnostics. Machine learning models can overcome many of the above limitations. Such models are trained on a set of genomes with known phenotypes, and the models learn which genes or single-nucleotide polymorphisms (SNPs) are responsible for particular resistance phenotypes. The model is then validated, ideally on a different set of genomes. Machine learning models can learn interactions between loci and can be trained to weight different loci, since not all loci contribute equally to the phenotype [
14].
Machine learning models were used to uncover genetic mechanisms for resistance to 17 antibiotics in
Clostridium difficile,
Streptococcus pneumoniae,
Mycobacterium tuberculosis, and
Pseudomonas aeruginosa [
16]. Machine learning can be used to understand new resistance mechanisms and discover shared resistance mechanisms between different antibiotics, potentially helping to identify appropriate combination therapies [
16,
17]. Machine learning algorithms can reveal previously unknown associations and epistatic interactions between resistance determinants and can perform well even when resistance mechanisms are not well understood, as shown for
M. tuberculosis [
17,
18]. Models have also been recently applied to examine both whole-genome and transcriptome sequencing data to account for differential expression of genes [
19]. Gene expression and genome sequencing data from 414 clinical isolates were used to build highly sensitive predictive models for resistance to four drugs for
P. aeruginosa, an organism that exhibits many environment-driven gene expression changes [
19].
Machine learning algorithms may use different inputs—genes, contigs, or k-mers. Models have been applied to pan-genomes to identify resistance determinants in
E. coli [
20,
21],
M. tuberculosis [
17], and
Elizabethkingia [
22]. The pan-genome refers to the shared features of all strains in a species; the pan-genome may be classified into a core set that clusters genes present in all the genomes, and an accessory set that clusters all the remaining genes that are present in only one or a few genomes. Her and Wu showed that only 61% of the
E. coli resistance genes were in the accessory set (61%), suggesting that there are intrinsic resistance genes shared across all
E. coli, and the accessory set would serve to differentiate between resistance phenotypes of different strains. They designed an algorithm that selected gene clusters from the accessory set and trained it to predict resistance to four drugs, and this out-performed the genes shown in the literature to predict resistance [
20].
An alternative to using genes as inputs is k-mers, which are small overlapping sub-sequences within a contig. The model uses each k-mer as a feature, and it identifies the k-mers that contribute to antibiotic resistance. The use of k-mers reduces biases that occur when using reference genomes. Models that used k-mers have successfully predicted resistance in several species:
Neisseria gonorrhea [
23],
Klebsiella [
24], non-typhoidal
Salmonella [
25],
C. difficile,
M. tuberculosis,
P. aeruginosa, and
Streptococcus pneumoniae [
16].
Pesesky et al. compared rules-based approaches to machine learning models in predicting antimicrobial resistance from whole-genome data from 78
Enterobacteriaceae species that spanned a range of phenotypes. Rules-based models failed because of incomplete genome assemblies and new variants of known resistance markers. Machine learning models were confounded by low-frequency resistance markers that occurred very rarely or not at all in the training set [
15]. Although both models performed overall with similar accuracies, the researchers predict that machine learning models will prevail in the future because they perform similarly with any pathogen, rather than just well-characterized species. While rules-based models need databases with constant curation, updating machine learning algorithms will be simple. Once the training set is sufficiently large, machine learning algorithms will be able to uncover new resistance mechanisms that could not have been detected by traditional means. Machine learning models can also be trained to predict the MICs of antibiotics, which would be more relevant for guiding treatment [
15]. Models could predict MICs for 15 antibiotics in non-typhoidal
Salmonella [
25], for 20 antibiotics in
Klebsiella [
24], for 5 antibiotics in
N. gonorrhea [
26], and 6 antibiotics in
S. pneumoniae [
27,
28]. One study that compared machine learning models and rules-based models for prediction of MICs in
S. pneumoniae showed that rules-based models had higher false negatives [
27]. Models are not a “one size fits all” solution. In
M. tuberculosis, where mutations within genes often determine resistance, seven machine learning algorithms performed better than a rules-based approach in predicting resistance against eight drugs. However, each drug was predicted best by a different model and a different subset of mutations [
18].
It Is unreasonable to expect 100% accuracy from sequence-based diagnostics, even with machine learning models. There are non-genomic determinants of resistance, such as biofilm formation and DNA methylation patterns, that are difficult to predict [
21]. Rare mutations, of which we have incomplete knowledge, can also cause resistance. For example, among Staphylococcus aureus clinical isolates, 93% of rifampin resistance was caused by 8 mutations in rpoB, but the remaining 8% was caused by 72 rare mutations [
29]. That said, several steps can be taken to improve machine learning predictions. We need more genomic databases that are matched with phenotypic AST data. The training set used should be as large and diverse as possible. Researchers often use conveniently accessible databases, but these may not be varied geographically or temporally. Sampling—the temporal range, the geographic range, and the sampling approach—can affect the model. For example, the RpoB I491F mutation accounts for <5% of TB resistance to rifampicin in most countries, but in Swaziland, it accounted for 30% [
30]. Hicks et al. showed that model performance varies with the antibiotic, the dataset, the resistance metric used, and the species [
31]. For instance, in
N. gonorrhea, machine learning models accurately predicted ciprofloxacin resistance, which is mediated by mutations in a single gene (gyrA), but not resistance to azithromycin, which is mediated by various means [
31]. Many studies use datasets from highly resistant organisms, which does not help a balanced model. Nguyen et al. used a very diverse and balanced dataset of >5000 Salmonella genomes collected over 15 years. Their model could predict resistances in newer strains and possibly help with future outbreaks [
25].
Better databases for antibiotic resistance genes will enhance the performance of sequence-based AST. Most resistance gene prediction tools use a “best hit” approach, which results in these tools having low false-positive rates but high false-negative rates. Deep machine learning models were used to look at similarity distributions of sequences in the antibiotic resistance gene database across 30 antibiotic resistance categories to predict whether a gene was a resistance gene or not [
32]. The resulting Deep-ARG database has expanded the known repertoire of ARGs.
4. Machine Learning in Clinical Decision Making
In this section, we look at the various ways in which machine learning models have helped improve different stages of clinical decision-making—reducing clinical workload, reducing unnecessary hospital visits, and improving diagnosis and antibiotic prescriptions.
During laboratory diagnosis of UTI, urine culture is often performed, but 70–80% of the results are negative, mixed, or ambiguous cultures, and therefore unhelpful. Appropriate pre-processing of urine samples, such that only true microbially infected samples are further analyzed, will reduce clinical workload and improve efficiency [
33]. Machine learning algorithms can help in this pre-processing where positive urine cultures continue to be detected, but negative samples are ruled out before culture. One such algorithm was built from urine analysis data collected over a year in a single clinical laboratory servicing multiple hospitals [
33]. This algorithm incorporated the microscopy thresholds of white blood cell and bacterial counts, and other factors such as patient age, pregnancy, red blood cell count, and if the patient was pre-operative, immunocompromised, or an in-patient. The model worked best—with 95% sensitivity and 41% workload reduction—with separate decision trees for pregnant patients, for children below 11 years, and all others [
33].
Another study used data and urinalysis results from 59 patients; the algorithms developed were able to differentiate between cystitis and non-specific urethritis from only three parameters—erythrocyte count, suprapubic pain, and frequent urination [
34]. Using these models could eliminate the need for expensive laboratory tests.
An unsupervised machine learning model was used to detect UTIs in patients with dementia [
35]. People with dementia are at an increased risk for physical health-related factors, with UTIs being one of the top five reasons for hospital visits. Unfortunately, patients with dementia tend to receive poorer-quality care at hospitals and less favorable outcomes. Therefore, machine learning models may be useful, both to avoid unnecessary hospital visits for these patients and to provide early detection of UTI. The model used environmental and physiological in-home sensory data to analyze the patient’s daily routines and patterns to identify any anomalies or routine changes. Time-sensitive movement patterns, such as an increase in bathroom use frequency, along with an increase in body temperature, would generate a UTI alert, which would be followed up by healthcare practitioners [
35].
Another area where machine learning offers great potential is in emergency department visits. Diagnosis and treatment prescriptions in emergency departments are based on symptoms and physical findings, rather than urine culture; consequently, misdiagnoses and antibiotic overuse are frequent. In one study, machine learning models were built with a large dataset of >80,000 emergency department visits with UTI symptoms and urine culture results [
36]. Initial models used 211 input variables, but the models performed similarly with a curated list of just ten variables—age, gender, urine analysis (white blood cell count, nitrites, epithelial cells, leukocytes, blood, and bacteria), dysuria, and history of UTIs. The diagnoses provided by the models had significantly higher accuracy than the healthcare provider’s diagnoses (87% vs. 53%). When the model was applied retrospectively, 1 in 4 patients were reclassified from false positive to true negative, and 1 in 11 were reclassified from false negative to true positive [
36].
A study in Israel used machine learning on a 10-year longitudinal dataset of community- and retirement-home-acquired UTI patients to deliver personalized drug-specific predictions of resistance [
37]. This study found that the risk of an infection being resistant to antibiotics correlated with patient demographics (age, gender, pregnancy, and retirement home living), past patient history, past antibiotic use, and antibiotic resistance in past UTIs. When their model was applied retrospectively over a one-year test period, the number of mismatched prescriptions significantly decreased. Thus, machine learning algorithms can guide UTI treatments without urine culture [
37]. A similar study examined machine learning models to guide empiric antibiotic therapy in a children’s hospital in Cambodia. They found that the time from admission to culture, age of the patient, age-adjusted weight score, and if the infection was hospital- vs. community-acquired were the most important predictors [
38]. Thus, in resource-limited areas, readily available patient data can also be used to prescribe antibiotics appropriately. Another study developed machine learning models with lab test results from suspected UTI patients and linked administration data (individual and household background, employment, hospitalization, and prescription histories) from a large medical laboratory in Denmark. The resulting model succeeded in lowering antibiotic prescriptions for UTIs by 7.4% [
39].
Machine learning models have been used to predict the risk of infection in hospitalized patients. One such model identified patients at risk for drug-resistant
P. aeruginosa infections; the model was trained with ~3000 intensive care unit (ICU) admissions over a 13-year dataset from a hospital in Spain. The model performed well with data on date of culture, antibiotic used, the clinical origin of patient, disease reason for ICU admission, the time between admission and culture, and APACHE II (Acute physiology and chronic health evaluation) and SAPS 3 (Simplified acute physiology score) scores [
40]. Another model was developed with routinely available blood parameters (C-reactive protein, white cell count, bilirubin, creatinine, ALT, and alkaline phosphatase) to predict infection/sepsis among patients upon admission to a hospital [
41].
5. Summary and Future Prospects
We have discussed here the various ways in which machine learning is poised to aid emerging diagnostic technologies and enhance clinical decision-making. New methods of pathogen identification and pheno-molecular AST rely on machine learning models to deliver rapid results. Machine learning models are enabling clinical laboratories to reduce workload and healthcare providers to make better decisions regarding diagnosis and treatments. A large number of machine-learning-based studies in the last three years have advanced the possibilities of sequence-based ASTs, making it likely to be a clinically implemented tool soon. Furthermore, growing databases of pathogenic strains that are collected from around the world are continuously being characterized (whole-genome sequencing, AST/MIC tests, culture) and provided as a public resource for use in machine learning training sets and algorithmic development (examples include the CDC & FDA Antimicrobial Resistance Isolate Bank, the Active Bacterial Core Surveillance Isolate Bank, and the Antibacterial Resistance Leadership Group Virtual Repository). Since isolates are tested in the same parameters within a clinical diagnostic setting (for example, by CLSI guidelines), these datasets are robust for machine learning and algorithm development for pathogen detection and antibiotic resistance. Additionally, the antibiotic drug resistance profiles are available for more than 100+ drugs, inclusive of dosing concentrations and sensitive/intermediate/resistance (SIR) nomenclature. Such datasets hasten the development of automated algorithms from real-world evidence of antibiotic resistance strains of multiple genera and species. Machine learning can enable diagnostic development, new antibiotic drug discovery, perform safety and efficacy profiles of antibiotics, discovery of novel pathogenic mechanisms, and detection of new and unusual global public health antimicrobial resistance threats.
Another molecular diagnostics area where machine learning may offer potential value is in the use of multiplex PCR panels. Currently, such diagnostic panels exist for certain diseases, such as panels for respiratory pathogens [
42], gastrointestinal pathogens [
43], and meningitis/encephalitis pathogens [
44]. It is conceivable to build multiplex PCR panels that deliver both pathogen identification and tests for common antibiotic resistance markers. Machine learning may be employed to predict AST based on qPCR detection calls. This could provide a faster alternative to sequence-based diagnostics. Advances in multiplexing capabilities in modern real-time PCR instruments enable multi-target detection via coupling of multiple fluorophore probes. This further enables polymicrobial detection capabilities which may be amenable to machine learning discovery diagnostics for interactions between pathogens and resistance markers. Multiplex PCR offers high-throughput, cost-efficiency, and molecular algorithm development opportunities to stratify uropathogens and respective antibiotic resistance markers for UTI management.
Machine learning offers the potential for rapid diagnostics and accelerated discovery of translational research applications. The ability for machines to exponentiate intelligence and guide clinical treatment decisions is important for reducing overall healthcare burden. Antibiotic resistance is a growing public health concern. The ability for machine learning to enable judicious selection of antibiotic therapies encourages antibiotic stewardship for global public health. Continuous learning and complex stratification algorithms can enable targeted antibiotic precision medicine and enhanced predictive power for recurrent and emerging infectious states. For example, machine learning may aid in stratification of UTI patient populations who can be divested into out-patient care (less expensive for external clinics) vs. in-patient care (significantly more expensive for emergency room visits). This can enable a significant reduction in economic waste, better patient management and clinical decision-making, and early detection diagnostics coupled to efficient monitoring protocols for better patient care. Therefore, machine learning offers promise to identify the right patient at the right time for the right drug in the context of antibiotic resistance and UTIs, which is analogous to the paradigm set in oncology diagnostics today.