Correlation between Biomarkers and Severity of Clinical Categories in COVID-19 Patients: A Hospital-Based Study in Arunachal Pradesh, India
Abstract
1. Introduction
Regional Significance of the Study
2. Methodology
2.1. Study Design and Population
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Ethical Considerations
- (a)
- Mild category consisting of non-severe patients with symptoms of sore throat, fever, and cough.
- (b)
- Moderate category consisting of patients with symptoms of mild category along with the symptom of breathlessness with oxygen saturation (SPO2) of 90% to <93%.
- (c)
- Severe category, which included patients with symptoms of septic shock, breathlessness, SPO2 < 90%, respiratory failure, and/or multiple organ dysfunction.
2.5. Data Extraction and Quality Assessment
2.5.1. Biochemical Analysis
2.5.2. Statistical Analysis
3. Results
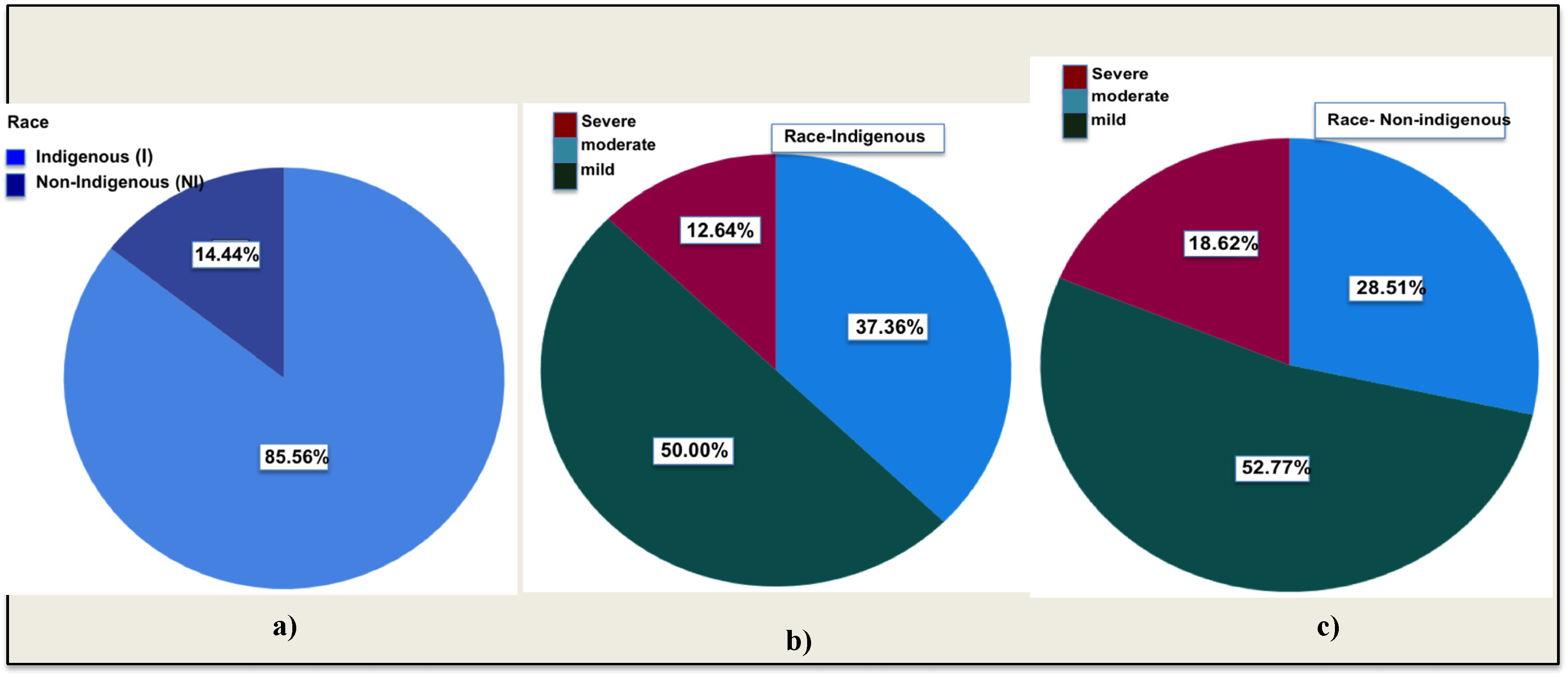
3.1. Study Population
3.2. Disease Severity
3.3. Analyzing Biomarkers: Assessing COVID-19 Severity across Indigenous Population
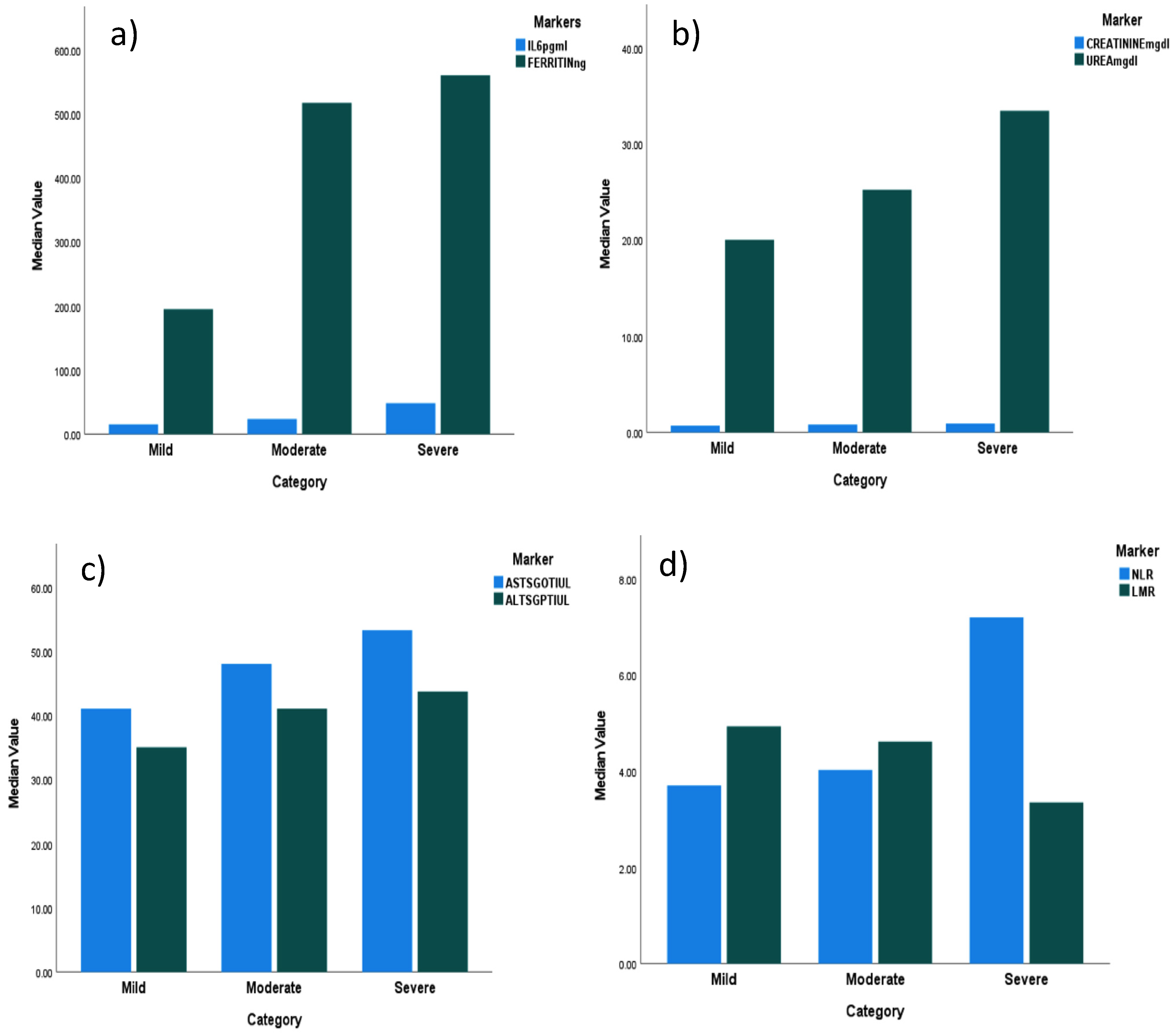
3.3.1. Inflammatory Marker Levels (Figure 2a–c)

3.3.2. Neutrophil-to-Lymphocyte Ratio (NLR) and Lymphocyte-to-Monocyte Ratio (LMR) (Figure 2d,e)
3.3.3. D-DIMER Level
3.3.4. Renal and Hepatic Function Biomarker Levels
3.3.5. Hepatic Markers
3.4. Comparative Analysis of Different Biomarker Levels across COVID-19 Clinical Outcomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci. Total Environ. 2020, 730, 138996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jaljaa, A.; Caminada, S.; Tosti, M.E.; D’Angelo, F.; Angelozzi, A.; Isonne, C.; Marchetti, G.; Mazzalai, E.; Giannini, D.; Turatto, F.; et al. Risk of SARS-CoV-2 infection in migrants and ethnic minorities compared with the general population in the European WHO region during the first year of the pandemic: A systematic review. BMC Public Health 2022, 22, 143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, D.; Sze, S.; Martin, C.A.; Nazareth, J.; Woolf, K.; Baggaley, R.F.; Hollingsworth, T.D.; Khunti, K.; Nellums, L.B.; Pareek, M. COVID-19 and ethnicity: We must seek to understand the drivers of higher transmission. BMJ 2021, 375, n2709. [Google Scholar] [CrossRef] [PubMed]
- Sze, S.; Pan, D.; Nevill, C.R.; Gray, L.J.; Martin, C.A.; Nazareth, J.; Minhas, J.S.; Divall, P.; Khunti, K.; Abrams, K.R.; et al. Ethnicity and clinical outcomes in COVID-19: A systematic review and meta-analysis. eClinicalMedicine 2020, 29, 100630. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Browne, A.J.; Varcoe, C.; Lavoie, J.; Smye, V.; Wong, S.T.; Krause, M.; Tu, D.; Godwin, O.; Khan, K.; Fridkin, A. Enhancing health care equity with Indigenous populations: Evidence-based strategies from an ethnographic study. BMC Health Serv. Res. 2016, 16, 544. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Khanijahani, A.; Iezadi, S.; Gholipour, K.; Azami-Aghdash, S.; Naghibi, D. A systematic review of racial/ethnic and socioeconomic disparities in COVID-19. Int. J. Equity Health 2021, 20, 248. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tjendra, Y.; Al Mana, A.F.; Espejo, A.P.; Akgun, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Predicting disease severity and outcome in COVID-19 patients: A review of multiple biomarkers. Arch. Pathol. Lab. Med. 2020, 144, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Ghazanfari, T.; Salehi, M.R.; Namaki, S.; Arabkheradmand, J.; Rostamian, A.; Chenary, M.R.; Ghaffarpour, S.; Ardestani, S.K.; Edalatifard, M.; Naghizadeh, M.M.; et al. Interpretation of hematological, biochemical, and immunological findings of COVID-19 disease: Biomarkers associated with severity and mortality. Iran. J. Allergy Asthma Immunol. 2021, 20, 46–66. [Google Scholar] [CrossRef] [PubMed]
- Copaescu, A.; Smibert, O.; Gibson, A.; Phillips, E.J.; Trubiano, J.A. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020, 146, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, J.; Gauba, K.; Gupta, S.; Purohit, P.; Mitra, P.; Garg, M.; Misra, S.; Sharma, P.; Banerjee, M. Interleukin-6 perpetrator of the COVID-19 cytokine storm. Indian J. Clin. Biochem. 2021, 36, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.; PVSN, K.K.; Tomo, S.; Sankanagoudar, S.; Charan, J.; Purohit, A.; Nag, V.; Bhatia, P.; Singh, K.; Dutt, N.; et al. Association of iron-related biomarkers with severity and mortality in COVID-19 patients. J. Trace Elem. Med. Biol. 2022, 74, 127075. [Google Scholar] [CrossRef] [PubMed]
- Kappert, K.; Jahić, A.; Tauber, R. Assessment of serum ferritin as a biomarker in COVID-19: Bystander or participant? Insights by comparison with other infectious and non-infectious diseases. Biomarkers 2020, 25, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Celaya-Padilla, J.M.; Villagrana-Bañuelos, K.E.; Oropeza-Valdez, J.J.; Monárrez-Espino, J.; Castañeda-Delgado, J.E.; Oostdam, A.S.H.-V.; Fernández-Ruiz, J.C.; Ochoa-González, F.; Borrego, J.C.; Enciso-Moreno, J.A.; et al. Kynurenine and hemoglobin as sex-specific variables in COVID-19 patients: A machine learning and genetic algorithms approach. Diagnostics 2021, 11, 2197. [Google Scholar] [CrossRef] [PubMed]
- Stringer, D.; Braude, P.; Myint, P.K.; Evans, L.; Collins, J.T.; Verduri, A.; Quinn, T.J.; Vilches-Moraga, A.; Stechman, M.J.; Pearce, L.; et al. The role of C-reactive protein as a prognostic marker in COVID-19. Int. J. Epidemiol. 2021, 50, 420–429. [Google Scholar] [CrossRef]
- Yao, Y.; Cao, J.; Wang, Q.; Shi, Q.; Liu, K.; Luo, Z.; Chen, X.; Chen, S.; Yu, K.; Huang, Z.; et al. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: A case control study. J. Intensive Care 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Battaglini, D.; Lopes-Pacheco, M.; Castro-Faria-Neto, H.C.; Pelosi, P.; Rocco, P.R. Laboratory biomarkers for diagnosis and prognosis in COVID-19. Front. Immunol. 2022, 13, 857573. [Google Scholar] [CrossRef]
- Qin, C.; Wei, Y.; Lyu, X.; Zhao, B.; Feng, Y.; Li, T.; Cao, H.; Yang, X.; Zhou, X.; Wang, W.; et al. High aspartate aminotransferase to alanine aminotransferase ratio on admission as risk factor for poor prognosis in COVID-19 patients. Sci. Rep. 2020, 10, 16496. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Nayak, S.; Tripathy, S.; Bhuniya, S.; Mangaraj, M.; Ramadass, B.; Sahu, S.; Bandyopadhyay, D.; Dash, P.; Saharia, G.K. Analysis of Biochemical and Inflammatory Markers for Predicting COVID-19 Severity: Insights from a Tertiary Healthcare Institution of Eastern India. Cureus 2023, 15, e33893. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069, Erratum in JAMA 2021, 325, 1113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Henry, B.M.; de Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zeng, F.; Huang, Y.; Guo, Y.; Yin, M.; Chen, X.; Xiao, L.; Deng, G. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int. J. Infect. Dis. 2020, 96, 467–474. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sarraf, S.; Singapurwala, M.; Jain, H.; Singh, R.; Julka, A. Role of Inflammatory Markers in Predicting Severity in COVID-19 Patients at Tertiary Care Hospital, Ujjain (M.P.). J. Pulmonol. Respir. Res. 2023, 7, 4–9. [Google Scholar]
- Pujani, M.; Raychaudhuri, S.; Singh, M.; Kaur, H.; Agarwal, S.; Jain, M.; Chandoke, R.K.; Singh, K.; Sidam, D.; Chauhan, V. An analysis of hematological, coagulation and biochemical markers in COVID-19 disease and their association with clinical severity and mortality: An Indian outlook. Am. J. Blood Res. 2021, 11, 580. [Google Scholar] [PubMed]
- Sen, A.; Nigam, A.; Vachher, M. Role of polypeptide inflammatory biomarkers in the diagnosis and monitoring of COVID-19. Int. J. Pept. Res. Ther. 2022, 28, 59. [Google Scholar] [CrossRef] [PubMed]
- Rowaiye, A.B.; Okpalefe, O.A.; Onuh Adejoke, O.; Ogidigo, J.O.; Oladipo, O.H.; Ogu, A.C.; Oli, A.N.; Olofinase, S.; Onyekwere, O.; Abubakar, A.R.; et al. Attenuating the effects of novel COVID-19 (SARS-CoV-2) infection-induced cytokine storm and the implications. J. Inflamm. Res. 2021, 14, 1487–1510. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.; Rankawat, G.; Singh, A.; Wadhwani, D.; Patel, B. Evaluation of interleukin-6 and its association with the severity of disease in COVID-19 patients. Indian J. Med. Spec. 2020, 11, 132. [Google Scholar]
- Gill, G.; Devra, A.G. Role of interleukin-6 levels in predicting COVID severity: A single-center experience. Indian J. Med. Spec. 2023, 14, 178–179. [Google Scholar]
- Tsegay, Y.G.; Bitew, M.; Workneh, T.; Atlaw, A.; Aragaw, M.; Gemechu, M.; Brhane, N. The level of liver and renal function biomarker abnormalities among hospitalized COVID-19 patients in Ethiopia. medRxiv 2022, 22271010. [Google Scholar] [CrossRef]
- Singh, A.; Bhadani, P.P.; Surabhi Sinha, R.; Bharti, S.; Kumar, T.; Nigam, J.S. Significance of immune-inflammatory markers in predicting clinical outcome of COVID-19 patients. Indian J. Pathol. Microbiol. 2023, 66, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Shobeiri, P.; Kulasinghe, A.; Rezaei, N. Novel Systemic Inflammation Markers to Predict COVID-19 Prognosis. Front. Immunol. 2021, 12, 741061. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 20, 269–270. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gupta, D.; Jain, A.; Chauhan, M.; Dewan, S. Inflammatory Markers as Early Predictors of Disease Severity in COVID-19 Patients Admitted to Intensive Care Units: A Retrospective Observational Analysis. Indian J. Crit. Care Med. 2022, 26, 482–486. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sakthivadivel, V.; Bohra, G.K.; Maithilikarpagaselvi, N.; Khichar, S.; Meena, M.; Palanisamy, N.; Gaur, A.; Garg, M.K. Association of Inflammatory Markers with COVID-19 Outcome among Hospitalized Patients: Experience from a Tertiary Healthcare Center in Western India. Maedica 2021, 16, 620–627. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Côté, A.; Ternacle, J.; Pibarot, P. Early prediction of the risk of severe coronavirus disease 2019: A key step in therapeutic decision making. eBioMedicine 2020, 59, 102948. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Indigenous | Non-Indigenous | p-Value | ||
|---|---|---|---|---|---|
| N | Median | N | Median | ||
| Age (years) | 987 | 40 | 127 | 50 | <0.001 |
| Urea (mg/dL) | 896 | 24.2 | 80 | 26.45 | 0.225 |
| Creatinine (mg/dL) | 895 | 0.7 | 79 | 0.8 | 0.052 |
| Sodium (mEq/L) | 823 | 137.4 | 75 | 136.5 | 0.036 |
| Pottasium (mEq/L) | 823 | 3.9 | 75 | <0.001 | |
| Chloride (mEq/L) | 821 | 100.2 | 75 | 99.2 | 0.007 |
| Total bilirubin (mg/dL) | 898 | 0.7 | 80 | 0.8 | 0.057 |
| Direct bilirubin (mg/dL) | 896 | 0.4 | 80 | 0.4 | 0.329 |
| AST/SGOT (U/L) | 895 | 46 | 80 | 42.55 | 0.714 |
| ALT/SGPT (U/L) | 893 | 39.8 | 80 | 40 | 0.724 |
| TP Total Protein (g/dL) | 894 | 6.2 | 80 | 6.3 | 0.475 |
| Albumin (g/dL) | 897 | 3.7 | 80 | 3.8 | 0.264 |
| Globulin (g/dL) | 892 | 2.5 | 80 | 2.5 | 0.594 |
| CRP (ng/mL) | 384 | 49,580.58 | 27 | 42,708.6 | 0.254 |
| Ferritin (ng/mL) | 430 | 363.85 | 30 | 555.45 | 0.542 |
| IL6 (pg/mL) | 413 | 22.68 | 28 | 27.665 | 0.359 |
| NLR | 900 | 4.18 | 83 | 4.67 | 0.456 |
| LMR | 900 | 4.59 | 82 | 4.87 | 0.962 |
| PLR | 889 | 0.09 | 81 | 0.08 | 0.61 |
| Study Target Group (Indigenous Population from Arunachal Pradesh, India) | Study Group (Non-Indigenous Population from Eastern India) by Suchitra Kumari et al. [18]. | p-Value for Spearman Rank Correlation Test | r-Value | ||||
|---|---|---|---|---|---|---|---|
| Parameters (n-592) | Median (IQR) | Parameters (n-7395) | Median (IQR) | ||||
| Sex | Male | 348 (58.78%) | Sex | Male | 4656 (62.96%) | <0.0001 | 0.9876 |
| Female | Female | 2739 (37.04%) | |||||
| Age (years) | 41 (31–54) | Age | 48 (32–60) | ||||
| Urea (mg/dL) | 23.00 (17.10–32.70) | Urea (mg/dL) | 25 (18–40) | ||||
| Creatinine (mg/dL) | 0.8 (0.7–1.0) | Creatinine (mg/dL) | 0.9 (0.7–1.2) | ||||
| Sodium (mEq/L) | 137.2 (135.2–139.0) | Sodium (mEq/L) | 135 (132–137) | ||||
| Pottasium (mEq/L) | 3.9 (3.6–4.3) | Pottasium (mEq/L) | 4.23 (3.89–4.61) | ||||
| Chloride (mEq/L) | 100.4 (98.2–102.0) | Chloride (mEq/L) | 100 (96–103) | ||||
| Total bilirubin (mg/dL) | 0.7 (0.6–0.9) | Total bilirubin (mg/dL) | 0.5 (0.3–0.7) | ||||
| Direct bilirubin (mg/dL) | 0.4 (0.3–0.6) | Direct bilirubin (mg/dL) | 0.17 (0.1–0.3) | ||||
| AST/SGOT (U/L) | 46.25 (43–49.8) | AST/SGOT (U/L) | 36 (24–64) | ||||
| ALT/SGPT (U/L) | 40 (38–44) | ALT/SGPT (U/L) | 32 (19–57) | ||||
| TP Total Protein (g/dL) | 6.4 (6.3–6.5) | TP Total Protein (g/dL) | 7 (6.4–7.6) | ||||
| Albumin (g/dL) | 3.8 (3.4–4.0) | Albumin (g/dL) | 3.6 (3.1–4.1) | ||||
| Globulin (g/dL) | 2.3 (1.9–2.7) | Globulin (g/dL) | 3.4 (3–3.8) | ||||
| CRP (ng/mL) | 44.95 (39.2–50.8) | CRP (ng/mL) | 55.15 (14–138.9) | ||||
| Ferritin (ng/mL) | 366.0 (306.7–447.5) | Ferritin (ng/mL) | 371 (128.7–955.6) | ||||
| IL6 (pg/mL) | 20.65 (16.36–24.7) | IL6 (pg/mL) | 19.8 (5.9–60) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nobin, H.; Paley, T.; Anu, R.; Yami, H.; Tago, N.; Saikia, N.J.; Nyodu, R. Correlation between Biomarkers and Severity of Clinical Categories in COVID-19 Patients: A Hospital-Based Study in Arunachal Pradesh, India. COVID 2024, 4, 1157-1171. https://doi.org/10.3390/covid4080081
Nobin H, Paley T, Anu R, Yami H, Tago N, Saikia NJ, Nyodu R. Correlation between Biomarkers and Severity of Clinical Categories in COVID-19 Patients: A Hospital-Based Study in Arunachal Pradesh, India. COVID. 2024; 4(8):1157-1171. https://doi.org/10.3390/covid4080081
Chicago/Turabian StyleNobin, Hage, Tamar Paley, Rubu Anu, Hibu Yami, Nani Tago, Naba Jyoti Saikia, and Rajni Nyodu. 2024. "Correlation between Biomarkers and Severity of Clinical Categories in COVID-19 Patients: A Hospital-Based Study in Arunachal Pradesh, India" COVID 4, no. 8: 1157-1171. https://doi.org/10.3390/covid4080081
APA StyleNobin, H., Paley, T., Anu, R., Yami, H., Tago, N., Saikia, N. J., & Nyodu, R. (2024). Correlation between Biomarkers and Severity of Clinical Categories in COVID-19 Patients: A Hospital-Based Study in Arunachal Pradesh, India. COVID, 4(8), 1157-1171. https://doi.org/10.3390/covid4080081





