Evaluation of an Antigen Detection Rapid Diagnostic Test for Detection of SARS-CoV-2 in Clinical Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Test Evaluation
2.3. Virus Isolates
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schalk, A.F.; Hawn, M.C. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931, 78, 413–423. [Google Scholar]
- Kendall, E.J.C.; Bynoe, M.L.; Tyrrell, D.A.J. Virus Isolations from Common Colds Occurring in a Residential School. BMJ 1962, 2, 82–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965, 1, 1467–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, P.S. The Molecular Biology of Coronaviruses; Elsevier: Amsterdam, The Netherlands, 2006; pp. 193–292. [Google Scholar]
- Brian, D.A.; Baric, R.S. Coronavirus Genome Structure and Replication; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–30. [Google Scholar]
- ICTV. Virus Taxonomy: 2020 Release. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 1 November 2021).
- Gorbalenya, A.; Baker, S.; Baric, R.; de Groot, R.; Drosten, C.; Gulyaeva, A.; Haagmans, B.; Lauber, C.; Leontovich, A.; Neuman, B.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.; Sridhar, S.; Chiu, K.H.; Hung, D.L.; Li, X.; Hung, I.F.; Tam, A.R.; Chung, T.W.; Chan, J.F.; Zhang, A.J.; et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg. Microbes. Infect. 2021, 10, 507–535. [Google Scholar] [CrossRef]
- WHO. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int./ (accessed on 1 November 2021).
- Hellewell, J.; Abbott, S.; Gimma, A.; Bosse, N.I.; Jarvis, C.I.; Russell, T.W.; Munday, J.D.; Kucharski, A.J.; Edmunds, W.J.; Funk, S.; et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob. Health 2020, 8, e488–e496. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [Green Version]
- Hung, I.F.-N.; Cheng, V.C.-C.; Li, X.; Tam, A.R.; Hung, D.L.-L.; Chiu, K.H.-Y.; Yip, C.C.-Y.; Cai, J.-P.; Ho, D.T.-Y.; Wong, S.-C.; et al. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: A case series. Lancet Infect. Dis. 2020, 20, 1051–1060. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- McCullough, P.A.; Kelly, R.J.; Ruocco, G.; Lerma, E.; Tumlin, J.; Wheelan, K.R.; Katz, N.; Lepor, N.E.; Vijay, K.; Carter, H.; et al. Pathophysiological Basis and Rationale for Early Outpatient Treatment of SARS-CoV-2 (COVID-19) Infection. Am. J. Med. 2021, 134, 16–22. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef]
- WHO. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021, 3, CD013705. [Google Scholar] [CrossRef]
- Chan, K.H.; Sridhar, S.; Zhang, R.R.; Chu, H.; Fung, A.Y.; Chan, G.; Chan, J.F.; To, K.K.; Hung, I.F.; Cheng, V.C.; et al. Factors affecting stability and infectivity of SARS-CoV-2. J. Hosp. Infect. 2020, 106, 226–231. [Google Scholar] [CrossRef]
- Yip, C.C.-Y.; Sridhar, S.; Cheng, A.K.-W.; Leung, K.-H.; Choi, G.K.-Y.; Chen, J.H.-K.; Poon, R.W.-S.; Chan, K.-H.; Wu, A.K.-L.; Chan, H.S.-Y.; et al. Evaluation of the commercially available LightMix® Modular E-gene kit using clinical and proficiency testing specimens for SARS-CoV-2 detection. J. Clin. Virol. 2020, 129, 104476. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yip, C.C.-Y.; To, K.K.-W.; Tang, T.H.-C.; Wong, S.C.-Y.; Leung, K.-H.; Fung, A.Y.-F.; Ng, A.C.-K.; Zou, Z.; Tsoi, H.-W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef] [Green Version]
- MacKay, M.J.; Hooker, A.C.; Afshinnekoo, E.; Salit, M.; Kelly, J.; Feldstein, J.V.; Haft, N.; Schenkel, D.; Nambi, S.; Cai, Y.; et al. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat. Biotechnol. 2020, 38, 1021–1024. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Bullard, J.; Dust, K.; Funk, D.; Strong, J.E.; Alexander, D.; Garnett, L.; Boodman, C.; Bello, A.; Hedley, A.; Schiffman, Z.; et al. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin. Infect. Dis. 2020, 71, 2663–2666. [Google Scholar] [CrossRef] [PubMed]
- Maltezou, H.C.; Raftopoulos, V.; Vorou, R.; Papadima, K.; Mellou, K.; Spanakis, N.; Kossyvakis, A.; Gioula, G.; Exindari, M.; Froukala, E.; et al. Association Between Upper Respiratory Tract Viral Load, Comorbidities, Disease Severity, and Outcome of Patients With SARS-CoV-2 Infection. J. Infect. Dis. 2021, 223, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Yokota, I.; Sakurazawa, T.; Sugita, J.; Iwasaki, S.; Yasuda, K.; Yamashita, N.; Fujisawa, S.; Nishida, M.; Konno, S.; Teshima, T. Performance of Qualitative and Quantitative Antigen Tests for SARS-CoV-2 Using Saliva. Infect. Dis. Rep. 2021, 13, 742–747. [Google Scholar] [CrossRef]
- Khandker, S.S.; Nik Hashim, N.H.H.; Deris, Z.Z.; Shueb, R.H.; Islam, M.A. Diagnostic Accuracy of Rapid Antigen Test Kits for Detecting SARS-CoV-2: A Systematic Review and Meta-Analysis of 17,171 Suspected COVID-19 Patients. J. Clin. Med. 2021, 10, 3493. [Google Scholar] [CrossRef]
- Peto, T.; Affron, D.; Afrough, B.; Agasu, A.; Ainsworth, M.; Allanson, A.; Allen, K.; Allen, C.; Archer, L.; Ashbridge, N.; et al. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021, 36, 100924. [Google Scholar] [CrossRef]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Liotti, F.M.; Menchinelli, G.; Lalle, E.; Palucci, I.; Marchetti, S.; Colavita, F.; La Sorda, M.; Sberna, G.; Bordi, L.; Sanguinetti, M.; et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin. Microbiol. Infect. 2021, 27, 487–488. [Google Scholar] [CrossRef]
- Weitzel, T.; Legarraga, P.; Iruretagoyena, M.; Pizarro, G.; Vollrath, V.; Araos, R.; Munita, J.M.; Porte, L. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med. Infect. Dis. 2021, 39, 101942. [Google Scholar] [CrossRef]
- Mertens, P.; De Vos, N.; Martiny, D.; Jassoy, C.; Mirazimi, A.; Cuypers, L.; Van den Wijngaert, S.; Monteil, V.; Melin, P.; Stoffels, K.; et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020, 7, 225. [Google Scholar] [CrossRef]
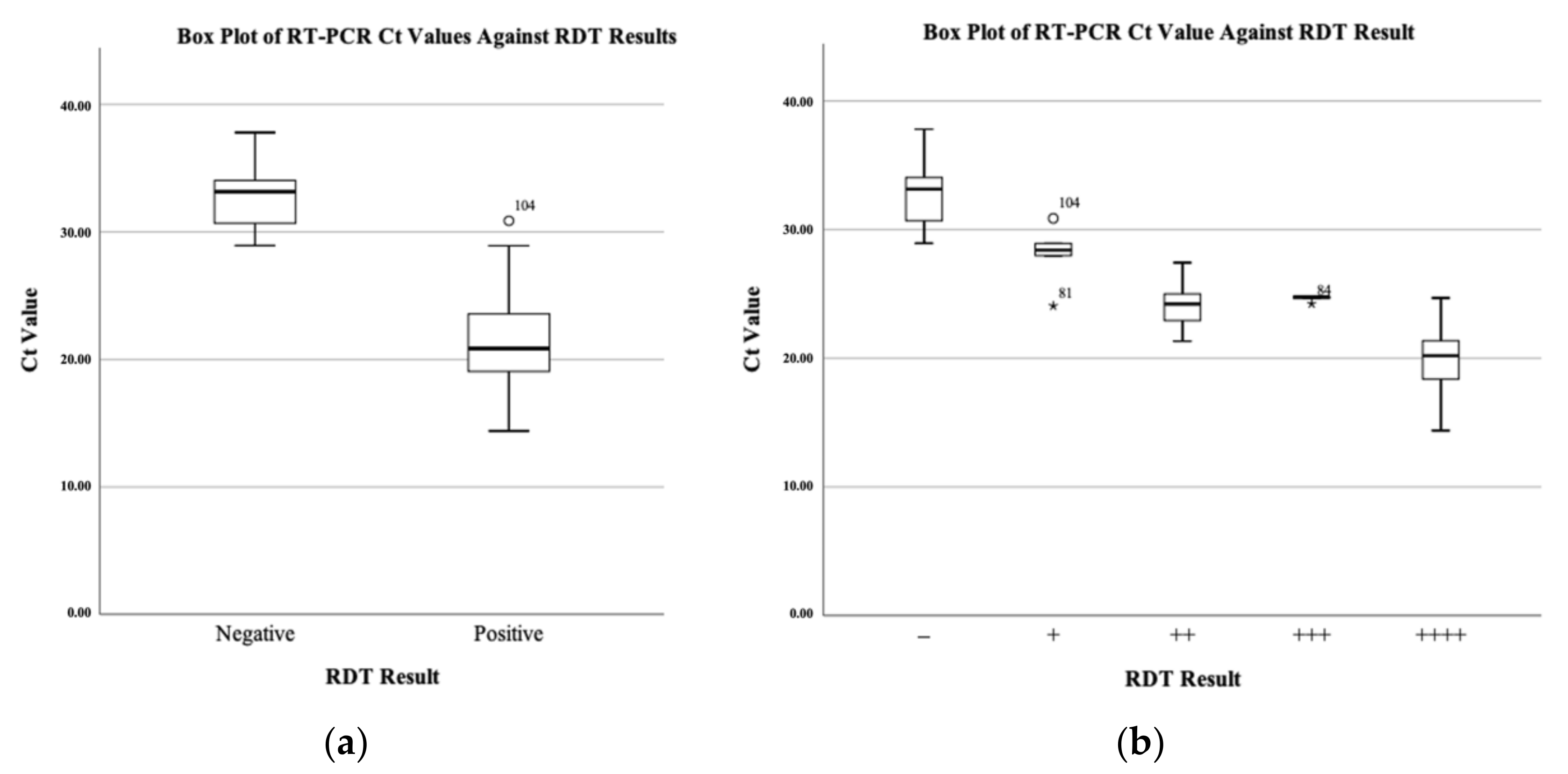
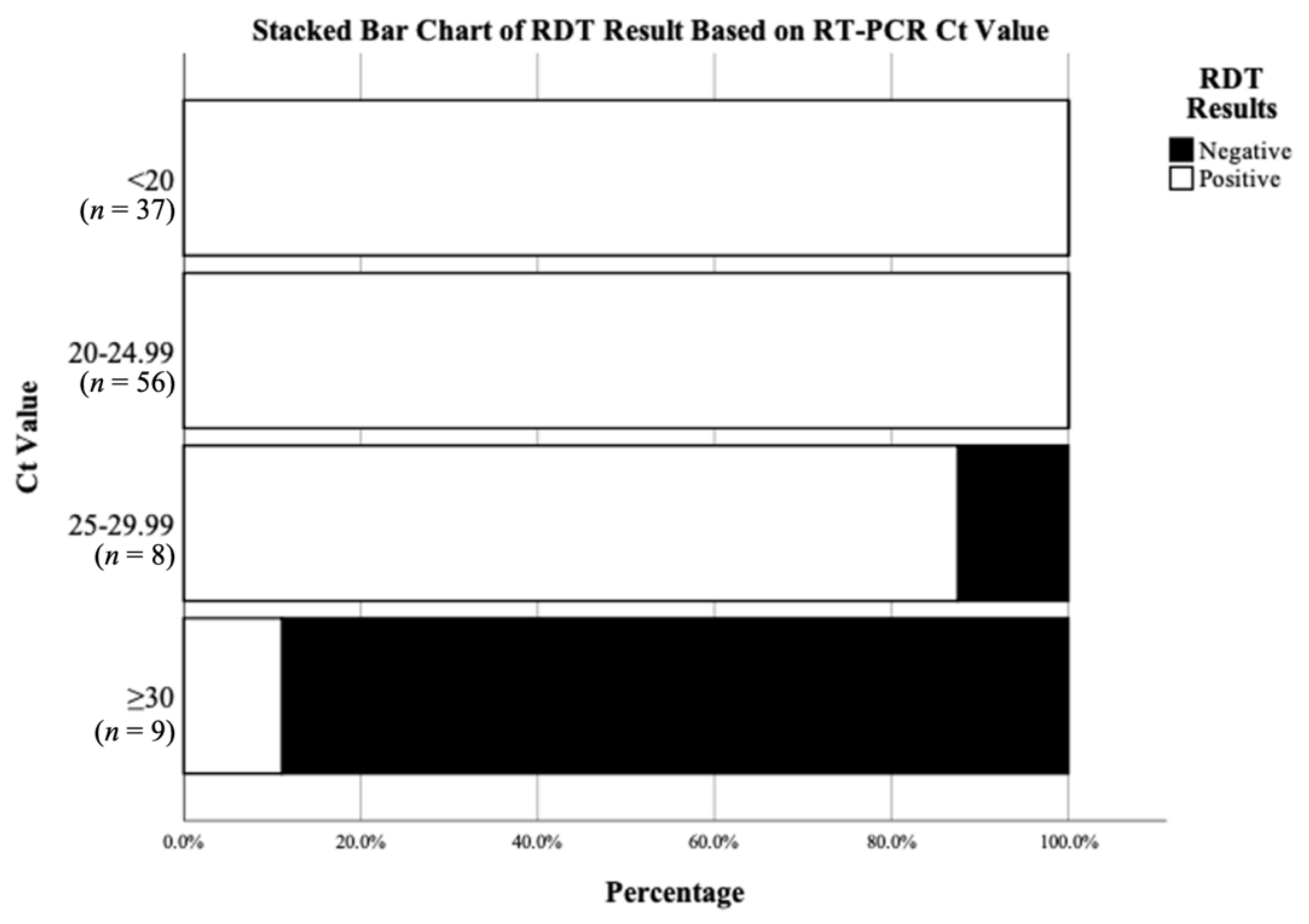
| Parameters | Results [95% Confidence Interval (CI)] |
|---|---|
| Overall Sensitivity | 92% (85–96%) |
| Sensitivity in Ct < 20 samples | 100% (91–100%) |
| Sensitivity in 20 ≤ Ct < 25 samples | 100% (94–100%) |
| Sensitivity in 25 ≤ Ct < 30 samples | 88% (47–100%) |
| Sensitivity in Ct ≥ 30 samples | 11% (0–48%) |
| Specificity | 97% (85–100%) |
| Positive predictive value (PPV) | 99% (94–100%) |
| Negative predictive value (NPV) | 79% (66–87%) |
| Youden index | 0.89 |
| Variant | Sample Type | Ct Value | RDT Result |
|---|---|---|---|
| Alpha | Deep throat saliva | 18.1 | ++++ |
| Alpha | Deep throat saliva | 19.9 | ++++ |
| Alpha | Deep throat saliva | 21.0 | ++++ |
| Alpha | Deep throat saliva | 35.0 | – |
| Beta | Deep throat saliva | 21.0 | ++++ |
| Gamma | Deep throat saliva | 15.0 | ++++ |
| Delta | Deep throat saliva | 16.3 | ++++ |
| Delta | Deep throat saliva | 18.1 | ++++ |
| Delta | NPS | 21.3 | +++ |
| Eta | NPS | 25.0 | +++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, A.H.-Y.; Leung, K.-Y.; Zhang, R.; Liu, D.; Fan, Y.; Tam, A.R.; Yip, C.C.-Y.; Cheng, V.C.-C.; Yuen, K.-Y.; Hung, I.F.-N.; et al. Evaluation of an Antigen Detection Rapid Diagnostic Test for Detection of SARS-CoV-2 in Clinical Samples. COVID 2021, 1, 775-783. https://doi.org/10.3390/covid1040062
Lam AH-Y, Leung K-Y, Zhang R, Liu D, Fan Y, Tam AR, Yip CC-Y, Cheng VC-C, Yuen K-Y, Hung IF-N, et al. Evaluation of an Antigen Detection Rapid Diagnostic Test for Detection of SARS-CoV-2 in Clinical Samples. COVID. 2021; 1(4):775-783. https://doi.org/10.3390/covid1040062
Chicago/Turabian StyleLam, Athene Hoi-Ying, Ka-Yi Leung, Ruiqi Zhang, Danlei Liu, Yujing Fan, Anthony Raymond Tam, Cyril Chik-Yan Yip, Vincent Chi-Chung Cheng, Kwok-Yung Yuen, Ivan Fan-Ngai Hung, and et al. 2021. "Evaluation of an Antigen Detection Rapid Diagnostic Test for Detection of SARS-CoV-2 in Clinical Samples" COVID 1, no. 4: 775-783. https://doi.org/10.3390/covid1040062
APA StyleLam, A. H.-Y., Leung, K.-Y., Zhang, R., Liu, D., Fan, Y., Tam, A. R., Yip, C. C.-Y., Cheng, V. C.-C., Yuen, K.-Y., Hung, I. F.-N., & Chan, K.-H. (2021). Evaluation of an Antigen Detection Rapid Diagnostic Test for Detection of SARS-CoV-2 in Clinical Samples. COVID, 1(4), 775-783. https://doi.org/10.3390/covid1040062








