The Potential Functions of Protein Domains during COVID Infection: An Analysis and a Review
Abstract
:1. Introduction
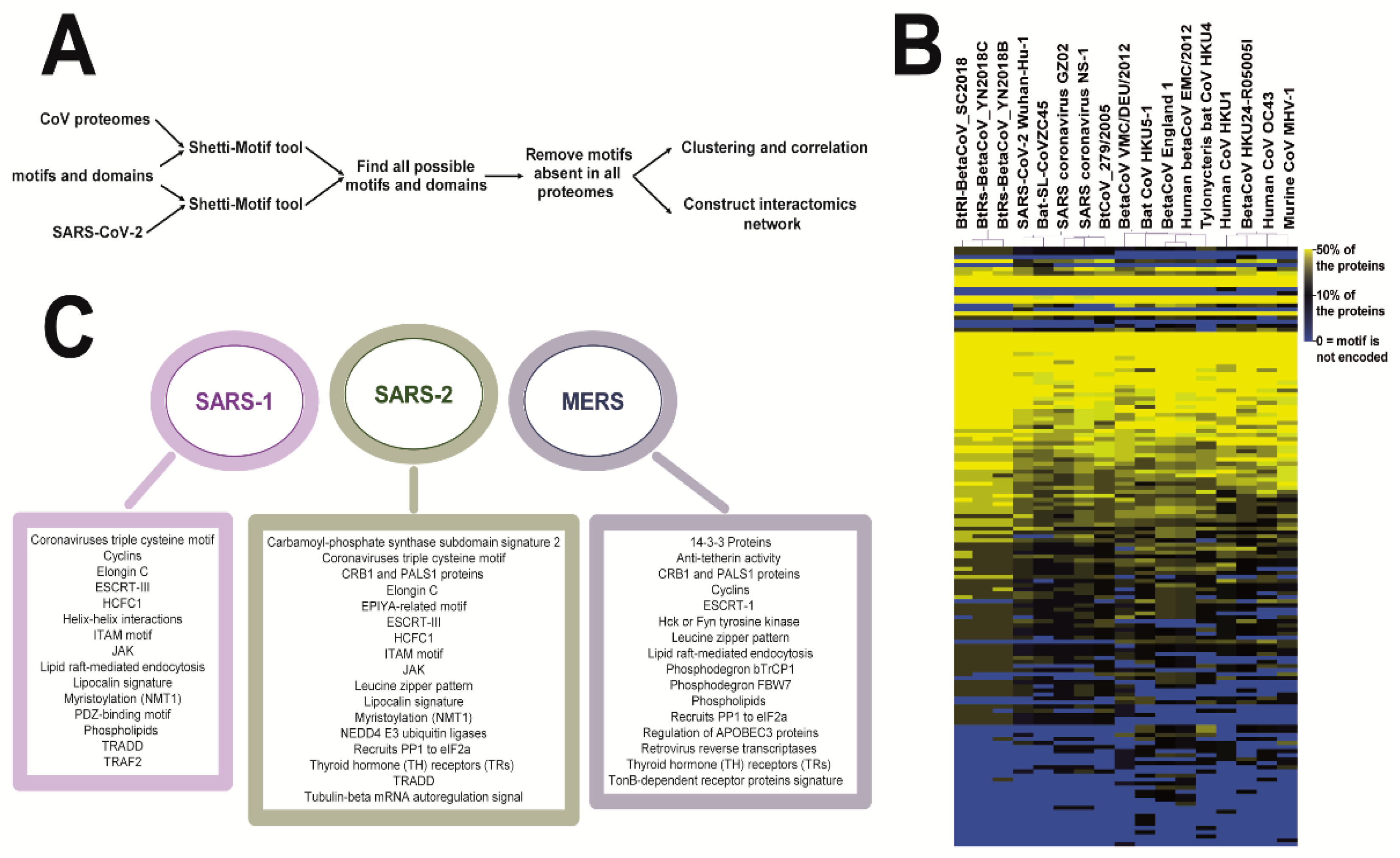
2. Materials and Methods
3. Results
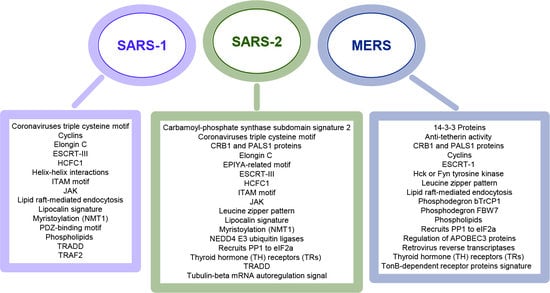
3.1. Coronaviruses Encode Wide-Range of Functional Motifs
3.2. The Potential CoV-Human Protein Interactions Pathways Based on Functional Motif
3.3. The SARS-CoV-2 Nsp1 Protein Encodes for a Tubulin MREL Motif
4. Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, K.; Huang, C.; Makino, S. Sars coronavirus accessory proteins. Virus Res. 2008, 133, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From sars to mers, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Sobhy, H. Virophages and their interactions with giant viruses and host cells. Proteomes 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobhy, H. A bioinformatics pipeline to search functional motifs within whole-proteome data: A case study of poxviruses. Virus Genes 2017, 53, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobhy, H. A review of functional motifs utilized by viruses. Proteomes 2016, 4, 3. [Google Scholar] [CrossRef] [Green Version]
- Davey, N.E.; Trave, G.; Gibson, T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hraber, P.; O’Maille, P.E.; Silberfarb, A.; Davis-Anderson, K.; Generous, N.; McMahon, B.H.; Fair, J.M. Resources to discover and use short linear motifs in viral proteins. Trends Biotechnol. 2020, 38, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, R.L.; Urbe, S. The emerging shape of the escrt machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Sette, P.; Jadwin, J.A.; Dussupt, V.; Bello, N.F.; Bouamr, F. The escrt-associated protein alix recruits the ubiquitin ligase nedd4-1 to facilitate hiv-1 release through the lypxnl l domain motif. J. Virol. 2010, 84, 8181–8192. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Madara, J.J.; Liu, Y.; Liu, W.; Ruthel, G.; Freedman, B.D.; Harty, R.N. Alix rescues budding of a double ptap/ppey l-domain deletion mutant of ebola vp40: A role for alix in ebola virus egress. J. Infect. Dis. 2015, 212, S138–S145. [Google Scholar] [CrossRef]
- Wolff, S.; Ebihara, H.; Groseth, A. Arenavirus budding: A common pathway with mechanistic differences. Viruses 2013, 5, 528–549. [Google Scholar] [CrossRef]
- Conrad, K.P. Might proton pump or sodium-hydrogen exchanger inhibitors be of value to ameliorate SARS-CoV-2 pathophysiology? Physiol. Rep. 2021, 8, e14649. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Lu, H.; Wang, J.; He, X.; Liu, Y.; Ye, C.; Lin, W.; Hu, J.; Ji, J.; et al. The e protein is a multifunctional membrane protein of SARS-CoV. Genom. Proteom. Bioinform. 2003, 1, 131–144. [Google Scholar] [CrossRef] [Green Version]
- Okada, H.; Yoshida, S.; Hara, A.; Ogura, S.; Tomita, H. Vascular endothelial injury exacerbates coronavirus disease 2019: The role of endothelial glycocalyx protection. Microcirculation 2020, 28, e12654. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ace2. Cell 2020, 183, 1043–1057.e1015. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Liu, D.X. Proteolytic activation of the spike protein at a novel rrrr/s motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009, 83, 8744–8758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, S.M.; Karki, R.; Kanneganti, T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017, 277, 61–75. [Google Scholar] [CrossRef] [Green Version]
- Nainu, F.; Shiratsuchi, A.; Nakanishi, Y. Induction of apoptosis and subsequent phagocytosis of virus-infected cells as an antiviral mechanism. Front. Immunol. 2017, 8, 1220. [Google Scholar] [CrossRef]
- Sobocinska, J.; Roszczenko-Jasinska, P.; Ciesielska, A.; Kwiatkowska, K. Protein palmitoylation and its role in bacterial and viral infections. Front. Immunol. 2017, 8, 2003. [Google Scholar] [CrossRef] [Green Version]
- Corse, E.; Machamer, C.E. The cytoplasmic tail of infectious bronchitis virus e protein directs golgi targeting. J. Virol. 2002, 76, 1273–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Yuan, Q.; Torres, J.; Tam, J.P.; Liu, D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 2006, 349, 264–275. [Google Scholar] [CrossRef]
- Lopez, L.A.; Riffle, A.J.; Pike, S.L.; Gardner, D.; Hogue, B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008, 82, 3000–3010. [Google Scholar] [CrossRef] [Green Version]
- Udenwobele, D.I.; Su, R.C.; Good, S.V.; Ball, T.B.; Varma Shrivastav, S.; Shrivastav, A. Myristoylation: An important protein modification in the immune response. Front. Immunol. 2017, 8, 751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Zuckermann, F.A.; Yoo, D. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth. Virus Res. 2010, 147, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Lechuga, G.C.; Souza-Silva, F.; Sacramento, C.Q.; Trugilho, M.R.O.; Valente, R.H.; Napoleão-Pêgo, P.; Dias, S.S.G.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Carels, N.; et al. SARS-CoV-2 proteins bind to hemoglobin and its metabolites. Int. J. Mol. Sci. 2021, 22, 9035. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Javorsky, A.; Humbert, P.O.; Kvansakul, M. Structural basis of coronavirus e protein interactions with human pals1 pdz domain. Commun. Biol. 2021, 4, 724. [Google Scholar] [CrossRef]
- Chai, J.; Cai, Y.; Pang, C.; Wang, L.; McSweeney, S.; Shanklin, J.; Liu, Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein pals1. Nat. Commun. 2021, 12, 3433. [Google Scholar] [CrossRef]
- Teoh, K.T.; Siu, Y.L.; Chan, W.L.; Schluter, M.A.; Liu, C.J.; Peiris, J.S.; Bruzzone, R.; Margolis, B.; Nal, B. The sars coronavirus e protein interacts with pals1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell 2010, 21, 3838–3852. [Google Scholar] [CrossRef] [Green Version]
- Pelka, P.; Ablack, J.N.; Fonseca, G.J.; Yousef, A.F.; Mymryk, J.S. Intrinsic structural disorder in adenovirus e1a: A viral molecular hub linking multiple diverse processes. J. Virol. 2008, 82, 7252–7263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreon, J.C.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural basis for subversion of cellular control mechanisms by the adenoviral e1a oncoprotein. Proc. Natl. Acad. Sci. USA 2009, 106, 13260–13265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansieau, S.; Leutz, A. The conserved mynd domain of bs69 binds cellular and oncoviral proteins through a common pxlxp motif. J. Biol. Chem. 2002, 277, 4906–4910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef]
- Gasic, I.; Mitchison, T.J. Autoregulation and repair in microtubule homeostasis. Curr. Opin. Cell Biol. 2019, 56, 80–87. [Google Scholar] [CrossRef]
- Min, Y.-Q.; Mo, Q.; Wang, J.; Deng, F.; Wang, H.; Ning, Y.-J. SARS-CoV-2 nsp1: Bioinformatics, potential structural and functional features, and implications for drug/vaccine designs. Front. Microbiol. 2020, 11, 7317. [Google Scholar] [CrossRef]
- Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Nieto-Torres, J.L.; DeDiego, M.L.; Castano-Rodriguez, C.; Fernandez-Delgado, R.; Perlman, S.; Enjuanes, L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015, 11, e1005215. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobhy, H. The Potential Functions of Protein Domains during COVID Infection: An Analysis and a Review. COVID 2021, 1, 384-393. https://doi.org/10.3390/covid1010032
Sobhy H. The Potential Functions of Protein Domains during COVID Infection: An Analysis and a Review. COVID. 2021; 1(1):384-393. https://doi.org/10.3390/covid1010032
Chicago/Turabian StyleSobhy, Haitham. 2021. "The Potential Functions of Protein Domains during COVID Infection: An Analysis and a Review" COVID 1, no. 1: 384-393. https://doi.org/10.3390/covid1010032
APA StyleSobhy, H. (2021). The Potential Functions of Protein Domains during COVID Infection: An Analysis and a Review. COVID, 1(1), 384-393. https://doi.org/10.3390/covid1010032