Simultaneous Detection of Seven Human Coronaviruses by Multiplex PCR and MALDI-TOF MS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Target Gene Selection
2.2. Primer Design
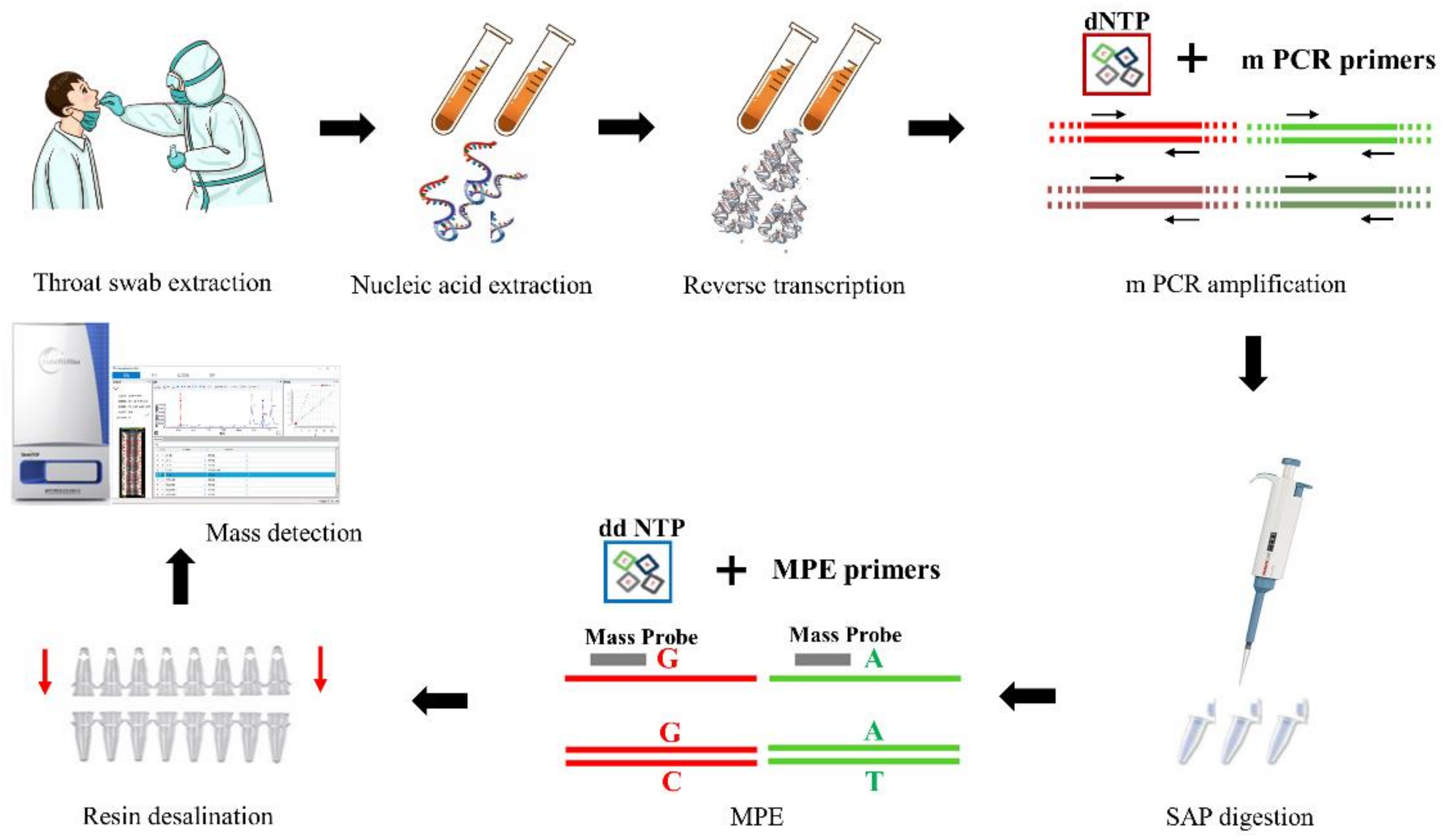
2.3. HCoV-MS Method Establishment
2.4. Protocol for the HCoV-MS Method
2.5. Testing the HCoV-MS Method
2.6. Application of the HCoV-MS Method
2.7. Comparison of the HCoV-MS and Real-Time PCR Methods
3. Results
3.1. Performance of the Hcov-MS Method
3.2. Specificity of the HCoV-MS Method
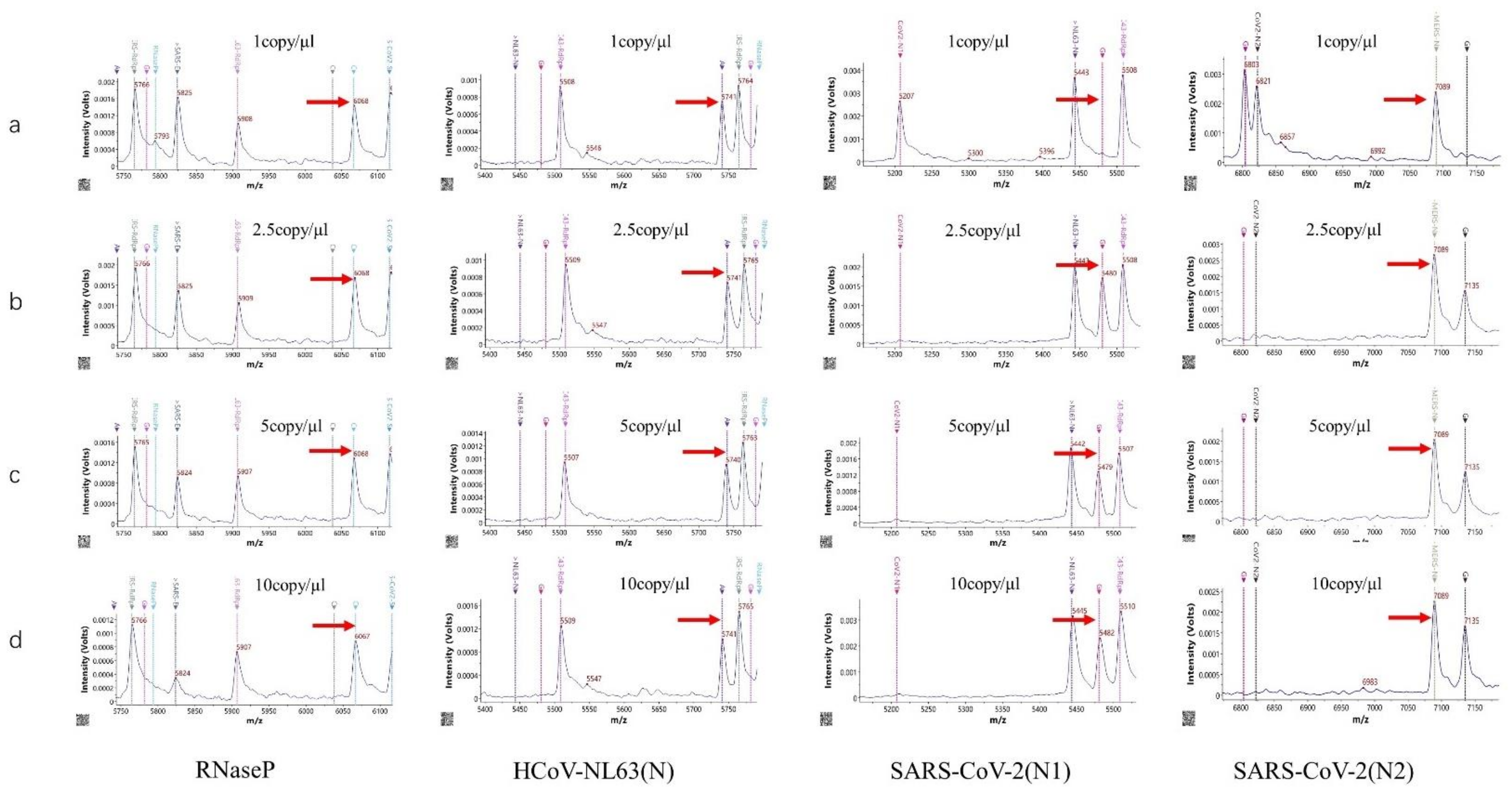
3.3. Sensitivity of the HCoV-MS Method
3.4. Screening Clinical Samples by the HCoV-MS Method
3.5. Sensitivity Comparisons of HCoV-MS and RT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 2017, 8, 1510. [Google Scholar] [CrossRef] [PubMed]
- Ezhilan, M.; Suresh, I.; Nesakumar, N. SARS-CoV, MERS-CoV and SARS-CoV-2: A Diagnostic Challenge. Measurement 2021, 168, 108335. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Miller, K.; McGrath, M.E.; Hu, Z.; Ariannejad, S.; Weston, S.; Frieman, M.; Jackson, W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy 2020, 16, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Younes, N.; Al-Sadeq, D.W.; Al-Jighefee, H.; Younes, S.; Al-Jamal, O.; Daas, H.I.; Yassine, H.M.; Nasrallah, G.K. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2. Viruses 2020, 12, 582. [Google Scholar] [CrossRef]
- Xiao-Shuang, Z.; Zeng, L.-P.; Yang, X.-L.; Ge, X.-Y.; Zhang, W.; Lin-Fa, W.; Xie, J.-Z.; Dong-Sheng, L.; Zhang, Y.-Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Tang, Y.-W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef]
- Gao, J.; Quan, L. Current Status of Diagnostic Testing for SARS-CoV-2 Infection and Future Developments: A Review. Med. Sci. Monit. 2020, 26. [Google Scholar] [CrossRef]
- Appak, Ö.; Duman, M.; Belet, N.; Sayiner, A.A. Viral respiratory infections diagnosed by multiplex polymerase chain reaction in pediatric patients. J. Med. Virol. 2019, 91, 731–737. [Google Scholar] [CrossRef]
- Gilsenan-Reed, C.; Higgins, G.; Langlois, N. Determining a sampling regime for PCR detection of respiratory tract viral infection at coronial post-mortem examinations. Forensic Sci. Med. Pathol. 2020, 16, 457–462. [Google Scholar] [CrossRef]
- Pabbaraju, K.; Wong, A.A.; Ma, R.; Zelyas, N.; Tipples, G.A. Development and validation of a multiplex reverse transcriptase-PCR assay for simultaneous testing of influenza A, influenza B and SARS-CoV-2. J. Virol. Methods 2021, 293, 114151. [Google Scholar] [CrossRef]
- Li, J.; Mao, N.-Y.; Zhang, C.; Yang, M.-J.; Wang, M.; Xu, W.-B.; Ma, X.-J. The development of a GeXP-based multiplex reverse transcription-PCR assay for simultaneous detection of sixteen human respiratory virus types/subtypes. BMC Infect. Dis. 2012, 12, 189. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Muenker, M.C.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef]
- Nörz, D.; Hoffmann, A.; Aepfelbacher, M.; Pfefferle, S.; Lütgehetmann, M. Clinical evaluation of a fully automated, laboratory-developed multiplex RT-PCR assay integrating dual-target SARS-CoV-2 and influenza A/B detection on a high-throughput platform. J. Med. Microbiol. 2021, 70, 001295. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Corsetti, A. The use of multiplex PCR to detect and differentiate food- and beverage-associated microorganisms: A review. J. Microbiol. Methods 2007, 69, 1–22. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, L.; Cai, G.; Zhang, D.; Lin, J. Establishment of a simultaneous detection method for ten duck viruses using MALDI-TOF mass spectrometry. J. Virol. Methods 2019, 273, 113723. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, J.; Wang, X.; Liu, L.; Gong, J.; Zhai, Z.; He, L.; Meng, F.; Xiao, D. A multisite SNP genotyping and macrolide susceptibility gene method for Mycoplasma pneumoniae based on MALDI-TOF MS. iScience 2021, 24, 102447. [Google Scholar] [CrossRef]
- Zhang, C.; Xiao, Y.; Du, J.; Ren, L.; Wang, J.; Peng, J.; Jin, Q. Application of Multiplex PCR Coupled with Matrix-Assisted Laser Desorption Ionization–Time of Flight Analysis for Simultaneous Detection of 21 Common Respiratory Viruses. J. Clin. Microbiol. 2015, 53, 2549–2554. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.C.-M.; Chow, V.C.-Y.; Lee, M.K.-P.; Tang, K.P.-S.; Li, D.K.-C.; Lai, R.W.-M. Evaluation of the Xpert Xpress SARS-CoV-2/Flu/RSV Assay for Simultaneous Detection of SARS-CoV-2, Influenza A and B Viruses, and Respiratory Syncytial Virus in Nasopharyngeal Specimens. J. Clin. Microbiol. 2021, 59, e02965-20. [Google Scholar] [CrossRef]
- Paradis, S.; Lockamy, E.; Cooper, C.K.; Young, S. Clinical evaluation of the molecular-based BD SARS-CoV-2/flu for the BD MAX™ System. J. Clin. Virol. 2021, 143, 104946. [Google Scholar] [CrossRef]
- Xi, Y.; Xu, C.-Z.; Xie, Z.-Z.; Zhu, D.-L.; Dong, J.-M.; Xiao, G. Development of a reverse transcription recombinase polymerase amplification assay for rapid detection of human respiratory syncytial virus. Mol. Cell. Probes 2019, 45, 8–13. [Google Scholar] [CrossRef]
- Zhang, W.S.; Pan, J.; Li, F.; Zhu, M.; Xu, M.; Zhu, H.; Yu, Y.; Su, G. Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Facile and Highly Sensitive Colorimetric SARS-CoV-2 Detection. Anal. Chem. 2021, 93, 4126–4133. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Fu, Y.; Xu, M.; Su, L.; Cao, L.; Xu, J.; Cheng, X. Evaluation of a PCR/ESI-MS platform to identify respiratory viruses from nasopharyngeal aspirates. J. Med Virol. 2015, 87, 1867–1871. [Google Scholar] [CrossRef]
- Aoki, A.; Mori, Y.; Okamoto, Y.; Jinno, H. Development of a genotyping platform for SARS-CoV-2 variants using high-resolution melting analysis. J. Infect. Chemother. 2021, 27, 1336–1341. [Google Scholar] [CrossRef]
- Lope, P.; Maribel, H.; Egma, M.; Henri, B.; Carlos, P. Characterization of influenza A(H1N1)pdm09 isolates of Peru using HRM, a post PCR molecular biology method. Bioinformation 2019, 15, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, M.; Li, X.; Zhang, D.; Chen, C.; Feng, Z.; Ma, X. Clinical evaluation of the isothermal amplification assays for the detection of four common respiratory viruses in children with pneumonia. Arch. Virol. 2017, 162, 1311–1318. [Google Scholar] [CrossRef]
- Chau, C.H.; Strope, J.; Figg, W.D. COVID-19 Clinical Diagnostics and Testing Technology. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2020, 40, 857–868. [Google Scholar] [CrossRef] [PubMed]
- U.S. CDC. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 15 December 2021).
- ECDC. SARS-CoV-2 Variants of Concern as of 15 December 2021. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 15 December 2021).

| Target Gene | mPCR Primer (F) 1 | mPCR Primer (R) 1 | MPE Primer |
|---|---|---|---|
| Human RNase P | AGATTTGGACCTGCGAGCG | GAGCGGCTGTCTCCACAAGT | CAGTAGCTGTTTCTGAACT |
| HCoV-229E-N | ACGGTGTTAGGCGCAAGAAT | AGGAGCACGGGAGTCAGGTT | CCAACCAGAGATACCACACTTCAA |
| HCoV-229E-RdRp | GGACCACGAGCAGTCCATGT | GTTCTGCCCTCATGCCAAGT | CCCCATGTATAACTTACTTAAAGG |
| HCoV-NL63-N | GTTGCTGCTGTTACTTTGGCT | CCCTGGGTTGAGAAAGAGGCT | TACCAGTCGAAGTCACCT |
| HCoV-NL63-RdRp | CACTTGTTACAACTGCTGGTT | TGTCAACCTAACTGARTGTGT | GAAAGCAATTAGGTTTGGT |
| HCoV-OC43-N | AGCAACCAGGCTGATGTCAA | GGCGGAAACCTAGTCGGAAT | ATTCGCTACTTGGGTCCCGAT |
| HCoV-OC43-RdRp | CCCAGGATGTGGTGTTGCTAT | CGCAATCCAATGCATGACACA | CGCATGACACATGGTCAG |
| HCoV-HKU1-N | ACTCCCGGTCATYATGCTGG | TTCGYTCAGATTGGTCARCC | GGAGAAGTTTTCTTGAGGATT |
| HCoV-HKU1-RdRp | ACACACCGYTATCGTTTGTCT | CAAGCAGAGCACTAGCAGATG | GGGTGCATAGCAGGATCTGCTGCATA |
| MERS-CoV-N | TTGGCGGAGACAGGACAGAA | GGAATGGGAGTGCTGCTTCG | CCAAAATTAATACCGGGAATGGA |
| MERS-CoV-E | ACACTCTTGGTGTGTATGGCT | GCGGGCTGAACTAACAGGGTA | CCGGCTACTAGATTATGTGTGCAAT |
| MERS-CoV-RdRp | GGAGAACGTGTACGCCAAGC | AGCACACCGACTAAACCAGC | AGCCAAGCTATCTTAAACA |
| MERS-CoV-ORF1b | GCTGCTCTTCTTGCCGGTTC | TGGTCAAGGGCTGTGCATCA | CCCCACAGGGTCATCAACAAT |
| SARS-CoV-N | TGATGAAGCTCAGCCTTTGCC | AATCATCCATGTCAGCCGCAG | GGGCAGAGACAAAAGAAGCAGCCC |
| SARS-CoV-E | GCCATCCTTACTGCGCTTCG | ACGCGAGTAGACGTAAACCG | TCTTGTTAACGTGAGTTTA |
| SARS-CoV-ORF1b | AGAAACGCCCGTAATGGTGT | CTAGCTTGTGCTGGTCCCTT | GGGTTTTAATAACAGAAGGTTCAGT |
| SARS-CoV2-ORF1b | TGCTGTAGATGCTGCTAAAGC | GCCTGACCAGTACCAGTGTG | CCCATCTTAACACAATTAGTGATTGG |
| SARS-CoV2-N1 | AGAATGGAGAACGCAGTGGG | CGGTGAACCAAGACGCAGTA | CGACGTTGTTTTGATCG |
| SARS-CoV2-N2 | CAACTCCAGGCAGCAGTAGG | TGTCAAGCAGCAGCAAAGCA | TCTGGCTGGCAATGGCGGTGAT |
| SARS-CoV2-S | ACAGGCACAGGTGTTCTTACT | TGGATCACGGACAGCATCAGT | CTCAATTTGGCAGAGACATT |
| Assays | Target | Detection Limit (Copies/Reaction) |
|---|---|---|
| Human RNase P | Human RNase P | 1 |
| HCoV-NL63 | N | 1 |
| RdRp | 1 | |
| HCoV-229E | N | 2.5 |
| RdRp | 2.5 | |
| HCoV-OC43 | N | 2.5 |
| RdRp | 2.5 | |
| HCoV-HKU1 | N | 1 |
| RdRp | 2.5 | |
| MERS-CoV | N | 1 |
| RdRp | 2.5 | |
| E | 2.5 | |
| ORF1b | 2.5 | |
| SARS-CoV | E | 5 |
| N | 5 | |
| ORF1b | 5 | |
| SARS-CoV-2 | N1 | 2.5 |
| N2 | 2.5 | |
| S | 2.5 | |
| ORF1b | 2.5 |
| Name | HCoV-MS | RT-PCR | ||||
|---|---|---|---|---|---|---|
| pos 1 | F-pos 2 | neg 3 | F-neg 4 | pos | neg | |
| HCoV-229E | 1 | 0 | 0 | 0 | 1 | 0 |
| HCoV-NL63 | 1 | 0 | 0 | 0 | 1 | 0 |
| SARS-CoV | 2 | 0 | 0 | 0 | 2 | 0 |
| SARS-CoV-2 | 95 | 0 | 0 | 0 | 95 | 0 |
| —— | - | - | 64 | - | - | 64 |
| Sample | Gradient | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||||||
| RT-PCR * | HCoV-MS * | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | |
| 1 | No Ct/No Ct | − 1 | No Ct/No Ct | − | 40.74/No Ct | + 2 | 36.87/No Ct | + | 34.17/No Ct | + |
| 2 | No Ct/No Ct | − | No Ct/No Ct | − | 40.65/No Ct | − | 36.30/No Ct | + | 34.36/No Ct | + |
| 3 | 35.94/No Ct | − | 32.84/No Ct | − | 33.35/No Ct | − | 32.56/No Ct | + | 30.16/No Ct | + |
| 4 | 36.88/34.01 | + | 35.96/32.03 | + | 32.92/29.77 | + | 31.24/27.63 | + | 28.52/25.92 | + |
| 5 | 39.64/33.68 | + | 37.01/32.01 | + | 34.79/30.05 | + | 33.03/27.92 | + | 30.6/25.83 | + |
| 6 | 37.33/35.04 | + | 36.67/32.81 | + | 35.45/31.17 | + | 34.28/29.21 | + | 32.9/27.14 | + |
| 7 | 40.63/34.79 | + | 40.11/32.60 | + | 38.66/30.97 | + | 35.83/28.46 | + | 33.83/26.86 | + |
| 8 | No Ct/No Ct | − | No Ct/39.02 | − | 39.35/36.45 | − | 36.3/34.71 | + | 35.15/32.30 | + |
| 9 | No Ct/No Ct | − | No Ct/No Ct | − | No Ct/No Ct | − | 39.33/36.50 | + | 36.32/34.60 | + |
| 10 | No Ct/No Ct | − | 37.23/37.43 | − | 37.06/34.76 | + | 35.16/32.90 | + | 33.24/30.99 | + |
| 11 | 39.02/36.78 | − | 36.35/33.84 | + | 34.35/31.44 | + | 32.36/30.16 | + | 30.66/28.16 | + |
| 12 | No Ct/No Ct | + | No Ct/No Ct | + | No Ct/38.39 | + | 35.95/35.15 | + | 34.61/32.06 | + |
| 13 | No Ct/No Ct | + | No Ct/No Ct | + | No Ct/No Ct | + | 34.99/No Ct | + | 33.94/38.01 | + |
| 14 | No Ct/No Ct | − | No Ct/No Ct | − | No Ct/38.16 | + | 37.12/35.66 | + | 34.52/33.22 | + |
| 15 | No Ct/No Ct | + | 37.15/36.10 | + | 37.04/33.67 | + | 35.26/31.76 | + | 33.01/30.57 | + |
| 16 | No Ct/No Ct | + | 40.81/No Ct | + | No Ct/38.45 | + | 39.89/36.62 | + | 37.84/35.04 | + |
| 17 | No Ct/No Ct | − | No Ct/39.73 | − | 39.31/37.82 | + | 37.87/36.36 | + | 36.43/34.64 | + |
| 18 | No Ct/No Ct | − | No Ct/40.08 | − | No Ct/39.48 | − | 42.18/38.98 | + | 40.41/36.18 | + |
| 19 | No Ct/No Ct | + | 37.22/36.00 | + | 36.90/34.46 | + | 34.81/33.69 | + | 32.92/31.26 | + |
| 20 | 37.80/32.63 | + | 36.22/30.56 | + | 33.91/28.28 | + | 32.24/26.46 | + | 29.53/23.98 | + |
| 21 | 39.93/34.69 | + | 36.10/32.38 | + | 32.28/30.01 | + | 30.51/27.60 | + | 27.69/25.37 | + |
| 22 | No Ct/No Ct | − | No Ct/38.56 | + | 38.56/39.32 | + | 37.10/34.75 | + | 36.13/33.54 | + |
| 23 | No Ct/No Ct | − | 35.15/No Ct | − | 34.93/No Ct | − | 34.15/38.72 | − | 30.79/35.09 | + |
| 24 | No Ct/No Ct | − | 39.17/36.95 | + | 36.61/34.35 | + | 35.95/31.70 | + | 33.32/29.73 | + |
| 25 | No Ct/No Ct | − | No Ct/No Ct | + | No Ct/38.88 | + | 36.66/37.20 | + | 33.94/35.70 | + |
| 26 | No Ct/No Ct | − | 36.46/No Ct | + | 34.97/38.94 | + | 35.39/35.67 | + | 33.65/34.16 | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Kang, L.; Li, Y.; Huang, J.; Guo, Z.; Xu, J.; Hu, Y.; Zhai, Z.; Kang, X.; Jiang, T.; et al. Simultaneous Detection of Seven Human Coronaviruses by Multiplex PCR and MALDI-TOF MS. COVID 2022, 2, 5-17. https://doi.org/10.3390/covid2010002
Liu T, Kang L, Li Y, Huang J, Guo Z, Xu J, Hu Y, Zhai Z, Kang X, Jiang T, et al. Simultaneous Detection of Seven Human Coronaviruses by Multiplex PCR and MALDI-TOF MS. COVID. 2022; 2(1):5-17. https://doi.org/10.3390/covid2010002
Chicago/Turabian StyleLiu, Tingting, Lin Kang, Yanwei Li, Jing Huang, Zishuo Guo, Jinglin Xu, Yi Hu, Zhixiang Zhai, Xiaoping Kang, Tao Jiang, and et al. 2022. "Simultaneous Detection of Seven Human Coronaviruses by Multiplex PCR and MALDI-TOF MS" COVID 2, no. 1: 5-17. https://doi.org/10.3390/covid2010002
APA StyleLiu, T., Kang, L., Li, Y., Huang, J., Guo, Z., Xu, J., Hu, Y., Zhai, Z., Kang, X., Jiang, T., Li, H., Song, H., Wang, J., Gao, S., Li, J., Zhou, X., Yuan, Y., Zhao, B., Wang, J., & Xin, W. (2022). Simultaneous Detection of Seven Human Coronaviruses by Multiplex PCR and MALDI-TOF MS. COVID, 2(1), 5-17. https://doi.org/10.3390/covid2010002






