Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can spread to the environment through several routes and persist for a more extended period. Therefore, we reviewed pertinent literature to understand the transmission dynamics of SARS-CoV-2 and genomic epidemiology of emerging variants of concern (VOCs) in the environment, their inactivation strategies, and the impact of COVID-19 on the ecosystem. The fallouts of the reviewed studies indicate that SARS-CoV-2 transmits through air and fomite, contaminated surfaces, biomedical wastes, and stool, which contaminates the environment through wastewater. As a result, multiple VOCs of SARS-CoV-2 were circulating in the environment. Genomic epidemiology revealed that the most prevalent VOC was Delta (B.1.617.2; 44.24%), followed by Omicron (B.1.1.529; 43.33%), in the environment. Phylogenetic analysis showed that environmental strains are clustered with a likeness of the human strains of the same or nearby countries, emphasizing the significance of continued environmental surveillance to track the emergence of the new variant. Thus, we should reduce viral dispersion in the environment through rapid and appropriate disinfection strategies. Moreover, the increased production and use of macro and microfiber plastic products should be brought under strict legislation with integrated waste management to control the unrelenting propagation of viral RNA. Finally, a comprehensive understanding of the environmental transmission pathways of SARS-CoV-2 is crucial for forecasting outbreak severity in the community, allowing us to prepare with the correct tools to control any impending pandemic. We recommend wastewater-based SARS-CoV-2 surveillance and air particulates to track the emerging VOCs of SARS-CoV-2 spread in the environment.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected human health regardless of geographical boundaries, with the first cases reported at the end of December 2019 in Wuhan, Hubei Province, China. Even with vaccination doses of approximately 3.4 billion worldwide, approximately 186.8 million confirmed cases and 4.0 million deaths were reported by the second week of July 2021 [1,2]. Infection is characterized by the presence of high fever with coughing, breathing difficulty, and fatigue with evidence of acute respiratory distress, which may cause the death of an individual. Initial evidence on airborne transmission of the virus through air fomites of ≤5 μm-sized particles suggests maintaining a 1 m social distance policy [3]. Furthermore, a list of factors related to the environment and human behaviors are considered responsible for transmitting SARS-CoV-2 between individuals. Although there is rare evidence of the potent virus in feces and contact with the coronavirus disease 2019 (COVID-19) via aerosolization from feces, transmission via contact with a contaminated surface to the mucous membrane of the mouth, nose, and eyes was confirmed [3]. Consequently, environmental factors such as temperature, humidity, sustainability of fomites, aeration, and filtering systems in households, hospitals, and other mass gathering places may influence the viral spread that we need to dig out.
Transmission of the virus at the community level is mainly responsible for converting the outbreak into a pandemic. Though it has been proven that the virus can be transmitted directly through an infected person’s cough, oral and nasal secretion, and interceded contaminated droplets [4], the indirect route of virus transmission is still unreported or poorly understood [5]. Therefore, the existence of the virus in environmental samples indicates that the virus is present in the community. In spite of having a higher reproduction number [6] and low incubation period [7], one study has reported the non-infectiousness of SARS-CoV-2 RNA recovered from wastewater to humans [8]. However, further studies are required to target the survivability of the virus in water and wastewater under different environmental conditions to detect whether the contaminated wastewater is an emerging concern for transmission of SARS-CoV-2 to humans [9].
Earlier, SARS-CoV-1 was transmitted from feces to air and the environment [10]. The “virus-laden droplets” that occur through bathroom ventilation in the room can be a source of airborne spread [11]. Similarly, the SARS-CoV-2 droplets may spread through the wastewater sanitation arrangement of a building’s different floors and air by cross-contamination [12]. Further, one study has already reported that the environment is a potential medium of transmission of SARS-CoV-2 after detecting positive samples from the toilet bowl, sink, and swab samples of air exhaust outlets of COVID-19 patient rooms [13].
Overall, the COVID-19 pandemic has a detrimental effect on public health caused by environmental risk factors [1,14,15]. So, the safe management of domestic and household waste could be critical during the ongoing COVID-19 pandemic. Further, there is a knowledge gap about other possible environmental transmission routes, such as air fomites and surface-level contamination, which have come across due to the steady increase in infection rates. Since SARS-CoV-2 is considered highly infectious among the coronavirus family, it is essential to unveil the pattern and possible environmental transmission pathways and inactivation.
Therefore, we conducted this review to understand the transmission pathways, persistence, and inactivation of SARS-CoV-2 in environmental contact surfaces and the impact of plastic waste pollution globally. We have highlighted sewage tracking to surveil COVID-19 in this pandemic situation and identify the credibility of the existing wastewater-based epidemiology (WBE) for SARS-CoV-2 surveillance and monitoring in different geographical regions.
2. Methodology
We screened published literature containing the following information: 1. SARS-CoV-2 transmission pathways from humans to the environment; 2. persistence of SARS-CoV-2 in different environments and surfaces; 3. the inactivation strategies of SARS-CoV-2; and 4. global plastic waste pollution due to SARS-CoV-2. We used the Google scholar, PubMed, Scopus, and the Web of Science databases accessed through Hinary (https://www.who.int/hinari/en/; accessed on 25 September 2021). We developed Boolean words under descriptive, outcome, and population terms (Table 1) for searching the literature.

Table 1.
Boolean operator to search databases.
For the emerging variant epidemiology, we retrieved all the complete genome sequences of environmental strains of different variants of SARS-CoV-2 from the GISAID on 22 September 2021. We calculated the percentage of each emerging variant as the number of each variant’s sequences over the total number of sequences. We illustrated the possible cyclic pathway of risk of transmission of the viral particles from infected human to the environment and genomic surveillance in Figure 1. We graphically showed the temporal distribution of emerging variants from environmental samples using MS-Excel 2015. We created the spatial distribution map for emerging variants of concerns (VOCs) using ArcGIS software [16].
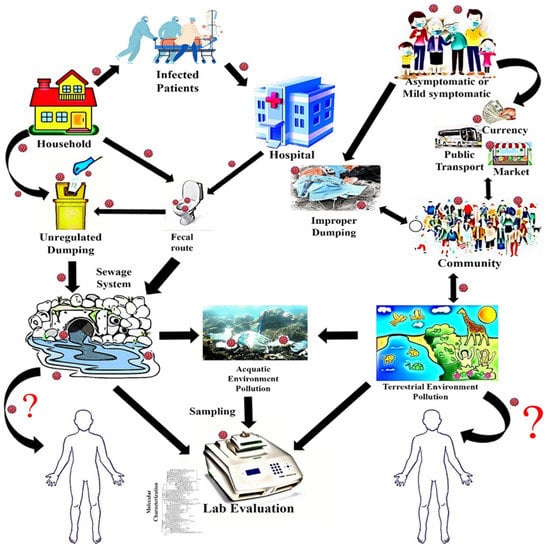
Figure 1.
Possible cyclic risk pathway and source of genomic surveillance.
We selected genomes (Supplementary File S1) for phylogenetic analysis because we verified their grouping quality for additional investigation in the GISAID dataset. In contrast, genome arrangements have >5% Network Neurobehavioral Scale (NNNS), and <29,000 nt were avoided considering poor-quality successions. The reference SARS-CoV-2 Wuhan genome (NC_045512) was utilized. Succession Dataset developer (SEDA; https://www.sing-group.org/seda/, accessed on 25 March 2022) was used to eliminate all interior stop codon containing arrangements. Moreover, we utilized numerous grouping arrangement program (MAFFT) order lines (https://mafft.cbrc.jp/arrangement/programming/, accessed on 25 March 2022) [14] to adjust all recovered genome successions over the reference succession. We used the MEGA 7 apparatus for the phylogenetic examination as portrayed by [6,15]. We constructed the phylogenetic trees using the neighbor-joining technique [17] and the Kimura–Nei method [18] for all the detailed developmental relationship examinations where the bootstrap test (1000 reproduces) is displayed close to the branches. During the essential decision making, we pondered declaring time, the geographical region close by human–environmental interface reports, unpredictable model combination dates, close by, and the significance between every plan and their uncovered pathogenic power. This phylogenetic tree addressed the developmental relationship of and delegates emerging variants of SARS-CoV-2 from both humans and the environment. Its fundamental design was to clarify the human–environment interfacial transformative relationship alongside the transmission dynamic of SARS-CoV-2.
3. Risk of Transmission Dynamics and Persistence of SARS-CoV-2 in the Environment
3.1. Risk of Transmission of SARS-CoV-2 through Stool
Human pathogenic viruses pass into the environment through fecal, urogenital, and oropharyngeal secretions, blood, and sweat (Figure 1) [19]. The overall SARS-CoV-2 shedding period varies from 2 to 10 days for symptomatic cases, extending to 20 days for immunocompromised patients [20,21]. Another study detected the RNA in COVID-19 patients’ stool after 10–30 days of onset of illness [22]. Liu et al. [23] detected SARS-CoV-2 RNA for up to 5 days in the urine of COVID-19 patients. The median lifespan of SARS-CoV-2 in stool specimens of infected patients was 22 days (17–31 days) [24], longer than that of SARS-CoV-1 (≤4 days) [21,25], higher than that of respiratory droplets (18 days, 13–29 days) and serum samples (16 days, 11–21 days) [24], which is alarming because in experimental setup, viral RNA isolated from a COVID-19-affected patient’s stool sample has demonstrated the infection ability of the African green monkey kidney cell (Vero) [26].
Evidence of fecal contamination was first identified in Macau, China [27], and simultaneously, several other studies in China detected viral RNA in feces (Table 2). Almost 66.67% (6/9) of SARS-CoV-2-positive fecal samples has been observed in Munich, Germany [28]. In Canada and England, the alpha variant was identified from sewage samples, which implies the possible transmission of the virus through fecal shedding and water contamination [29,30].
Although the current knowledge is aggregated based on a few studies, the findings are significant because of environmental contamination by releasing contaminated feces into affluent and onsite hygiene systems and open defecation [31]. The recent trend of the multifaceted SARS-CoV-2 transmission favors the probable fecal shedding and spread of the SARS-CoV-2 virus through water [32,33,34]. In addition, most of the studies identified a high concentration of SARS-CoV-2 RNA in fecal samples in infected patients, regardless of the infectivity of the virus. It is essential to conduct further research in environmental and laboratory setups to confirm the hypothesis of viral spread through fecal shedding [13].

Table 2.
Detection of SARS-CoV-2 RNA in stool.
Table 2.
Detection of SARS-CoV-2 RNA in stool.
| Country | Location | Detection Time | Detection Methods | PCR Target Regions | Positive Rate n/N (%) | Reference |
|---|---|---|---|---|---|---|
| China | Hubei, Shandong, and Beijing | 1 January to 17 February 2020 | rRT-PCR | Open reading frame 1ab gene | 44/153 (29%) | [35] |
| China | Jinhua | 27 January to 10 February 2020 | RT-PCR | Not found | 5/14 (35.7%) | [36] |
| China | Shanghai and Qingdao | Early February | RT-PCR | 1ab gene and nucleocapsid protein gene | 5/10 (50%) | [22] |
| China | Zhuhai | 1 to 14 February 2020 | rRT-PCR | Not found | 39/73 (53.4%) | [37] |
| China | Zhoushan | RT-PCR | N gene | 1/1 (100%) | [32] | |
| Singapore | Singapore City | January–February, 2020 | RT-PCR | Not found | 5/18 (27.8%) | [38] |
| China | Shanghai | 20 January to 10 February 2020 | RT-PCR | Not found | 11/66 (16.7%) | [39] |
| China | Guangdong | February 2020 | RT-PCR | N Gene | 5/6 (83.3%) | [40] |
| Singapore | Kallang | 13 February 2020 | rRT-PCR | N gene | 1/1 (100%) | [41] |
| China | Sichuan | January | RT-PCR | Not found | 8/9 (88.9%) | [42] |
| China | Macau | 21 January to 16 February 2020 | qRT-PCR | Not found | 10/10 (100%) | [27] |
| China | Zhuhai | 16 January to 15 March 2020 | RT-PCR | RdRp gene, N gene, E gene | 41/74 (55%) | [43] |
| China | Shandong | 17 January to 6 March 2020 | RT-PCR | Not found | 3/3 (100%) | [44] |
| China | Tianjin | 3 to 17 February 2020 | RT-PCR | N gene | 3/3 (100% | [45] |
| Korea | Seoul | April 2020 | RT-PCR | RdRp gene | 2/46 (4.34%) | [46] |
| China | Wuhan | 9 to 20 February 2020 | RT-PCR | Not found | 28/42 (66.67%) | [47] |
| USA | RT-PCR | S gene, N gene | 2/7 (28.57%) | [48] | ||
| USA | Illinois | Not found | RT-PCR | S gene | 2/2 (100%) | [49] |
| Germany | Munich | 23 January 2020 | RT–PCR | E gene | 6/9 (66.67) | [28] |
| France | Paris | Not found | RT–PCR | E gene | 2/5 (40%) | [50] |
| South Korea | Chungbuk | 25 February–5 March 2020 | qRT-PCR | SARS-CoV-2 RNA | 100% | [46] |
| China | Wuhan | 27 January–7 February 2020 | qRT-PCR | SARS-CoV-2 RNA | 12/28 | [47] |
| USA | Massachusetts | Not found | qRT-PCR | N1, N2, E, RdRp gene | 35/60 | [51] |
| Brazil | Jan to Jul 2020 | qRT-PCR | NSP3 segment and ORF1/2 junction region | 10/121 (8.3%) | [52] |
rRT-PCR: real-time reverse transcriptase-polymerase chain reaction; RT-PCR: reverse transcriptase-polymerase chain reaction.
3.2. Risk of Transmission Dynamics and Persistence of SARS-CoV-2 in Sewage
Long before the emergence of SARS-CoV-2, other coronaviruses were detected in the effluent of sewage treatment plants. Evidence suggests that the survival of different coronavirus strains depends on the nature and type of wastewater and temperature variation. Human coronaviruses (HCoVs) are inactivated rapidly in water, i.e., HCoV-229E survived only for seven days at 23 °C in water [53]. Temperature is a crucial factor in the persistence of the virus. HCoV-229E survives with a wide fluctuation of temperature variations as low as 4 °C to as high as 25 °C for 21 days. However, viral persistence also varied among different strains, such as transmissible gastroenteritis virus (TGEV) for 35 days on pasteurized sewage at 40 °C. Its persistence decreased to 21 days while the temperature increased to 25 °C [54]. Similarly, for SARS-CoV-1, the persistence depends on the temperature of domestic sewage [55]. After experiments on the primary and secondary effluent, one study reported that the persistence of HCoV-229E was similar for two days at 23 °C [53].
The usual phenomenon is that feces and urine of infected patients are discharged into sewer systems (Figure 1), ultimately finding their way into wastewater and sewage treatment systems/plants [34]. This is considered the primary route of SARS-CoV-2 transmission to water and wastewater [56]. Thus, there is a chance of SARS-CoV-2 spread via gasp of open toilet setup and filthy oropharyngeal drops from effluent, particularly in crowded domestic areas [12,57]. COVID-19 patients can shed the virus for a more extended period than asymptomatic humans. This may increase the transmission of the virus particles in the sewage for an extended period. These, in turn, will end up in water streams if no treatment facility is available in place.
SARS-CoV-2 droplets may spread through the wastewater sanitation arrangement of a building’s different floors and air by cross-contamination [12]. Further, one study has already reported that the environment is a potential medium of transmission of SARS-CoV-2 after detecting positive samples from the toilet bowl, sink, and swab samples of air exhaust outlets of COVID-19 patients’ rooms [13]. On the contrary, a recent study found that following culture, extracted RNA from the exterior surface of continuous positive airway pressure helmets has no cytopathic effect [58]. Again, SARS-CoV-2 has been identified in wastewater in almost all regions of Europe, America, Asia, and the Middle East, regardless of the country’s economic classification (Table 3). Eleven studies detected SARS-CoV-2 in the effluent Asia-Pacific region, namely Bangladesh, India, China, Australia, Pakistan, United Arab Emirates, and Japan [59]. Among the European countries, Italy, Spain, France, Germany, The Netherlands, Turkey, the Czech Republic, and Slovenia detected SARS-CoV-2 RNA in the wastewater [60,61,62,63,64,65,66,67,68,69] (Table 3).

Table 3.
Detection of SARS-CoV-2 RNA in wastewater.
3.3. Risk of Transmission of SARS-CoV-2 through Biomedical Wastage
Moreover, in Switzerland, wastewater-based surveillance reported the existence of both the alpha (B.1.1.7) and beta variant (501.V2) with a variant-specific signature mutation in sewer water [88]. The same study identified three co-occurring signature mutations of alpha variants from wastewater in Switzerland, suggesting a new strain of the SARS-CoV-2 virus in the community [88]. In the North and South American regions, eight studies have identified the existence of the virus in wastewater and raw sewage in the U.S., Brazil, and Ecuador [80,81,82,83,84,85,86]. Therefore, it is suggested to treat wastewater, raw sewage, and river water as potential environmental media for the dispersal of SARS-CoV-2 [9]. Nevertheless, no study determined the infectiousness of SARS-CoV-2 from different types of raw and treated wastewater. Thus, we recommend conducting further studies to determine the infectiousness of SARS-CoV-2 from wastewater, different sewage, and sludge at various stages of treatment plants. The absence of different enteric and respiratory viruses, such as other coronaviruses, noroviruses, hepatitis A virus, hepatitis E virus, adenovirus, and astrovirus, in treated wastewater or sewer indicates the better efficacy of the treatment plant [89,90,91,92]. Although enveloped and non-enveloped viruses act differently in the environment, the enveloped SARS-CoV-2 is also an indicator virus, detection of which in the treated wastewater or sewer determines that the treatment plant or system is not safe for public health [89,90,91,92]. However, no study determined the infectiousness of SARS-CoV-2 ribonucleic acid (RNA) detected from sewage samples. Further, SARS-CoV-2 is sensitive to free chlorine [10]. So, the infectiousness of SARS-CoV-2 in wastewater is still in question considering their vulnerability to disinfection processes.
Due to the pandemic, the demand and use of different personal protective equipment (PPE) have increased dramatically. A study estimated that approximately 2.3 billion face masks were used as of 31 July 2020 in 49 Asian countries [93]. Individual Asian countries produced more than 16 thousand tons of medical waste during this pandemic [93]—the amount of medical waste increased along with the rise of COVID-19 cases. From the beginning of the pandemic to May 2020, South Korea produced 2000 tons of COVID-19 waste [94]. In Malaysia, the generation of clinical waste has increased by up to 27% during the pandemic compared to pre-pandemic time [95]. Romania produced more than 4 thousand tons of medical waste during lockdown from 26 February to 15 June 2020 [96]. Indonesia was approximately 13 thousand tons in 60 days from the hospital and household settings [97].
Waste generated by COVID-19 patients treated in households or private hospitals and medical centers or individuals in quarantine increases the likelihood of infection transmission to the environment [98]. Biomedical waste produced from hospitals and clinics engaged in COVID-19 patients treatment is potentially a bearer of SARS-CoV-2 [99,100]. Other biomedical waste generated from households and hospitals, such as disposable gowns, face masks, hand gloves, goggles, and face shields, can easily be mixed with domestic and hospital waste (Figure 1) [101]. Studies on the persistence of SARS-CoV-2 on biomedical waste also support the risk of infection through both hospital and household waste. However, the SARS-CoV-2 virus can survive from hours to days of COVID-19 waste, including disposable gowns, masks (inner and outer layer), tissue paper, testing kits, and gloves, depending on the temperature [102]. COVID-19 medical waste dumped without being appropriately treated has the possibility of mixing with the environment through water, food, soil, air, and livestock. This puts the environment and human lives at risk [103,104,105].
3.4. Risk of Transmission of SARS-CoV-2 through Diverse Inanimate Environmental Surface Contact
The role of environmental factors has long been studied for different coronaviruses. Several studies have identified potential environmental pathways through inanimate surface contact before this pandemic (Figure 1). The persistence of other coronaviruses on different porous and non-porous surfaces, including steel, aluminum, paper, wood, metal, glassware, plastics, polyvinyl chloride (PVC), rubber and surgical gloves, onetime gowns, ceramic, Teflon, cloth, surgical masks, tissue paper, cardboard, polymer notes, paper, cotton, and vinyl has been presented in Table 4. Different coronaviruses can remain infectious on steel surfaces for 4 h to more than 28 days, mainly depending on temperature, humidity, and viral load [102,106]. Further, the virus can survive on the contaminated surface for 6 to 9 days post-contamination [107]. SARS-CoV-2 can persist for up to 9 days at room temperature [108].

Table 4.
Presence and persistence of different coronaviruses in the diverse environmental media.
Moreover, lower temperatures increase the duration of the tenacity of the virus. For instance, at 4 °C, the transmissible gastroenteritis virus (TGEV) remains active on steel for more than 28 days, but it is reduced to 4–96 h when the temperature increases to 40 °C. The earlier study corroborated that at 22 °C, SARS-CoV-2 may survive for four days on steel, one day on wood, 30 min on paper and tissue, two days on glass, one day on cloth, and 4–7 days on single used face masks [102].
The empirical evidence suggests that the persistence of HCoV, SARS-CoV, and MERS-CoV varied from 2 to 7 days at 21–25 °C on different surfaces (Table 4). Again, SARS-CoV-2 has been detected from various environmental samples collected from a light switch, bathroom doorknob, inner wall of the toilet, towel, sewer inlet, inner surface of washbowl, floor, bedside table surface, pillow, and duvet cover of a quarantine room by Hu et al. [117]. SARS-CoV-2 is more contagious compared to other coronaviruses [117]. The current data on SARS-CoV-2 persistence suggest that although there is still limited evidence to establish the idea of the virus spreading through surface contact, it is arguable to say that surface contamination may also increase the chance of infection [5,117].
3.5. Presence and Risk of Transmission of SARS-CoV-2 Virus through Air
A vital transmission route of SARS-CoV-2 is via respiratory droplets and close contact with aerosol particles [118]. The virus can be attached to any medium, i.e., respiratory droplets from humans, which will carry it to another human [119]. These droplets can spread or settle down on surfaces and subsequently infect humans. Humans can be infected after inhaling hundreds of virus particles, whereas a single droplet can become tens of thousands of virus particles [120]. Aerosols are usually less than 5 μm, and droplets are more significant than 5 μm [119]. The large particles settle down easily due to their weight; thus, there is less chance of the virus spreading to a wider area [121].
On the other hand, aerosols can remain suspended in the air from 1.0 to 15.0 s depending on the velocity and direction of wind [122]. The study reported that SARS-CoV-2 could travel via air up to 4 m [123].
Aerosol transmission depends on various criteria such as virus concentration in aerosols, virus’ survival time, and infective dose [124]. Nevertheless, all these parameters are still not known. Additionally, air circulation is less indoors than outdoors, and indoor air samples were more contaminated [125]. SARS-CoV-2 can remain viable for 3 h in aerosols in laboratory conditions detected in the half-life of SARS-CoV-2 at 1.1 to 1.2 h. However, the authors of [126] reported that in aerosols, SARS-CoV-2 could remain infective for 16 h although the viability of viruses decreases when the temperature [127] and humidity [128] increase.
Many researchers argue that airborne transmission can cause more infection and possibly spread in three ways—(i) through air circulation in confined compartments with infected patients; (ii) recirculating air in building ventilation systems; (iii) through ventilation, air conditioning, and heating systems’ connection with outside air of the health facilities [129]. Respiratory droplet transmission (>5 μ m) is the primary mode of spread for SARS-CoV-2. The persistence of the virus in the aerosols lasts for more than 3 h (<5 μm), which is infectious in humans [111,130,131]. Believing in aerosol-driven infections, several researchers showed that aerosol transmission of the disease in closed environments may cause community transmission [22,130,131,132,133,134,135]. Another study detected SARS-CoV-2 in the air within approximately 4 m of COVID-19 patients [136].
In addition, the identification and perseverance of SARS-CoV-2 on different porous and non-porous surfaces, water environment, and stool have long been documented, which may increase plausible air-fomite transmission. Moreover, researchers argued that air pollution and microfiber contamination (2.5 m-sized particles) are risk factors for the transmission and severity of SARS-CoV-2 infection [137] and regardless of allergy status, co-exposure to airborne pollen increases susceptibility to SARS-CoV-2 virus infections [138]. Although SARS-CoV-2 transmission through fomites is relatively low compared to sneezing or coughing droplets, microfiber or pollen may act as a vehicle for virus transfer at a high concentration, or the particles may injure the lungs when inhaled. As a result, the severity of SARS-CoV-2 increased dramatically. Moreover, the Centre for Disease Control (CDC) recommends practicing handwashing and sanitizing after contact with possible contaminated surfaces such as door handles, tables, gas pumps, shopping carts, or electronic cashier registers/screens, which are frequently touched by other people [139]. However, most of the stated studies were performed in experimental conditions. Therefore, the researcher should test the persistence of SARS-CoV-2 in a real-life environment to show airborne transmission effectiveness.
4. Genomic Epidemiology of Emerging Variants of SARS-CoV-2 in the Environment
By 15 April 2022, in GISAID, 5860 complete sequences of SARS-CoV-2 RNA collected from different environmental sources including wastewater, clinic material, and surfaces worldwide were deposited and are used in this paper (Supplementary File S1). Among them, 5013 sequences (Alpha n = 605, 12.07%; Beta n = 8, 0.16%; Gamma n = 9, 0.18; Delta n = 2218, 44.24%; MU n = 1, 0.02%; Omicron n = 2172, 43.33%) were of emerging variants of concern. The delta variant was reported from environmental samples in November 2020. However, from January 2021, the alpha variant was slowly increasing in environmental samples [140]. This trend persisted until June 2021. Nevertheless, delta replaced the alpha variant among the environmental samples (Figure 2). Emerging variants detected in several countries from environmental samples have been shown in Figure 2.
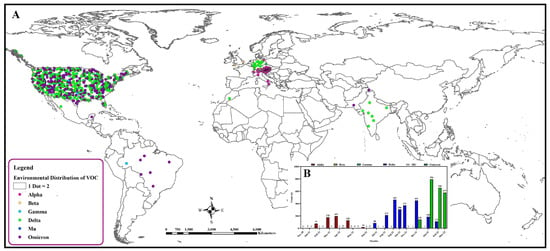
Figure 2.
Distribution of emerging variants of SARS-CoV-2 in environmental samples globally. (A) Spatial distribution. (B) Temporal distribution.
The phylogeny of environmental strains of SARS-CoV-2 is shown in Figure 3. Different VOCs formed a separate cluster in the tree, having close relations with human strains isolated from the same country (Figure 3). Another phylogeny for the omicron variant from environmental samples has been shown in Figure 4, whereas the phylogeny for the delta variant has been shown in Figure 5. This variant of the environment and humans in the same regions shows genetic resemblance [140]. For Omicron, the environmental strains from the USA are grouped with human strains from Italy, the USA, and Mexico; environmental strains from Austria are grouped with human strains from Belgium and Austria; environmental strains from Liechtenstein are grouped with human strains from Belgium, USA, and Austria. However, interestingly, strains from different countries were also grouped: environmental strains from Austria and Liechtenstein; human strains from the Netherlands and environmental strains from Austria; human strains from Belgium, USA, Germany, England, and environmental strains from Austria (Figure 5).
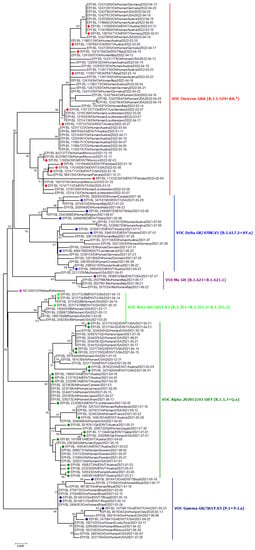
Figure 3.
Phylogenetic analysis of emerging variants from environmental samples. Green, red, violet, purple- and indigo-colored blocks represent alpha, beta, gamma, mu, and delta variants from the respective environment. The Fuchsia pink color indicates the reference sequence from Wuhan, China.
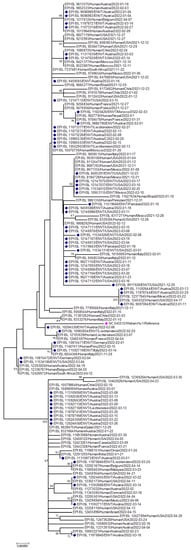
Figure 4.
Phylogenetic analysis of Omicron variants from environmental samples. Deep Indigo-colored blocks represent omicron variants from the environment, whereas the fuchsia pink color indicates the reference sequence from Wuhan, China.
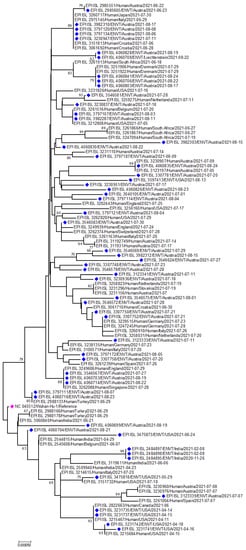
Figure 5.
Phylogenetic analysis of Delta variants from environmental samples. Indigo-colored blocks represent delta variants from the environment, whereas the fuchsia pink color indicates the reference sequence from Wuhan, China.
5. Inactivation Strategies of SARS-CoV-2 in Different Environmental Conditions
The virus may be inactivated using different methods such as ultraviolet (U.V.) rays, heat, and alcohol treatment [141], in water treatment plants, health care settings, and agricultural fields [142,143]. However, biocidal efficacy depends on various factors, including virus strain, titer, nature of the surface, and ambient conditions [144]. Below, described methods are followed to inactivate viruses in the environment.
5.1. Inactivation of SARS-CoV-2 Using Biocidal Agents
Alcohol-based disinfectant solutions such as isopropyl alcohol at different concentrations are widely used to inactivate different viruses in household and hospital settings. A comprehensive study on inactivation of different coronaviruses showed a wide variety of disinfectant such as 78–95% ethanol, 70–100% 2-propanol, 45% 2-propanol in combination with 30% 1-propanol, 0.5–2.5% glutardialdehyde, 0.7–1% formaldehyde and 0.23–7.5% povidone-iodine can be useful for readily inactivation of coronavirus infectivity at 4 log10 fold [145]. However, p-chloro-m-xylenol (PCMX) can inactivate the SARS-CoV-2 virus on glass surfaces within 0.5 to 10 min at ambient temperature [146]. In addition, 0.21% sodium hypochlorite and 0.5% hydrogen peroxide are also effective against SARS-CoV-2. Hospitals are using ethanol as a hand sanitizer. These alcohol-based disinfectants can be used on inanimate contact surfaces such as doorknobs, telephones, and lift buttons, reducing the chance of infection. Other products for solid surface decontamination include--quaternary ammonium compounds, peroxy compounds, sodium hypochlorite (NaClO), alcohol, and organic acids [147].
It is critical to inactivate the virus before they pollute the water bodies. In this regard, Hypochlorite (HClO) is the most efficient way to inactivate the pathogen of wastewater [148]. NaClO disinfection combined with U.V. rays for tertiary treatment can remove SARS-CoV-2 from wastewater [66]. Compared to other means such as U.V./Ozone, disinfections work more effectively for SARS-CoV-2 than chlorine-based solutions and offer several benefits, including lesser power consumption, lower toxicity, simple equipment, and setup [144]. Enveloped viruses such as SARS-CoV-2 can be easily removed from wastewater as they frequently adhere to organic biomass [149]. It was reported that moving bed biofilm reactors and sequencing batch reactors are efficient secondary treatment strategies to abolish the virus from wastewater [71]. Other efficient inactivation processes include activated sludge, biological nutrient removal, and algae bioreactors [150]. However, the aerosolization of viable viruses from water and wastewater may risk people being involved in treatment activities [151]. Open aerobic wastewater treatment plants, activities such as pumping wastewater, discharge, and flow are highly likely to participate in virus aerosolization.
5.2. Inactivation of SARS-CoV-2 Using Non-Biocidal Agents
Heat and U.V. irradiation-based inactivation techniques have been widely used in the hospital and biomedical sectors to sterilize medical equipment and apparatus. Both methods effectively kill SARS-CoV-2 from the surfaces [152]. Several types of research have been carried out to estimate the efficiency of UV-C irradiation on inanimate surfaces [153]. While other U.V. methods showed no significant inactivation for up to 15 min, UV-C increased the virus deactivation rate by 400 fold within 6 minutes [154]. In addition, this method was effective while stabilizing the virus from biomedical waste [145].
Furthermore, for decades, heat and thermal deactivation methods have been used for virus inactivation. The temperature’s effect on virus deactivation was an approximately 102-fold reduction within 12 days at 24 °C and a log4 unit reduction at 70 °C within 2.5 min for human/murine norovirus and a log3 fold reduction within two days at 71 °C for feline calicivirus [155]. In another experimental study, the virus was inactivated at 90% within 7 to 14 min in different culture media [156]. The dosages and methods of irradiation are crucial factors to ponder in their application as an essential means to battle the SARS-CoV-2 pandemic.
6. Environmental Pollution Is Due to the COVID-19 Pandemic
The main compounds of disposable masks are polypropylene, polyethylene, and other polymers such as polyesters, polyurethane, and polystyrene [157]. The surge in manufacturing and use of face masks and other PPE items raised a challenge for proper disposal in the environment [158]. Plastic molecules ultimately their way to the freshwater and marine environment [158]. Studies have reported that more than 200 masks enter Indonesia’s aquatic ecosystem per day [159]. Sea turtles and seabirds consume plastic litter [160] and this may obstruct their gastrointestinal tracts, resulting in debilitation and death. An adult Magellanic penguin (Spheniscus magellanicus) was found dead in Brazil due to ingesting a face mask [161].
Further, SARS-CoV-2 can survive in a surgical mask, gloves, and other plastic material for several days, and in developing countries, sewage waste goes to the ocean directly without any treatment, which may increase the chance of the virus migrating long distance [162,163]. Although SARS-CoV-2 has not yet been detected in aquatic mammals [163], previous research linked contaminated wastewater to SARS-CoV-2 reverse zoonotic transmission to wildlife [164]. Moreover, the scientific community expects that by highlighting the vulnerability and transmission of SARS-CoV-2 among wildlife [165], policy decisions about wastewater management worldwide will be shaped to help safeguard at-risk wild species and marine mammals that may be exposed to this coronavirus. Furthermore, other subfamily, gamma (ϒ) coronaviruses, are infective to aquatic mammals, mainly beluga whales and bottlenose dolphins and mammals of the Cetacea family [166].
Dedicated waste management legislation has been found in approximately 24% of world countries and approximately 33% of countries have general legislation, whereas 43% of countries are deprived of health care waste management legislation (Figure 6). Although North American countries have the highest dedicated legislation in contrast to all the countries globally, they face the challenge of gradually increasing contamination of SARS-CoV-2 in wastage. The global use of approximately 89 million face masks each month is due to COVID-19, which is recognized as plastic or plastic derivatives pollutants [167]. Face masks of polyethylene polymers [168,169] ultimately get in the way of dumpsites and water streams and pollute the aquatic and terrestrial environment [168]. Discarded face masks, gloves, and sanitizer bottles in the open environment such as parks, walkways, and even on main roads increase the challenges of environmental pollution, with adverse effects on the human and wildlife ecosystem. Waste and sewer treatment plants release their effluents into the water bodies and should be sensitive to the efficacy of their treatment [77]. Improperly treated effluents may increase environmental contamination and increase the possibility of exposure of the community population to SARS-CoV-2. This emphasizes the need for all of us to make better decisions and respond more quickly to infection spillover and the challenges posed by environmental contamination [170].
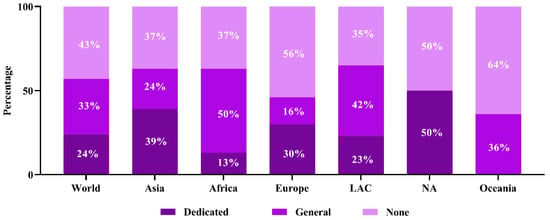
Figure 6.
Region-wise health care waste management legislative status.
Furthermore, the spread of ‘VOCs’ in the environment warns of the risk of SARS-CoV-2 establishing in the environment, and this could spill back to other animal species with significant population densities [171]. Indeed, Omicron’s genetic variants are sufficiently numerous that they could have been acquired by circulation in an animal reservoir, which is a plausible alternative explanation for its formation [170,171]. As a result, we urge strengthening the OneHealth approach of surveillance practice at the human–animal interface and in the environment to prevent future epidemics and pandemics.
7. Conclusions and Recommendations
SARS-CoV-2 has different transmission pathways, leading to environmental persistence and further spreading to remote areas. The spread of SARS-CoV-2 via wastewater and sewage has recently been a concern for the scientific community. Airborne transmission and transmission from contaminated surfaces are also seriously considered due to the considerable length of virus survival in air particles and inanimate surfaces. Moreover, the prevalent emerging VOCs, Delta and Omicron, are mostly circulating in the diverse environmental media which are genetically related to human strains of SARS-CoV-2. Thus, different methods must be adopted to inactivate the virus using potential agents. Human health, wildlife, and aquatic mammals are in danger due to environmental contamination of SARS-CoV-2 through household and medical wastage. The virus may spread to more expansive areas through environmental contaminants and have a much more expansive impact than we could imagine. Thus, wastewater surveillance may be an efficient tool to detect emerging variants circulating in the community and may act as an early warning system for public health mitigation. Wastewater surveillance and sequencing of SARS-CoV-2 variants are needed to integrate with public health initiatives. This review will help in preventing and controlling the environmental contamination of SARS-CoV-2 and help in understanding integrated medical waste management.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/covid2070067/s1, File S1: Environmental detection of SARS-CoV-2 metadata.
Author Contributions
Conceptualization, A.I.; methodology, A.I. and M.A.S.; formal analysis, A.I., M.A.S. and O.S.; writing—original draft preparation, A.I., M.A.S. and J.F.; writing—review and editing, A.I., M.A.S., M.A.K., J.F., S.S., J.A., S.I., S.D.C. and M.M.H.; visualization, A.I., M.A.S. and O.S.; supervision, A.I. and M.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
The authors did not receive any external funds to conduct this research. However, the research team was partially supported by NIH, National Institute of Allergy and Infectious Diseases (NIAID) Award U01AI153420 (PI Jonathan H Epstein) through EcoHealth Alliance.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Acknowledgments
We sincerely acknowledge all of the scientists from the originating laboratories who collected the samples, provided the SARS-CoV-2 genome data, and submitted them to the GISAID database, which has used for the analyses, displayed in this article (Supplementary File S1). The authors are thankful to the Institute of Epidemiology, Disease Control and Research (IEDCR), EcoHealth Alliance, the USA, and Chattogram Veterinary and Animal Sciences University (CVASU) for their continued support of our research team.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Ferdous, J.; Shano, S.; Choudhury, S.D.; Hassan, M.M. Spatiotemporal patterns and trends of community transmission of the pandemic COVID-19 in South Asia: Bangladesh as a case study. Biosaf. Health 2020, 3, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Anon. COVID-19 Vaccine Tracker: The Global Race to Vaccinate. Available online: https://ig.ft.com/coronavirus-vaccine-tracker/?areas=gbr&areas=isr&areas=usa&areas=eue&cumulative=1&populationAdjusted=1 (accessed on 12 May 2021).
- WHO. Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions. Scientific brief on 9 July 2020. Geneva. 2020. Available online: https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions (accessed on 12 May 2021).
- Liu, Y.; Ning, Z.; Chen, Y.; Guo, M.; Liu, Y.; Gali, N.K.; Sun, L.; Duan, Y.; Cai, J.; Westerdahl, D. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 2020, 582, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Roy, P.C.; Ferdaus, A.; Ibnat, H.; Alam, A.R.U.; Nigar, S.; Jahid, I.K.; Hossain, M.A. Prevalence and stability of SARS-CoV-2 RNA on Bangladeshi banknotes. Sci. Total Environ. 2021, 779, 146133. [Google Scholar] [CrossRef]
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Zamil, S.; Abedin, J.; Saha, O.; Hassan, M.M. Assessment of basic reproduction number (R0), spatial and temporal epidemiological determinants, and genetic characterization of SARS-CoV-2 in Bangladesh. Infect. Genet. Evol. 2021, 92, 104884. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chen, Q. Insight into 2019 novel coronavirus—An updated interim review and lessons from SARS-CoV and MERS-CoV. Int. J. Infect. Dis. 2020, 94, 119–124. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods-A scoping review. Water Res. 2020, 179, 115899. [Google Scholar] [CrossRef]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.-S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.; Sarmah, A.K.; Chao, H.-P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2020, 193, 110265. [Google Scholar] [CrossRef]
- McKinney, K.R.; Gong, Y.Y.; Lewis, T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health 2006, 68, 26. [Google Scholar]
- WHO. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS). Available online: https://apps.who.int/iris/bitstream/handle/10665/70863/WHO_CDS_CSR_GAR_2003.11_eng.pdf (accessed on 20 May 2021).
- Gormley, M.; Aspray, T.J.; Kelly, D.A. COVID-19: Mitigating transmission via wastewater plumbing systems. Lancet Glob. Health 2020, 8, e643. [Google Scholar] [CrossRef] [Green Version]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA 2020, 323, 1610–1612. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, O.; Islam, I.; Shatadru, R.N.; Rakhi, N.N.; Hossain, M.S.; Rahaman, M.M. Temporal landscape of mutational frequencies in SARS-CoV-2 genomes of Bangladesh: Possible implications from the ongoing outbreak in Bangladesh. Virus Genes 2021, 57, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Ferdous, J.; Islam, S.; Hassan, M.M. Geospatial dynamics of COVID-19 clusters and hotspots in Bangladesh. Transbound. Emerg. Dis. 2021, 68, 3643–3657. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Fong, T.-T.; Lipp, E.K. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005, 69, 357–371. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2020, 2, e13–e22. [Google Scholar] [CrossRef]
- Cai, J.; Xu, J.; Lin, D.; Xu, L.; Qu, Z.; Zhang, Y.; Zhang, H.; Jia, R.; Wang, X.; Ge, Y.; et al. A Case Series of Children With 2019 Novel Coronavirus Infection: Clinical and Epidemiological Features. Clin. Infect. Dis. 2020, 71, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, T.; Deng, Y.; Liu, S.; Zhang, D.; Li, H.; Wang, X.; Jia, L.; Han, J.; Bei, Z.; et al. Stability of SARS-CoV-2 on environmental surfaces and in human excreta. J. Hosp. Infect. 2021, 107, 105–107. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.Y.; Cheng, P.K.; Lim, W.W. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005, 41, e67–e71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, C.; Song, Y.; Zhu, S.; Wang, D.; Zhang, H.; Han, G.; Weng, Y.; Xu, J.; Xu, J. Excretion of SARS-CoV-2 through faecal specimens. Emerg. Microbes Infect. 2020, 9, 2501–2508. [Google Scholar] [CrossRef]
- Lo, I.L.; Lio, C.F.; Cheong, H.H.; Lei, C.I.; Cheong, T.H.; Zhong, X.; Tian, Y.; Sin, N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Bio. Sci. 2020, 16, 1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Wilton, T.; Bujaki, E.; Klapsa, D.; Fritzsche, M.; Mate, R.; Martin, J.J.m. Rapid increase of SARS-CoV-2 variant B. 1.1. 7 detected in sewage samples from England between October 2020 and January 2021. Msystems 2021, 6, e00353-21. [Google Scholar] [CrossRef]
- Landgraff, C.; Wang, L.Y.R.; Buchanan, C.; Wells, M.; Schonfeld, J.; Bessonov, K.; Ali, J.; Robert, E.; Nadon, C. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B. 1.1. 7 variant of concern from a Canadian municipality concurrent with an outbreak. medRxiv 2021. [Google Scholar] [CrossRef]
- Gwenzi, W. Leaving no stone unturned in light of the COVID-19 faecal-oral hypothesis? A water, sanitation and hygiene (WASH) perspective targeting low-income countries. Sci. Total Environ. 2020, 753, 141751. [Google Scholar] [CrossRef]
- Tang, A.N.; Tong, Z.D.; Wang, H.L.; Dai, Y.X.; Li, K.F.; Liu, J.N.; Wu, W.J.; Yuan, C.; Yu, M.L.; Li, P.; et al. Detection of novel coronavirus by rt-pcr in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. J. 2020, 26, 1337–1339. [Google Scholar] [CrossRef]
- Goh, G.K.-M.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. Shell disorder analysis predicts greater resilience of the SARS-CoV-2 (COVID-19) outside the body and in body fluids. Micro. Path. 2020, 144, 104177. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wang, S.; Xue, Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020, 92, 680–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroentero 2020, 158, 1831–1833. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef] [Green Version]
- Ling, Y.; Xu, S.-B.; Lin, Y.-X.; Tian, D.; Zhu, Z.-Q.; Dai, F.-H.; Wu, F.; Song, Z.-G.; Huang, W.; Chen, J. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020, 133, 1039–1043. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Kam, K.-q.; Yung, C.F.; Cui, L.; Tzer Pin Lin, R.; Mak, T.M.; Maiwald, M.; Li, J.; Chong, C.Y.; Nadua, K.; Tan, N.W.H. A well infant with coronavirus disease 2019 with high viral load. Clin. Infect. Dis. 2020, 71, 847–849. [Google Scholar] [CrossRef]
- Xie, C.; Jiang, L.; Huang, G.; Pu, H.; Gong, B.; Lin, H.; Ma, S.; Chen, X.; Long, B.; Si, G. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020, 93, 264–267. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastro. Hepa. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Xing, Y.; Ni, W.; Wu, Q.; Li, W.; Li, G.; Tong, J.; Song, X.; Xing, Q. Prolonged presence of SARS-CoV-2 in feces of pediatric patients during the convalescent phase. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Cui, X.; Zhao, X.; Wang, J.; Zheng, J.; Zheng, G.; Guo, W.; Cai, C.; He, S.; Xu, Y. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J. Med. Virol. 2020, 92, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.W.; Kim, S.-M.; Kim, H.-S.; Kim, Y.-I.; Kim, J.H.; Cho, J.Y.; Kim, S.-H.; Kang, H.; Kim, S.-G.; Park, S.-J.; et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1520–1524. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef] [PubMed]
- Isakbaeva, E.T.; Khetsuriani, N.; Beard, R.S.; Peck, A.; Erdman, D.; Monroe, S.S.; Tong, S.; Ksiazek, T.G.; Lowther, S.; Smith, I.P. SARS-associated coronavirus transmission, United States. Emerg. Infect. Dis. 2004, 10, 225. [Google Scholar] [CrossRef]
- Ghinai, I.; McPherson, T.D.; Hunter, J.C.; Kirking, H.L.; Christiansen, D.; Joshi, K.; Rubin, R.; Morales-Estrada, S.; Black, S.R.; Pacilli, M. First known person-to-person transmission of severe acute respiratory syndrome oronavirus 2 (SARS-CoV-2) in the USA. Lancet 2020, 395, 1137–1144. [Google Scholar] [CrossRef]
- Lescure, F.-X.; Bouadma, L.; Nguyen, D.; Parisey, M.; Wicky, P.-H.; Behillil, S.; Gaymard, A.; Bouscambert-Duchamp, M.; Donati, F.; Le Hingrat, Q. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020, 20, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, U.D.; Eng, G.; Farcasanu, M.; Avena, L.E.; Choudhary, M.C.; Triant, V.A.; Flagg, M.; Schiff, A.E.; Gomez, I.; Froehle, L.M. Fecal Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) RNA Is Associated With Decreased Coronavirus Disease 2019 (COVID-19) Survival. Clin. Infect. Dis. 2021, 74, 1081–1084. [Google Scholar] [CrossRef]
- Fumian, T.M.; Malta, F.C.; Dos Santos, D.R.; Pauvolid-Corrêa, A.; Fialho, A.M.; Leite, J.P.; Miagostovich, M.P. SARS-CoV-2 RNA detection in stool samples from acute gastroenteritis cases, Brazil. J. Med. Virol. 2021, 93, 2543–2547. [Google Scholar] [CrossRef]
- Gundy, P.M.; Gerba, C.P.; Pepper, I.L. Survival of Coronaviruses in Water and Wastewater. Food Environ. Viro. 2008, 1, 10. [Google Scholar] [CrossRef] [Green Version]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Li, J.-S.; Jin, M.; Zhen, B.; Kong, Q.-X.; Song, N.; Xiao, W.-J.; Yin, J.; Wei, W.; Wang, G.-J.; et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Collivignarelli, M.C.; Collivignarelli, C.; Miino, M.C.; Abbà, A.; Pedrazzani, R.; Bertanza, G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020, 143, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Huang, X.; Zhou, P.; Li, C.; Wu, A. Alert for SARS-CoV-2 infection caused by fecal aerosols in rural areas in China. Infect. Control Hosp. Epidemiol. 2020, 41, 987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colaneri, M.; Seminari, E.; Novati, S.; Asperges, E.; Biscarini, S.; Piralla, A.; Percivalle, E.; Cassaniti, I.; Baldanti, F.; Bruno, R. Severe acute respiratory syndrome coronavirus 2 RNA contamination of inanimate surfaces and virus viability in a health care emergency unit. Clin. Microbiol. Infect. 2020, 26, 1094.e1–1094.e5. [Google Scholar] [CrossRef]
- Ahmed, F.; Islam, M.A.; Kumar, M.; Hossain, M.; Bhattacharya, P.; Islam, M.T.; Hossen, F.; Hossain, M.S.; Islam, M.S.; Uddin, M.M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre through wastewater surveillance in Bangladesh. Sci. Total Environ. 2020, 776, 145724. [Google Scholar] [CrossRef]
- Balboa, S.; Mauricio-Iglesias, M.; Rodríguez, S.; Martínez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv 2020. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Mlejnkova, H.; Sovova, K.; Vasickova, P.; Ocenaskova, V.; Jasikova, L.; Juranova, E. Preliminary study of SARS-CoV-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health 2020, 17, 5508. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef] [PubMed]
- Kocamemi, B.A.; Kurt, H.; Sait, A.; Sarac, F.; Saatci, A.M.; Pakdemirli, B. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv 2020. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Wurtzer, S.; Marechal, V.; Mouchel, J.-M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv 2020. [Google Scholar] [CrossRef]
- Gonçalves, J.; Koritnik, T.; Mioč, V.; Trkov, M.; Bolješič, M.; Berginc, N.; Prosenc, K.; Kotar, T.; Paragi, M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2020, 755, 143226. [Google Scholar] [CrossRef]
- Baldovin, T.; Amoruso, I.; Fonzo, M.; Buja, A.; Baldo, V.; Cocchio, S.; Bertoncello, C. SARS-CoV-2 RNA detection and persistence in wastewater samples: An experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy). Sci. Total Environ. 2020, 760, 143329. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef]
- Arora, S.; Nag, A.; Sethi, J.; Rajvanshi, J.; Saxena, S.; Shrivastava, S.K.; Gupta, A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020, 82, 2823–2836. [Google Scholar] [CrossRef]
- Or, I.B.; Yaniv, K.; Shagan, M.; Ozer, E.; Erster, O.; Mendelson, E.; Mannasse, B.; Shirazi, R.; Kramarsky-Winter, E.; Nir, O. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: A proof-of-concept for quantitative environmental surveillance. medRxiv 2020. [Google Scholar] [CrossRef]
- Xu, B.; Kraemer, M.U.; Gutierrez, B.; Mekaru, S.; Sewalk, K.; Loskill, A.; Wang, L.; Cohn, E.; Hill, S.; Zarebski, A. Open access epidemiological data from the COVID-19 outbreak. Lancet Infect. Dis. 2020, 20, 534. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Ling, H.; Huang, X.; Li, J.; Li, W.; Yi, C.; Zhang, T.; Jiang, Y.; He, Y.; Deng, S. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of fangcang hospital. Sci. Total Environ. 2020, 741, 140445. [Google Scholar] [CrossRef] [PubMed]
- Albastaki, A.; Naji, M.; Lootah, R.; Almeheiri, R.; Almulla, H.; Almarri, I.; Alreyami, A.; Aden, A.; Alghafri, R. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: The use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci. Total Environ. 2020, 760, 143350. [Google Scholar] [CrossRef] [PubMed]
- Yaqub, T.; Nawaz, M.; Shabbir, M.Z.; Ali, M.A.; Altaf, I.; Raza, S.; Shabbir, M.A.B.; Ashraf, M.A.; Aziz, S.Z.; Cheema, S.Q. A longitudinal survey for genome-based identification of SARS-CoV-2 in sewage water in selected lockdown areas of Lahore city, Pakistan; a potential approach for future smart lockdown strategy. medRxiv 2020. [Google Scholar] [CrossRef]
- Tanhaei, M.; Mohebbi, S.R.; Hosseini, S.M.; Rafieepoor, M.; Kazemian, S.; Ghaemi, A.; Shamloei, S.; Mirjalali, H.; Aghdaei, H.A.; Zali, M.R. The first detection of SARS-CoV-2 RNA in the wastewater of Tehran, Iran. Environ. Sci. Pollut. Res. 2021, 28, 38629–38636. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Weber, F.-A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751, 141750. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef]
- Miyani, B.; Fonoll, X.; Norton, J.; Mehrotra, A.; Xagoraraki, I. SARS-CoV-2 in detroit wastewater. Environ. Eng. 2020, 146, 06020004. [Google Scholar] [CrossRef]
- Wu, F.; Xiao, A.; Zhang, J.; Gu, X.; Lee, W.L.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; Endo, N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv 2020. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Wilder, M.; Middleton, F.A.; Collins, M.; Fenty, A.; Gentile, K.; Kmush, B.; Zeng, T.; Larsen, D.A. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. medRxiv 2020. [Google Scholar] [CrossRef]
- Prado, T.; Fumian, T.M.; Mannarino, C.F.; Maranhão, A.G.; Siqueira, M.M.; Miagostovich, M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2020, 115. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process. Eng. 2021, 40, 101815. [Google Scholar] [CrossRef] [PubMed]
- Jahn, K.; Dreifuss, D.; Topolsky, I.; Kull, A.; Ganesanandamoorthy, P.; Fernandez-Cassi, X.; Bänziger, C.; Stachler, E.; Fuhrmann, L.; Jablonski, K.P. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv 2021. [Google Scholar] [CrossRef]
- Gerba, C.P.; Betancourt, W.Q.; Kitajima, M. How much reduction of virus is needed for recycled water: A continuous changing need for assessment? Water Res. 2017, 108, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeChevallier, M.W.; Mansfield, T.J.; Gibson, J.M. Protecting wastewater workers from disease risks: Personal protective equipment guidelines. Water Environ. Res. 2020, 92, 524–533. [Google Scholar] [CrossRef]
- Sinclair, R.G.; Choi, C.Y.; Riley, M.R.; Gerba, C.P. Pathogen surveillance through monitoring of sewer systems. J. Advan. Appl. Micro. 2008, 65, 249. [Google Scholar]
- Maal-Bared, R.; Brisolara, K.; Munakata, N.; Bibby, K.; Gerba, C.; Sobsey, M.; Schaefer, S.; Swift, J.; Gary, L.; Sherchan, S. Implications of SARS-CoV-2 on current and future operation and management of wastewater systems. Water Environ. Res. 2020, 93, 502–515. [Google Scholar] [CrossRef]
- Sangkham, S. Face Mask and Medical Waste Disposal during the Novel COVID-19 Pandemic in Asia. Case Stud. Chem. Environ. Eng. 2020, 2, 100052. [Google Scholar] [CrossRef]
- EZCAP. The Safe Waste Treatment for COVID-19: Lessons from the Republic of Korea. Available online: https://repository.unescap.org/handle/20.500.12870/878 (accessed on 10 August 2021).
- Agamuthu, P.; Barasarathi, J. Clinical waste management under COVID-19 scenario in Malaysia. Waste Manag. Res. 2020, 39, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Mihai, F.-C. Assessment of COVID-19 waste flows during the emergency state in romania and related public health and environmental concerns. Int. J. Environ. Res. Public Health 2020, 17, 5439. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Iwasaki, F.; Johannes, H.P.; Edita, E.P. Strengthening Waste Management Policies to Mitigate the COVID-19 Pandemic. Available online: https://think-asia.org/handle/11540/12206 (accessed on 5 July 2021).
- Di Maria, F.; Beccaloni, E.; Bonadonna, L.; Cini, C.; Confalonieri, E.; La Rosa, G.; Milana, M.R.; Testai, E.; Scaini, F. Minimization of spreading of SARS-CoV-2 via household waste produced by subjects affected by COVID-19 or in quarantine. Sci. Total Environ. 2020, 743, 140803. [Google Scholar] [CrossRef] [PubMed]
- Ramteke, S.; Sahu, B.L. Novel coronavirus disease 2019 (COVID-19) pandemic: Considerations for the biomedical waste sector in India. J. Case Stud. Chem. Environ. Eng. 2020, 2, 100029. [Google Scholar] [CrossRef]
- Islam, A.; Kalam, M.; Sayeed, M.; Shano, S.; Rahman, M.; Islam, S.; Ferdous, J.; Choudhury, S.D.; Hassan, M.M. Escalating SARS-CoV-2 circulation in environment and tracking waste management in South Asia. Environ. Sci. Pollut. Res. 2021, 28, 61951–61968. [Google Scholar] [CrossRef]
- Zambrano-Monserrate, M.A.; Ruano, M.A.; Sanchez-Alcalde, L. Indirect effects of COVID-19 on the environment. Sci. Total Environ. 2020, 728, 138813. [Google Scholar] [CrossRef]
- Chin, A.W.; Chu, J.T.; Perera, M.R.; Hui, K.P.; Yen, H.-L.; Chan, M.C.; Peiris, M.; Poon, L.L. Stability of SARS-CoV-2 in different environmental conditions. Lancet Micro. 2020, 1, e10. [Google Scholar] [CrossRef]
- Kerdsuwan, S.; Laohalidanond, K. Efficiency improvement for medical waste management. Handb. Clean Energy Syst. 2015, 1–11. [Google Scholar] [CrossRef]
- Mahmood, S.; ud Din, N.; Mohsin, J.; Javed, H. Practices regarding hospital waste management at public and private sector hospitals of Lahore. Ann. King Edw. Med. Univ. 2011, 17, 113. [Google Scholar]
- Mohankumar, S.; Kottaiveeran, K. Hospital waste management and environmental problems in India. Framework 2007, 2, 1621–1626. [Google Scholar]
- Riddell, S.; Goldie, S.; Hill, A.; Eagles, D.; Drew, T.W. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol. J. 2020, 17, 145. [Google Scholar] [CrossRef] [PubMed]
- Marquès, M.; Domingo, J.L. Contamination of inert surfaces by SARS-CoV-2: Persistence, stability and infectivity. A review. Environ. Res. 2020, 193, 110559. [Google Scholar] [CrossRef] [PubMed]
- Fathizadeh, H.; Maroufi, P.; Momen-Heravi, M.; Dao, S.; Ganbarov, K.; Pagliano, P.; Esposito, S.; Kafil, H.S. Protection and disinfection policies against SARS-CoV-2 (COVID-19). Infez. Med. 2020, 28, 185–191. [Google Scholar]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Munster, V. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eurosurveillance 2013, 18, 20590. [Google Scholar] [CrossRef] [Green Version]
- Warnes, S.L.; Little, Z.R.; Keevil, C.W. Human coronavirus 229E remains infectious on common touch surface materials. MBio 2015, 6, e01697-15. [Google Scholar] [CrossRef] [Green Version]
- Sizun, J.; Yu, M.; Talbot, P. Survival of human coronaviruses 229E and OC43 in suspension and after drying onsurfaces: A possible source ofhospital-acquired infections. J. Hosp. Infect. 2000, 46, 55–60. [Google Scholar] [CrossRef]
- Dong, J.H.X.-P. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed. Environ. Sci. 2003, 16, 246–255. [Google Scholar]
- Chan, K.-H.; Peiris, J.M.; Lam, S.; Poon, L.L.; Yuen, K.-Y.; Seto, W.H. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011, 2011, 734690. [Google Scholar] [CrossRef]
- Rabenau, H.; Cinatl, J.; Morgenstern, B.; Bauer, G.; Preiser, W.; Doerr, H. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. 2005, 194, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Ni, W.; Wang, Z.; Ma, G.; Pan, B.; Dong, L.; Gao, R.; Jiang, F. The distribution of SARS-CoV-2 contamination on the environmental surfaces during incubation period of COVID-19 patients. Ecotoxicol. Environ. Saf. 2020, 208, 111438. [Google Scholar] [CrossRef] [PubMed]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.; Elshabrawy, H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, Y.; Nielsen, P.V.; Wei, J.; Jensen, R.L. Short-range airborne transmission of expiratory droplets between two people. Indoor Air 2017, 27, 452–462. [Google Scholar] [CrossRef]
- Watanabe, T.; Bartrand, T.A.; Weir, M.H.; Omura, T.; Haas, C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010, 30, 1129–1138. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.; Li, Y. Removal of exhaled particles by ventilation and deposition in a multibed airborne infection isolation room. Indoor Air 2010, 20, 284–297. [Google Scholar] [CrossRef]
- Feng, Y.; Marchal, T.; Sperry, T.; Yi, H. Influence of wind and relative humidity on the social distancing effectiveness to prevent COVID-19 airborne transmission: A numerical study. J. Aerosol Sci. 2020, 147, 105585. [Google Scholar] [CrossRef]
- Chang, L.; Yan, Y.; Wang, L. Coronavirus disease 2019: Coronaviruses and blood safety. Transfus. Med. Rev. 2020, 34, 75–80. [Google Scholar] [CrossRef]
- Contini, D.; Costabile, F. Does air pollution influence COVID-19 outbreaks? Atmosphere 2020, 11, 377. [Google Scholar] [CrossRef] [Green Version]
- Santarpia, J.L.; Rivera, D.N.; Herrera, V.; Morwitzer, M.J.; Creager, H.; Santarpia, G.W.; Crown, K.K.; Brett-Major, D.; Schnaubelt, E.; Broadhurst, M.J. Aerosol and surface transmission potential of SARS-CoV-2. medRxiv 2020, 10, 1–8. [Google Scholar]
- Fears, A.C.; Klimstra, W.B.; Duprex, P.; Hartman, A.; Weaver, S.C.; Plante, K.; Mirchandani, D.; Plante, J.; Aguilar, P.V.; Fernandez, D. Comparative dynamic aerosol efficiencies of three emergent coronaviruses and the unusual persistence of SARS-CoV-2 in aerosol suspensions. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Joonaki, E.; Hassanpouryouzband, A.; Heldt, C.L.; Areo, O. Surface chemistry can unlock drivers of surface stability of SARS-CoV-2 in variety of environmental conditions. Chem 2020, 6, 2135–2146. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Liu, J.; He, X.; Wang, B.; Fu, S.; Yan, J.; Niu, J.; Zhou, J.; Luo, B. Effects of temperature variation and humidity on the death of COVID-19 in Wuhan, China. Sci. Total Environ. 2020, 724, 138226. [Google Scholar] [CrossRef] [PubMed]
- Correia, G.; Rodrigues, L.; Silva, M.; Gonçalves, T. Airborne route and bad use of ventilation systems as non-negligible factors in SARS-CoV-2 transmission. Med. Hypotheses 2020, 141, 109781. [Google Scholar] [CrossRef] [PubMed]
- Luongo, J.C.; Fennelly, K.P.; Keen, J.A.; Zhai, Z.J.; Jones, B.W.; Miller, S.L. Role of mechanical ventilation in the airborne transmission of infectious agents in buildings. Indoor Air 2016, 26, 666–678. [Google Scholar] [CrossRef]
- Yu, I.T.; Li, Y.; Wong, T.W.; Tam, W.; Chan, A.T.; Lee, J.H.; Leung, D.Y.; Ho, T. Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 2004, 350, 1731–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, C.J.; Milton, D.K. Airborne Transmission of Communicable Infection-the Elusive Pathway. Available online: https://apps.dtic.mil/sti/pdfs/ADA429398.pdf (accessed on 12 May 2021).
- Shiu, E.Y.; Leung, N.H.; Cowling, B.J. Controversy around airborne versus droplet transmission of respiratory viruses: Implication for infection prevention. Curr. Opin. Infect. Dis. 2019, 32, 372–379. [Google Scholar] [CrossRef]
- Judson, S.D.; Munster, V.J. Nosocomial transmission of emerging viruses via aerosol-generating medical procedures. Viruses 2019, 11, 940. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Diao, M.; Yu, W.; Pei, L.; Lin, Z.; Chen, D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int. J. Infect. Dis. 2020, 93, 201–204. [Google Scholar] [CrossRef]
- Hirota, K. Air contamination with SARS-CoV-2 in the operating room. J. Anesth. 2020, 35, 333–336. [Google Scholar] [CrossRef]
- SanJuan-Reyes, S.; Gómez-Oliván, L.M.; Islas-Flores, H. COVID-19 in the environment. Chemosphere 2021, 263, 127973. [Google Scholar] [CrossRef] [PubMed]
- Damialis, A.; Gilles, S.; Sofiev, M.; Sofieva, V.; Kolek, F.; Bayr, D.; Plaza, M.P.; Leier-Wirtz, V.; Kaschuba, S.; Ziska, L.H. Higher airborne pollen concentrations correlated with increased SARS-CoV-2 infection rates, as evidenced from 31 countries across the globe. Proc. Natl. Acad. Sci. USA 2021, 118, e2019034118. [Google Scholar] [CrossRef] [PubMed]
- CDC. When and How to Wash Your Hands. Available online: https://www.cdc.gov/handwashing/when-how-handwashing.html (accessed on 19 July 2021).
- Islam, A.; Sayeed, M.A.; Kalam, M.A.; Ferdous, J.; Rahman, M.K.; Abedin, J.; Islam, S.; Shano, S.; Saha, O.; Shirin, T.; et al. Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally. Microorganisms 2021, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Hijnen, W.; Beerendonk, E.; Medema, G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo) cysts in water: A review. Water Res. 2006, 40, 3–22. [Google Scholar] [CrossRef]
- Pfaender, S.; Brinkmann, J.; Todt, D.; Riebesehl, N.; Steinmann, J.; Steinmann, J.; Pietschmann, T.; Steinmann, E. Mechanisms of methods for hepatitis C virus inactivation. Appl. Environ. Microbiol. 2015, 81, 1616–1621. [Google Scholar] [CrossRef] [Green Version]
- Macinga, D.R.; Sattar, S.A.; Jaykus, L.-A.; Arbogast, J.W. Improved inactivation of nonenveloped enteric viruses and their surrogates by a novel alcohol-based hand sanitizer. Appl. Environ. Microbiol. 2008, 74, 5047–5052. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Mazumder, P.; Mohapatra, S.; Thakur, A.K.; Dhangar, K.; Taki, K.; Mukherjee, S.; Patel, A.K.; Bhattacharya, P.; Mohapatra, P. A Chronicle of SARS-CoV-2: Seasonality, Environmental Fate, Transport, Inactivation, and Antiviral Drug Resistance. J. Hazard. Mater. 2020, 405, 124043. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Ijaz, M.K.; Nims, R.W.; Zhou, S.S.; Whitehead, K.; Srinivasan, V.; Kapes, T.; Fanuel, S.; Epstein, J.H.; Daszak, P.; Rubino, J.R. Microbicidal actives with virucidal efficacy against SARS-CoV-2 and other beta-and alpha-coronaviruses and implications for future emerging coronaviruses and other enveloped viruses. Sci. Rep. 2021, 11, 5626. [Google Scholar] [CrossRef]
- EPA. United States Environmental Protection Agency. List N: Disinfectants for Use against SARS-CoV-2. Available online: https://www.epa.gov/coronavirus/disinfectant-use-and-coronaviruscovid-19 (accessed on 10 August 2021).
- Wang, J.; Shen, J.; Ye, D.; Yan, X.; Zhang, Y.; Yang, W.; Li, X.; Wang, J.; Zhang, L.; Pan, L. Disinfection technology of hospital wastes and wastewater: Suggestions for disinfection strategy during coronavirus Disease 2019 (COVID-19) pandemic in China. Environ Pollut. 2020, 262, 114665. [Google Scholar] [CrossRef]
- Saawarn, B.; Hait, S. Occurrence, fate and removal of SARS-CoV-2 in wastewater: Current knowledge and future perspectives. J. Environ. Chem. Eng. 2021, 9, 104870. [Google Scholar] [CrossRef] [PubMed]
- Delanka-Pedige, H.M.; Munasinghe-Arachchige, S.P.; Zhang, Y.; Nirmalakhandan, N. Bacteria and virus reduction in secondary treatment: Potential for minimizing post disinfectant demand. Water Res. 2020, 177, 115802. [Google Scholar] [CrossRef] [PubMed]
- Wartecki, A.; Rzymski, P. On the Coronaviruses and Their Associations with the Aquatic Environment and Wastewater. Water 2020, 12, 1598. [Google Scholar] [CrossRef]
- Loveday, E.K.; Hain, K.S.; Kochetkova, I.; Hedges, J.F.; Robison, A.; Snyder, D.T.; Brumfield, S.K.; Young, M.J.; Jutila, M.A.; Chang, C.B. Effect of Inactivation Methods on SARS-CoV-2 Virion Protein and Structure. Viruses 2021, 13, 562. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Heat inactivation of different types of SARS-CoV-2 samples: What protocols for biosafety, molecular detection and serological diagnostics? Viruses 2020, 12, 735. [Google Scholar] [CrossRef]
- Darnell, M.E.; Subbarao, K.; Feinstone, S.M.; Taylor, D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods 2004, 121, 85–91. [Google Scholar] [CrossRef]
- Seo, K.; Lee, J.E.; Lim, M.Y.; Ko, G. Effect of temperature, pH, and NaCl on the inactivation kinetics of murine norovirus. J. Food Prot. 2012, 75, 533–540. [Google Scholar] [CrossRef]
- Ratnesar-Shumate, S.; Williams, G.; Green, B.; Krause, M.; Holland, B.; Wood, S.; Bohannon, J.; Boydston, J.; Freeburger, D.; Hooper, I. Simulated sunlight rapidly inactivates SARS-CoV-2 on surfaces. J. Infect. Dis. 2020, 222, 214–222. [Google Scholar] [CrossRef]
- Prata, J.C.; Silva, A.L.P.; Duarte, A.C.; Rocha-Santos, T. Disposable over Reusable Face Masks: Public Safety or Environmental Disaster? Environments 2021, 8, 31. [Google Scholar] [CrossRef]
- Joo, S.H.; Choi, H. Field grand challenge with emerging superbugs and the novel coronavirus (SARS-CoV-2) on plastics and in water. J. Environ. Chem. Eng. 2021, 9, 104721. [Google Scholar] [CrossRef]
- Kemp, K.; Griffiths, J.; Campbell, S.; Lovell, K. An exploration of the follow-up up needs of patients with inflammatory bowel disease. J. Crohn’s Colitis 2013, 7, e386–e395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühn, S.; Rebolledo, E.L.B.; Van Franeker, J.A. Deleterious effects of litter on marine life. In Marine Anthropogenic Litter; Springer Open: New York, NY, USA, 2015; pp. 75–116. [Google Scholar]
- Neto, H.G.; Bantel, C.G.; Browning, J.; Della Fina, N.; Ballabio, T.A.; de Santana, F.T.; e Britto, M.d.K.; Barbosa, C.B. Mortality of a juvenile Magellanic penguin (Spheniscus magellanicus, Spheniscidae) associated with the ingestion of a PFF-2 protective mask during the COVID-19 pandemic. Mar. Pollut. Bull. 2021, 166, 112232. [Google Scholar] [CrossRef] [PubMed]
- Van Bressem, M.-F.; Duignan, P.J.; Banyard, A.; Barbieri, M.; Colegrove, K.M.; De Guise, S.; Di Guardo, G.; Dobson, A.; Domingo, M.; Fauquier, D. Cetacean morbillivirus: Current knowledge and future directions. Viruses 2014, 6, 5145–5181. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Ferdous, J.; Islam, S.; Sayeed, M.; Dutta Choudhury, S.; Saha, O.; Hassan, M.M.; Shirin, T. Evolutionary Dynamics and Epidemiology of Endemic and Emerging Coronaviruses in Humans, Domestic Animals, and Wildlife. Viruses 2021, 13, 1908. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Stoddart, A.K.; Gagnon, G.A.; Dellaire, G. Pandemic danger to the deep: The risk of marine mammals contracting SARS-CoV-2 from wastewater. Sci. Total Environ. 2021, 760, 143346. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Ferdous, J.; Sayeed, M.A.; Islam, S.; Kaisar Rahman, M.; Abedin, J.; Saha, O.; Hassan, M.M.; Shirin, T. Spatial epidemiology and genetic diversity of SARS-CoV-2 and related coronaviruses in domestic and wild animals. PLoS ONE 2021, 16, e0260635. [Google Scholar] [CrossRef]
- Nabi, G.; Khan, S. Novel coronavirus transmission to water bodies; risk of COVID-19 pneumonia to aquatic mammals. Environ. Res. 2020, 188, 109732. [Google Scholar] [CrossRef]
- Schnurr, R.E.; Alboiu, V.; Chaudhary, M.; Corbett, R.A.; Quanz, M.E.; Sankar, K.; Srain, H.S.; Thavarajah, V.; Xanthos, D.; Walker, T.R. Reducing marine pollution from single-use plastics (SUPs): A review. Mar. Pollut. Bull 2018, 137, 157–171. [Google Scholar] [CrossRef]
- Fadare, O.O.; Okoffo, E.D. COVID-19 face masks: A potential source of microplastic fibers in the environment. Sci. Total Environ. 2020, 737, 140279. [Google Scholar] [CrossRef]
- Gereffi, G. What does the COVID-19 pandemic teach us about global value chains? The case of medical supplies. J. Int. Bus. Policy 2020, 3, 287–301. [Google Scholar]
- Islam, A.; Ferdous, J.; Islam, S.; Sayeed, M.A.; Rahman, M.K.; Saha, O.; Hassan, M.M.; Shirin, T. Transmission dynamics and susceptibility patterns of SARS-CoV-2 in domestic, farmed and wild animals: Sustainable One Health surveillance for conservation and public health to prevent future epidemics and pandemics. Transbound. Emerg. Dis. 2021, 69, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Montagutelli, X.; van der Werf, S.; Rey, F.A.; Simon-Loriere, E. SARS-CoV-2 Omicron emergence urges for reinforced One-Health surveillance. EMBO Mol. Med. 2022, 14, e15558. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).