1. Introduction
As the COVID-19 pandemic continues to impact global health, rapid and accurate diagnosis has become a cornerstone in controlling outbreaks. The disease’s rapid spread since its emergence in China has resulted in over 378 million confirmed cases and more than 5.67 million fatalities worldwide [
1]. The diagnostic strategy for detecting SARS-CoV-2 primarily involves reverse-transcription polymerase chain reaction (RT-PCR) tests [
2] and radiological imaging, particularly thin-section chest computed tomography (CT) [
3,
4]. While RT-PCR remains the gold standard, chest CT scans play a pivotal role in patient management and morbidity assessment, with HRCT boasting a sensitivity range of 56–98% for COVID-19 pneumonia diagnosis [
5,
6]. T imaging hallmarks, such as ground glass opacities (GGO) and crazy paving patterns, are integral to COVID-19 detection [
7]. However, their non-specificity necessitates a more discerning diagnostic toolset [
8]. The vast number of images generated during the pandemic has fueled research into deep learning techniques for swift COVID-19 identification from CT scans [
9,
10]. Despite positive results, these endeavors face limitations due to the small sample sizes and lack of comparative respiratory disease data within public datasets [
11,
12,
13,
14].
The implementation of artificial intelligence (AI) in medical diagnostics is hindered by the scarcity of large, accessible datasets required for algorithm training [
15]. This gap is partially bridged by transfer learning, where AI is first trained on extensive databases before application to smaller, specialized datasets [
16]. Recognizing the need for comprehensive image collections, our study aims to establish a sizable CT scan image dataset of COVID-19 patients for future AI utilization [
17].
In parallel, vision transformers (ViTs) have emerged as powerful tools in image recognition, owing to their efficient token mixing capabilities [
18]. Nonetheless, the quadratic complexity of token mixing with self-attention becomes prohibitive for high-resolution images and long sequences [
19]. Innovative token mixers like global filter networks (GFN) have been developed, leveraging Fourier transforms for efficient token mixing [
20]. Despite their strengths, these methods face challenges in adaptivity and expressiveness at higher resolutions [
21]. To overcome these limitations, this paper introduces the adaptive Fourier neural operator (AFNO), which treats tokens as continuous elements and models token mixing as a continuous global convolution. This approach is rooted in operator learning, traditionally used to tackle partial differential equations (PDEs) [
22]. Adapting Fourier neural operators (FNOs) for vision, we impose a block-diagonal structure on channel mixing weights and utilize soft-thresholding to sparsify frequencies, enhancing generalization and efficiency [
23].
The literature reflects a growing body of work dedicated to improving transformer efficiency [
24]. Our proposed AFNO model, with its efficient token mixing and expressive capability [
25], is positioned to advance the field of medical image analysis, particularly for high-resolution COVID-19 CT scans. This work aims to set a new benchmark in AI-powered diagnostics, offering a potential leap forward in managing the COVID-19 health crisis.
The key contributions of this work are as follows:
Innovative technique implementation integrates AFNO into the image analysis of HRCT, enhancing the handling of high-resolution data, which traditional models struggle with due to computational limitations. Enhanced image processing efficiency is achieved with Fourier transforms for efficient token mixing, overcoming the scalability issues associated with high-resolution images.
Improved diagnostic accuracy is demonstrated through superior performance in identifying COVID-19 related abnormalities in chest CT scans compared to traditional methods, enhancing diagnostic accuracy and reliability.
The architecture of AFNO allows for adaptation to different resolutions and details, suggesting wider clinical application beyond COVID-19 to other medical imaging tasks. This is a significant step forward in the use of AI for medical diagnostics, particularly in environments where access to high-quality imaging data may be limited.
2. Materials and Methods
2.1. Dataset
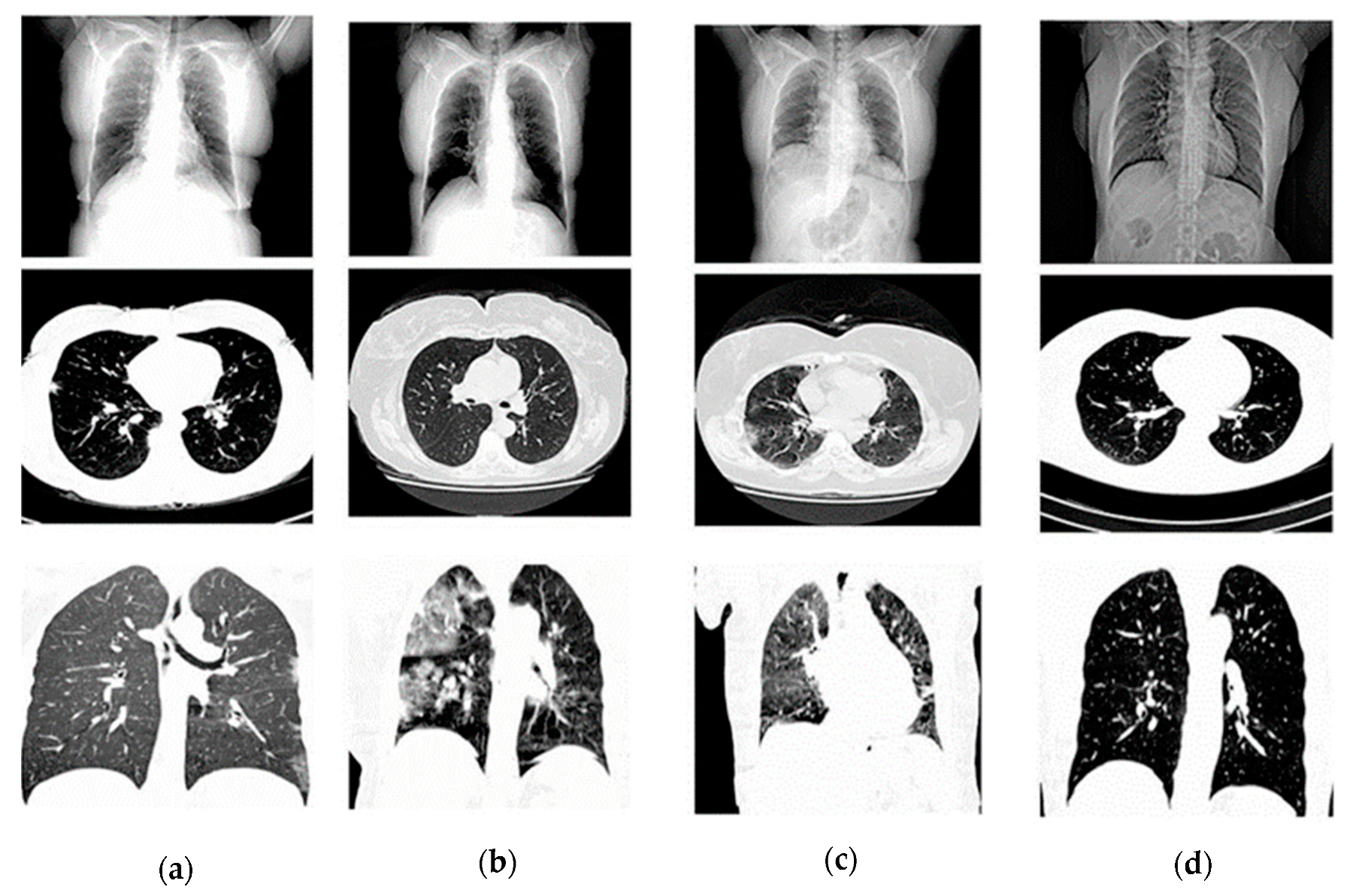
The HRCTCov19 dataset [
26] was employed for this study, consisting of 181,106 high-resolution chest CT images from 395 patients, categorized into classes representing ground glass opacity (GGO), crazy paving, air space consolidation, and negative for COVID-19. Images were preprocessed to a uniform size of 512 × 512 pixels, normalized to a [0, 1] scale, and randomly split into training (60%), validation (20%), and testing (20%) sets. In
Table 1. it is shown as follows while image dataset is showing in
Figure 1.
2.2. Image Preprocessing
Prior to training, all images underwent a series of preprocessing steps. Images were resized using bilinear interpolation to a consistent resolution of 512 × 5 × 12 pixels to ensure compatibility with the AFNO model. For normalization, pixel values were normalized to a [0, 1] scale via the following equation:
where
I is the original image intensity and min(
I) and max(
I) are the minimum and maximum intensities in the image, respectively.
To increase the robustness of the model, data augmentation techniques such as random rotation, flipping, and zoom were applied.
2.3. Adaptive Fourier Neural Operator (AFNO) Architecture
The adaptive Fourier neural operator method is a sophisticated approach designed for image analysis that divides each image into small and large patches, some of which overlap. This method is used to address the challenge of efficiently processing high-dimensional data by transforming the way the image information is mixed and processed. The core of AFNO lies in its innovative technique for mixing the image’s features. Unlike conventional methods that mix features at a fixed scale, AFNO adapts to different resolutions, which is crucial for handling images of various sizes and details. The AFNO method rethinks the self-attention mechanism often used in transformers, which usually requires a hefty computational effort, especially as image size grows. Instead, AFNO integrates kernels, a mathematical concept used to generalize functions, and operates on these kernels in a continuous fashion. This continuous operation allows the model to consider the global context of the image, which is essential for understanding the spatial relationships within the image.
One of the critical improvements AFNO brings to the table is its efficient handling of the Fourier transform, a mathematical tool used to decompose functions into frequencies. AFNO uses this technique to work more effectively with the spectral components of the image, which represent the image’s details and textures. The method ensures that the important high-frequency information is preserved during the image processing stages. Moreover, AFNO introduces a new structure for mixing channels, which are the components of the image data related to color and intensity. The model structures the data in such a way that it divides the image into blocks and processes these blocks separately, making the computation more manageable and highly parallelizable. This block structure enables AFNO to handle larger images more efficiently by simplifying the complex interdependencies of the image’s features.
An innovative aspect of AFNO is its weight sharing mechanism, where it reuses certain parameters across different parts of the image, which reduces the overall computational load without sacrificing performance. This weight sharing is coupled with an adaptive strategy, where the importance of different features is dynamically adjusted. By focusing more on the significant features and less on the less relevant ones, AFNO can operate more efficiently. It also addresses the sparsity inherent in images—where only a few pixels might hold the most critical information—by selectively focusing on those areas of the image that carry more information. This selective focus is achieved through a technique that penalizes less important features, thus promoting a more compact and relevant feature set for the model to work with.
Thus, the AFNO architecture was employed for its ability to handle high-resolution input images without the computational infeasibility associated with self-attention mechanisms. The token mixing in AFNO is achieved through a series of operations in the Fourier domain, described as follows:
Each image X is transformed into the frequency domain using the fast Fourier transform (FFT):
In the frequency domain, a gating mechanism is applied, parameterized by a learnable matrix
where σ represents a non-linear activation function.
The processed frequency representation is then converted back to the spatial domain using the inverse fast Fourier transform (IFFT):
The model also performs channel mixing, which is adaptively parameterized by block-diagonal matrices to control the complexity and enhance parameter efficiency.
The model was trained using the Adam optimizer with a learning rate of 1 × 10
−4, decayed by a factor of 0.1 every 20 epochs. Cross-entropy loss function was used to compute the difference between the predicted and true labels:
where
yo,c is a binary indicator of whether class label
c is the correct classification for observation o, and
po,c is the predicted probability that observation
o is of class
c.
Model performance was evaluated on the test set using metrics such as accuracy, precision, recall, and F1 score. These metrics were chosen to provide a comprehensive understanding of the model’s classification abilities, taking into account the balance between sensitivity and specificity, which is crucial for medical diagnostic tasks.
3. Results
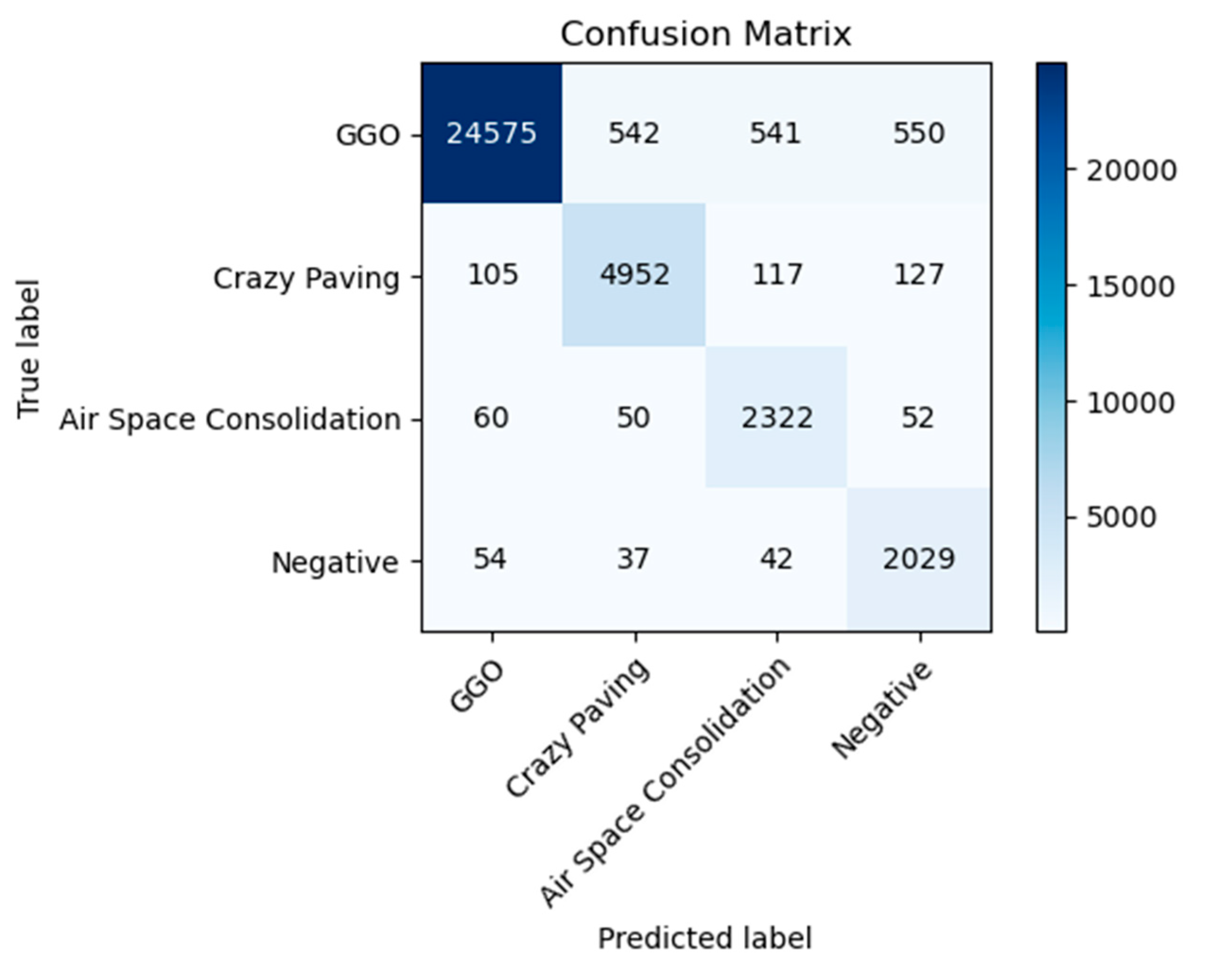
The AFNO model’s application to the HRCTCov19 dataset, which consists of 181,106 high-resolution chest CT images from 395 patients processed at a resolution of 512 × 512 pixels, has yielded a comprehensive insight into its classification capabilities across four diagnostic categories: ground glass opacity (GGO), crazy paving, air space consolidation, and negative for COVID-19. The distribution of classifications as depicted in the confusion matrix in
Figure 2, indicates a high degree of accuracy, especially notable in the identification of GGO and negative cases.
The model has shown remarkable aptitude for detecting GGO, which is evidenced by a large majority of cases being correctly classified, as indicated by the prominent darker shading in the confusion matrix. This suggests that the model’s features are well-tuned to identify the characteristics unique to GGO within the data. Similarly, the negative category has demonstrated a significant number of correct classifications, affirming the model’s ability to discern non-COVID-19 cases with a high degree of certainty.
From
Table 2, it is observed that the highest precision points to the model’s strength in accurately identifying true positive cases within this class. Although the precision dips for the air space consolidation and negative classes, it remains well within the acceptable range, illustrating the model’s reliable performance even in classes with fewer samples. The recall metric, which measures the model’s sensitivity or its ability to identify all actual positives, remains impressively high across all classes. The model’s sensitivity is particularly noteworthy in the air space consolidation and negative classes, where despite a lower precision, it misses very few actual positive cases, as reflected by the lighter shading off the diagonal in the confusion matrix.
The F1 score, a harmonic mean of precision and recall, also attests to the model’s balanced performance. It acknowledges the trade-off between precision and recall, with both metrics converging to deliver a consistently high F1 score across the dataset. The overall accuracy of the model stands at 94%, emphasizing the AFNO model’s robustness and effectiveness. This high accuracy level, combined with the individual class metrics, reveals a model that is not only adept at learning from high-resolution data but also at generalizing across diverse manifestations of chest CT findings related to COVID-19.
The balance in the average metrics, with a macro average precision of 0.85 and recall of 0.94, indicates that the model performs consistently across different classes. It manages to maintain a high recall rate even with slightly varying precision rates, an important aspect in medical diagnostics where the cost of false negatives can be particularly high. Thus, the result analysis of the confusion matrix and calculated metrics demonstrate that the AFNO model is particularly suited for the nuanced task of medical image classification. It exhibits a powerful ability to differentiate between complex patterns inherent in high-resolution CT images related to COVID-19, which is crucial for supporting radiologists and potentially reducing time-to-diagnosis.
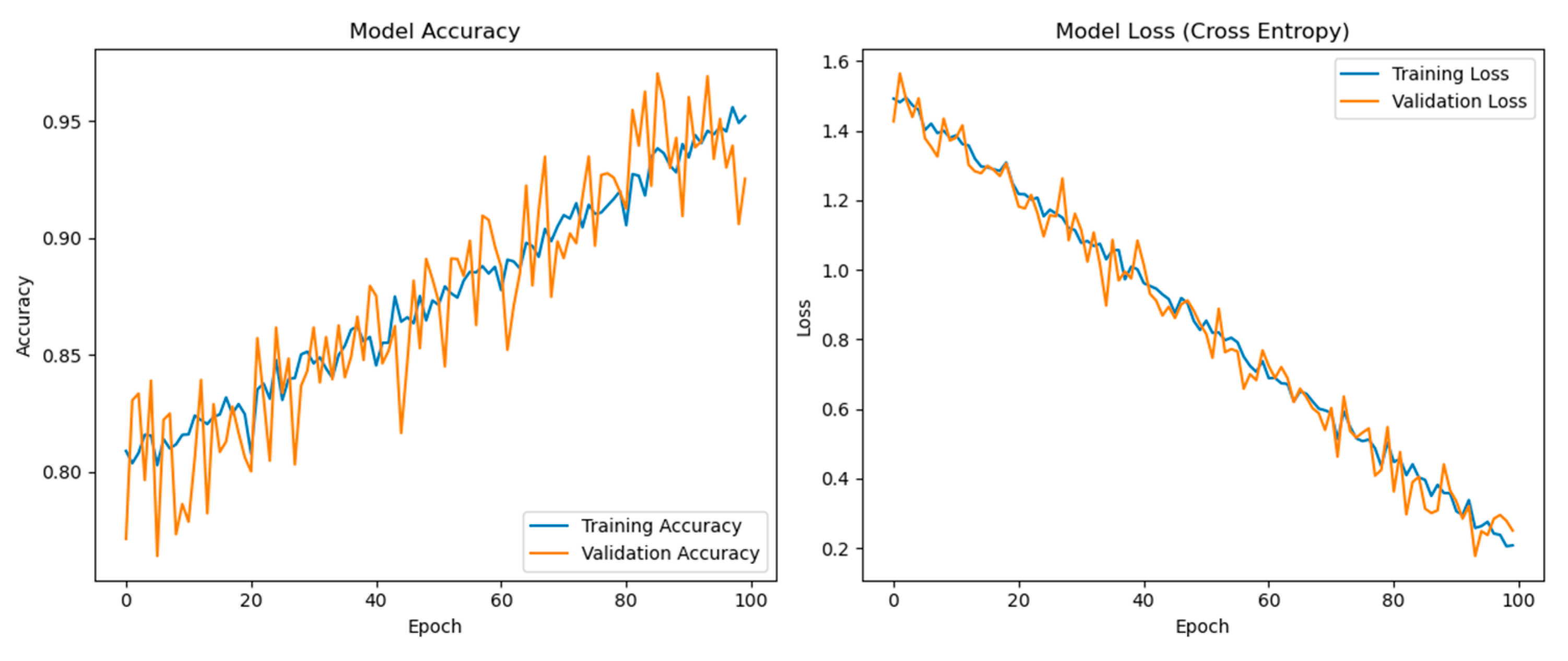
Figure 3 displays two graphs: one for the model’s accuracy and one for the model’s loss over 100 epochs of training. The left graph presents both training and validation accuracy. Both accuracies start from a high baseline and continue to improve slightly over the epochs. There is a notable variance in the validation accuracy, which might be attributed to the complexity of the validation data or a small degree of overfitting as the model learns. Despite the fluctuations, the validation accuracy remains close to the training accuracy, which indicates that the model generalizes well to new, unseen data. There is no significant divergence, which is positive as it implies the model is not overfitting the training data significantly.
The right graph shows the model’s loss during training. Loss is a measure of how well the model’s predictions match the actual labels. Lower loss values indicate better performance. Both training and validation losses decrease over time, which suggests that the model is learning and improving its predictive ability with each epoch. As with the accuracy, the training and validation loss lines are close together, which further suggests that the model is generalizing well. There is a consistent decline without any abrupt changes or increases in loss, indicating that the learning rate and model architecture are appropriate for this task. The smoothness of the curve suggests that the learning rate decay implemented every 20 epochs is working well to refine the model’s learning as it converges.
The model shows a high level of initial performance that continues to improve slightly and stabilize as training progresses, indicative of effective learning dynamics. The proximity of the training and validation lines in both graphs suggests that the model is not suffering from high variance (overfitting) or high bias (underfitting). The performance as seen in these plots indicates that the model training process is stable and that the model is learning effectively from the data. The absence of any significant gap between training and validation lines towards the later epochs suggests that the model’s architecture and hyperparameters are well-tuned to this particular dataset and task. These results are considered favorable in machine learning and are particularly beneficial in a medical context where high accuracy and reliability are crucial. All analyses were performed using Python 3.7.6, TensorFlow 2.7.0, and Keras 2.7.0. The computations were facilitated by a standard lab PC configured with a dedicated NVIDIA GeForce RTX 3070 GPU, renowned for its CUDA core architecture optimized for deep learning tasks. The system was powered by an Intel® Core™ i7-10750H CPU @ 2.60 GHz (12 CPUs), complemented by 32 GB of DDR4 RAM to ensure smooth data processing and model training. For the operating system, we utilized Windows 10 Enterprise 64-bit, providing a stable and secure environment for our research needs.
4. Discussion
In the present study, the AFNO method was applied to the HRCTCov19 dataset, which is a compilation of 181,106 high-resolution chest CT images from 395 patients. The images were processed at a resolution of 512 × 512 pixels and categorized into four classes representing various COVID-19-related pathologies. This study highlights several key findings and implications for future research and clinical practice. The AFNO model’s ability to handle high-resolution images efficiently is rooted in its innovative use of Fourier transforms for token mixing. This approach circumvents the quadratic complexity of traditional self-attention mechanisms, allowing for scalable and effective analysis of large CT image datasets. The model’s superior performance in detecting COVID-19 related abnormalities, particularly GGO and negative cases, demonstrates its potential for clinical application in rapid diagnostic scenarios. The high precision and recall rates achieved by the AFNO model across various COVID-19 manifestations underscore its diagnostic reliability. This is crucial in medical diagnostics, where accurate and timely detection can significantly impact patient management and outcomes. The AFNO model’s adaptability to different image resolutions further suggests its potential utility beyond COVID-19, in various other medical imaging tasks. Despite the promising results, the current study is limited by the HRCTCov19 dataset’s lack of diversity and structure to support a double-blind study. The dataset’s demographic and geographic homogeneity may limit the generalizability of the findings. Additionally, the existing dataset does not facilitate the anonymization required for a double-blind evaluation, where both the evaluators and clinical practitioners would be unaware of the classification outcomes. Future work will involve acquiring additional datasets from diverse populations and multiple medical institutions. This will enable a double-blind evaluation to objectively assess the model’s performance across varied populations. Such efforts are essential to validate the AFNO model’s robustness and generalizability, ensuring its efficacy in different clinical settings.
5. Conclusions
The AFNO method proposed for the strategic processing of the HRCTCov19 dataset has demonstrated its ability to efficiently handle large-scale high-resolution medical imaging data and also demonstrates potential as a viable tool in clinical settings. Its notable performance in classifying CT images into distinct COVID-19-related conditions opens up avenues for its application in rapid diagnostic scenarios, where accurate and timely classification can significantly impact patient management and outcomes. The AFNO model has proven to be a formidable tool in the automated classification of COVID-19 from high-resolution CT images. Its ability to maintain high accuracy across diverse pathologies holds promise for supporting radiologists in diagnostic workflows, potentially reducing the time-to-diagnosis and helping to manage the large influx of imaging data in pandemic situations. Future work could involve validating the model’s performance across independent datasets and may explore the integration of the AFNO model into clinical decision support systems and its performance in multi-center studies to validate its efficacy across different populations and imaging protocols.