Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Variant Annotation
2.2. SNP Analysis
2.3. Genomic Sequence Analysis and Epitopes Evaluation
2.4. Identification of Lineages and Phylogenetic Analyses
3. Results
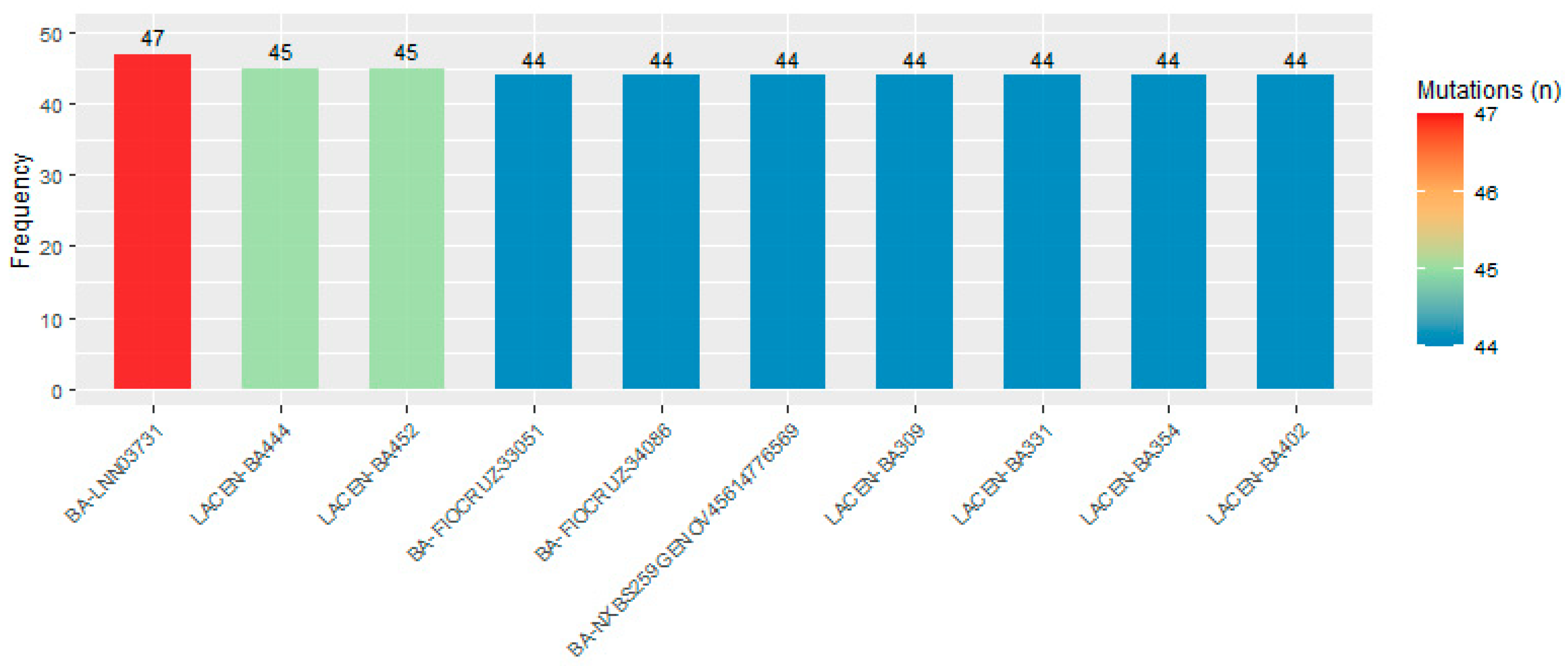
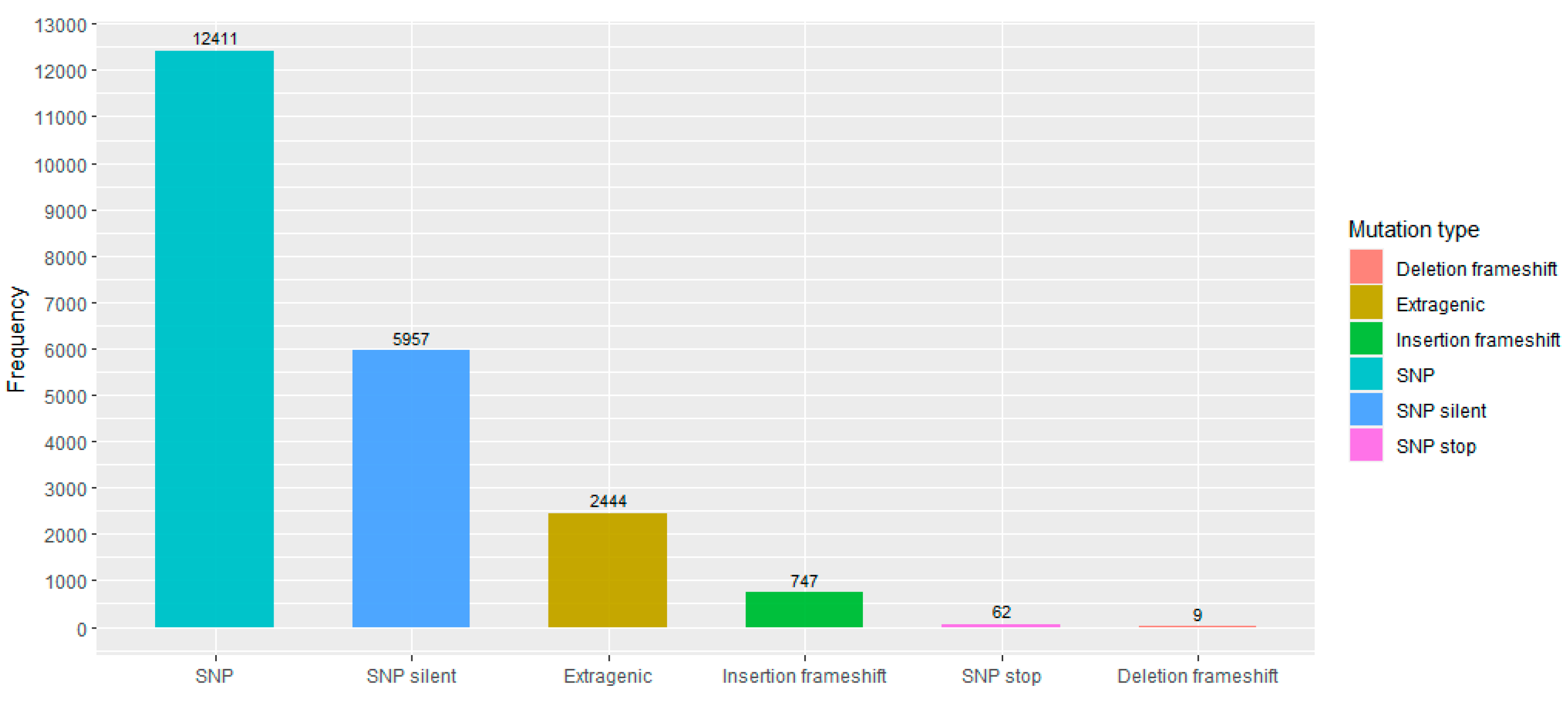
3.1. Variant Annotation
3.2. Structural and Functional Analysis
3.3. Epitope Evaluation
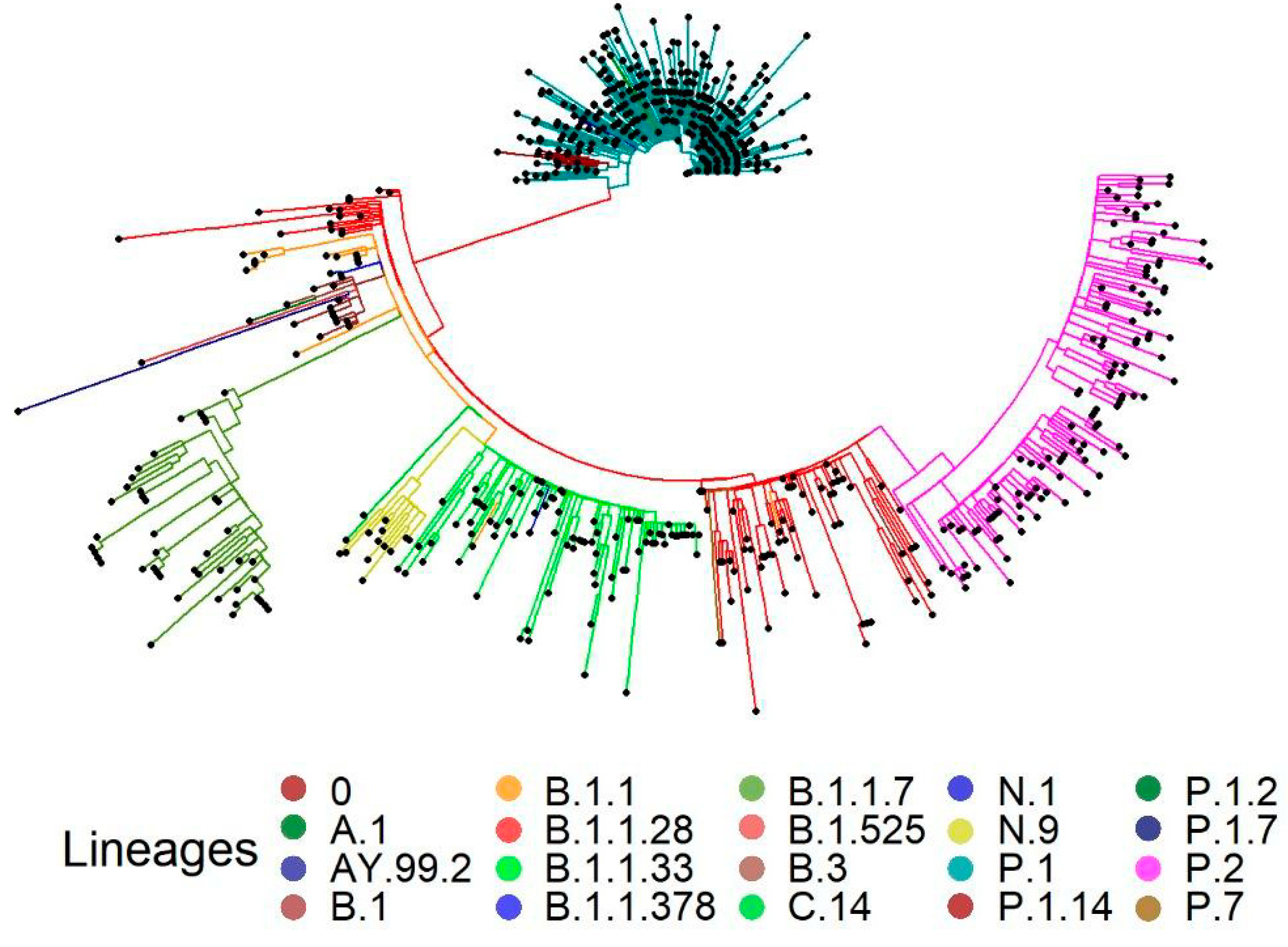
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rafiq, D.; Batool, A.; Bazaz, M.A. Three months of COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020, 30, e2113. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ma, X.; Bai, L.; Wang, M.; Deng, W.; Ning, N. A systematic review of etiology, epidemiology, clinical manifestations, image findings, and medication of 2019 Corona Virus Disease-19 in Wuhan, China. Medicine 2020, 99, e22688. [Google Scholar] [CrossRef]
- Khalil, O.A.K.; Khalil, S.S. SARS-CoV-2: Taxonomia, origem e constituição. Rev. Med. 2020, 99, 473–479. [Google Scholar] [CrossRef]
- Michelon, C.M. Main SARS-CoV-2 variants notified in Brazil. Rev. Bras. Anál. Clín. 2021, 53, 109–116. [Google Scholar] [CrossRef]
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; Dupleiss, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Dorp, L.V.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.S.; Boshier, F.A.T.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020, 83, 104351. [Google Scholar] [CrossRef]
- Morales, A.J.R.; Ramon, G.J.B.; Rabaan, A.A.; Sah, R.; Dhama, K.; Mondolfi, A.P.; Pagliano, P.; Esposito, S. Genomic Epidemiology and its importance in the study of the COVID-19 pandemic. Infez. Med. 2020, 28, 139–142. [Google Scholar]
- Zeiser, F.A.; Donida, B.; da Costa, C.A.; Ramos, G.O.; Scherer, J.N.; Barcellos, N.T.; Alegretti, A.P.; Ikeda, M.L.R.; Müller, A.P.W.C.; Bohn, H.C.; et al. First and second COVID-19 waves in Brazil: A cross-sectional study of patients’ characteristics related to hospitalization and in-hospital mortality. Lancet Reg. Health 2022, 6, 100–107. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Mercatteli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinform 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chiara, M.; Horner, D.S.; Gissi, C.; Pesole, G. Comparative Genomics Reveals Early Emergence and Biased Spatiotemporal Distribution of SARS-CoV-2. Mol. Biol. Evol. 2021, 38, 2547–2565. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.V.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Kumar, S.; Bansal, K. Cross-sectional genomic perspective of epidemic waves of SARS-CoV-2: A pan India study. Virus Res. 2022, 308, 198642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Kang, J.Y.; Chen, S.; He, Y.; Han, B.; Liu, M.F.; Lu, L.; Li, L.; Yi, Z.; et al. Potential transmission chains of variant B.1.1.7 and co-mutations of SARS-CoV-2. Cell Discov. 2021, 7, 44. [Google Scholar] [CrossRef]
- Kim, J.S.; Jang, J.H.; Kim, J.M.; Chung, Y.S.; Yoo, C.K.; Han, M.G. Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2. Osong Public Health Res. Perspect. 2020, 11, 101–111. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinform 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, P.; Niyogi, S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020, 148, e262. [Google Scholar] [CrossRef] [PubMed]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Goes, L.R.; Siqueira, J.D.; Garrido, M.M.; Alves, B.M.; Pereira, A.C.P.M.; Cicala, C.; Arthos, J.; Viola, J.P.B.; Soares, M.A. INCA COVID-19 Task Force. New infections by SARS-CoV-2 variants of concern after natural infections and post-vaccination in Rio de Janeiro, Brazil. Infect. Genet. Evol. 2021, 94, 104998. [Google Scholar] [CrossRef]
- Mahilkar, S.; Agrawal, S.; Chaudhary, S.; Parikh, S.; Sonkar, S.C.; Verma, D.K.; Chitalia, V.; Mehta, D.; Koner, B.C.; Vijay, N.; et al. SARS-CoV-2 variants: Impact on biological and clinical outcome. Front. Med. 2022, 9, 995960. [Google Scholar] [CrossRef]
- Al-Zyoud, W.; Haddad, H. Dynamics prediction of emerging notable spike protein mutations in SARS-CoV-2 implies a need for updated vaccines. Biochime 2021, 191, 91–103. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef]
- Pretti, M.A.M.; Galvani, R.G.; Vieira, G.F.; Bonomo, A.; Bonamino, M.H.; Boroni, M. Class I HLA Allele Predicted Restricted Antigenic Coverages for Spike and Nucleocapsid Proteins Are Associated With Deaths Related to COVID-19. Front. Immunol. 2020, 11, 565730. [Google Scholar] [CrossRef]
- Augusto, D.G.; Yusufali, T.; Peyser, N.D.; Butcher, X.; Marcus, G.M.; Olgin, J.E.; Pletcher, M.J.; Maiers, M.; Hollenbach, J.A. HLA-B*15: 01 is associated with asymptomatic SARS-CoV-2 infection. medRxiv 2021, 2021.05.13.21257065. [Google Scholar] [CrossRef]
- Avetyan, D.; Hakobyan, S.; Nikoghosyan, M.; Ghukasyan, L.; Khachatryan, G.; Sirunyan, T.; Muradyan, N.; Zakharyan, R.; Chavushyan, A.; Hayrapetyan, V.; et al. Molecular Analysis of SARS-CoV-2 Lineages in Armenia. Viruses 2022, 14, 1074–1087. [Google Scholar] [CrossRef]
- Gordiano, Y.F.; Anjos, M.B.; Nascimento, W.M.; Filho, R.A.A.B. A importância da vigilância genômica no contexto da pandemia de COVID-19 para o Estado do Amazonas. Conjecturas 2021, 21, 562–572. [Google Scholar] [CrossRef]
- Lao, B.; Lima, R.C.; Karnikowski, M.G.O.; Silva, I.C.R.; Azevedo, H.L.G. Variantes de atenção e/ou preocupação por sequenciamento genômico do COVID-19 no Brasil. REVISA 2021, 10, 783–787. [Google Scholar]
- Freitas, A.R.R.; Giovanetti, M.; Alcantara, L.C.J. Emerging variants of SARS-CoV-2 and its public health implications. Inter. Am. J. Med. Health 2021, 4. [Google Scholar] [CrossRef]
- Aleem, A.; Akbar Samad, A.B.; Vaqar, S. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; NCBI: Bethesda, ML, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK570580/ (accessed on 1 March 2023).


| HLA Loci | Total | Alleles |
|---|---|---|
| HLA-A | 2662 | *02:01, *02:06, *11:01, *24:02, *26:01, *31:01, *33:03. |
| HLA-B | 3266 | *03:01, *04:01, *06:01, *07:02, *11:01, *15:01, *16:02, *16:06, *20:03, *22:01, *35:01, *40:02, *40:06, *44:03, *46:01, *51:01, *52:01, *54:01. |
| HLA-C | 3599 | *01:02, *03:03, *03:04, *07:02, *08:01, *12:02, *14:02, *14:03. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, G.; Matias, G.; Chrisóstomo, L.; da Costa-Neto, J.; Sampaio, J.; Silva, A.; Cansanção, I. Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity. COVID 2023, 3, 655-663. https://doi.org/10.3390/covid3050047
Andrade G, Matias G, Chrisóstomo L, da Costa-Neto J, Sampaio J, Silva A, Cansanção I. Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity. COVID. 2023; 3(5):655-663. https://doi.org/10.3390/covid3050047
Chicago/Turabian StyleAndrade, Gabriela, Guilherme Matias, Lara Chrisóstomo, João da Costa-Neto, Juan Sampaio, Arthur Silva, and Isaac Cansanção. 2023. "Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity" COVID 3, no. 5: 655-663. https://doi.org/10.3390/covid3050047
APA StyleAndrade, G., Matias, G., Chrisóstomo, L., da Costa-Neto, J., Sampaio, J., Silva, A., & Cansanção, I. (2023). Genetic Analysis and Epitope Prediction of SARS-CoV-2 Genome in Bahia, Brazil: An In Silico Analysis of First and Second Wave Genomics Diversity. COVID, 3(5), 655-663. https://doi.org/10.3390/covid3050047











