Emodin as a Broad-Spectrum Inhibitor of QS-Regulated Pathogenicity and Biofilms: A Non-Antibiotic Strategy Against Microbial Virulence
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture, Growth Conditions, and Chemicals Used
2.2. Examination of Violacein in C. violaceum 12472
2.3. Assessment of P. aeruginosa PAO1 Virulence Factors
2.4. Examination of Prodigiosin Production in S. marcescens MTCC 97
2.5. Examination of Motility in S. marcescens MTCC 97
2.6. Determination of Cell Surface Hydrophobicity (CSH)
2.7. Examination of Exoprotease Activity
2.8. Biofilm Formation Assay
2.9. Molecular Docking
2.10. Molecular Dynamics and Simulation
2.11. Statistical Analysis
3. Results and Discussion
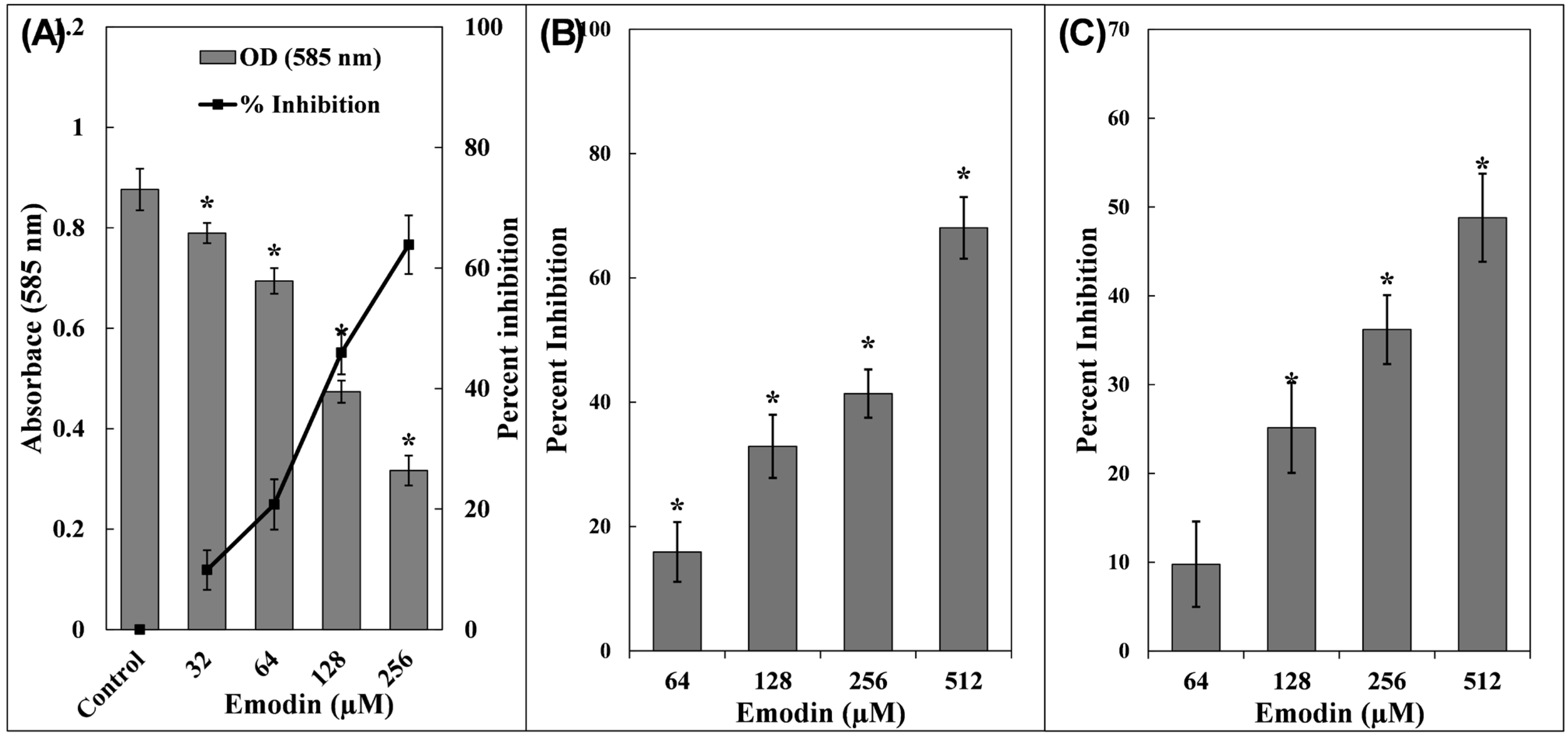
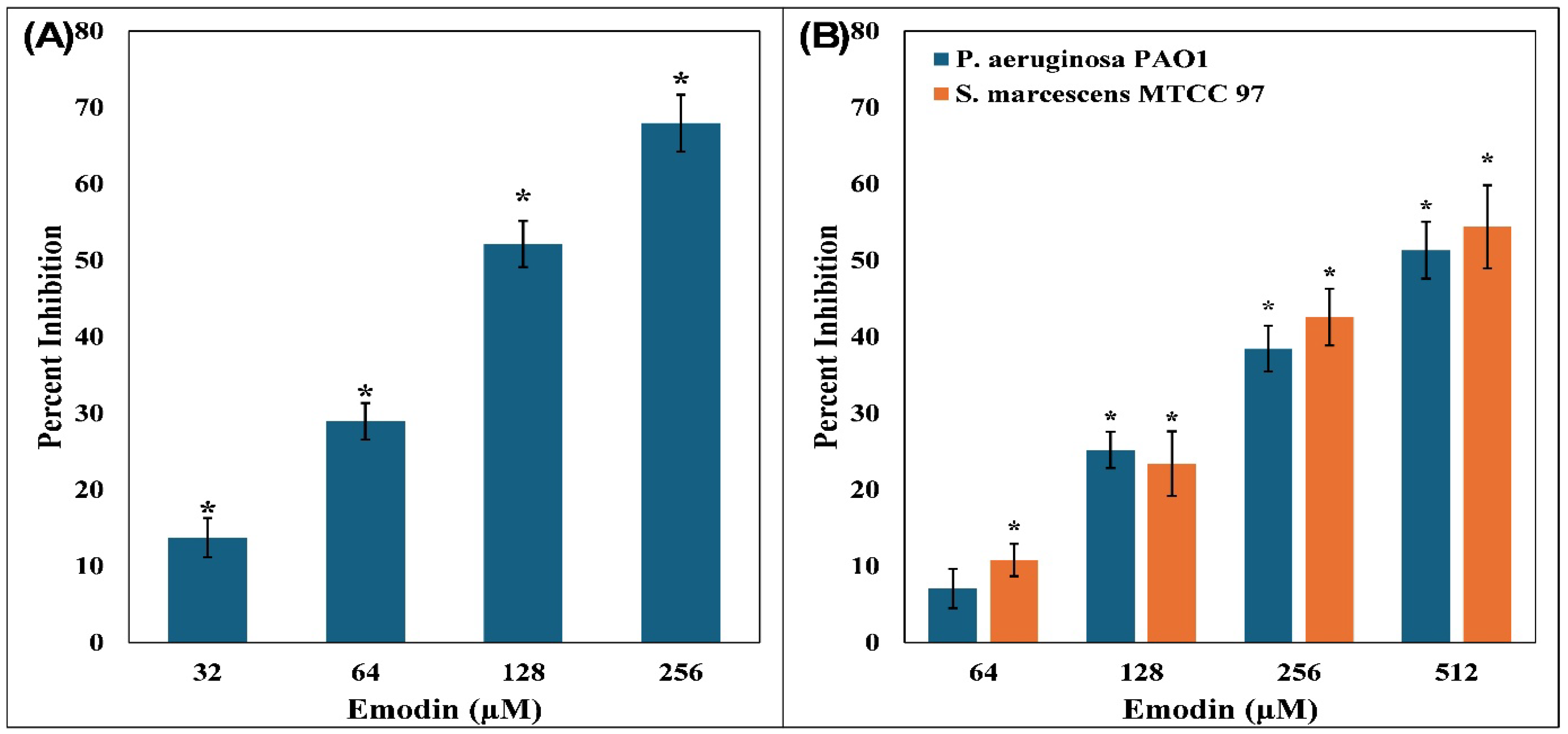
3.1. Inhibition of Violacein Pigment in C. violaceum 12472
3.2. Examination of P. aeruginosa PAO1 Virulence Factors
3.3. Examination of S. marcescens MTCC 97 Virulence Factors
3.4. Inhibition of Biofilm Formation by Emodin
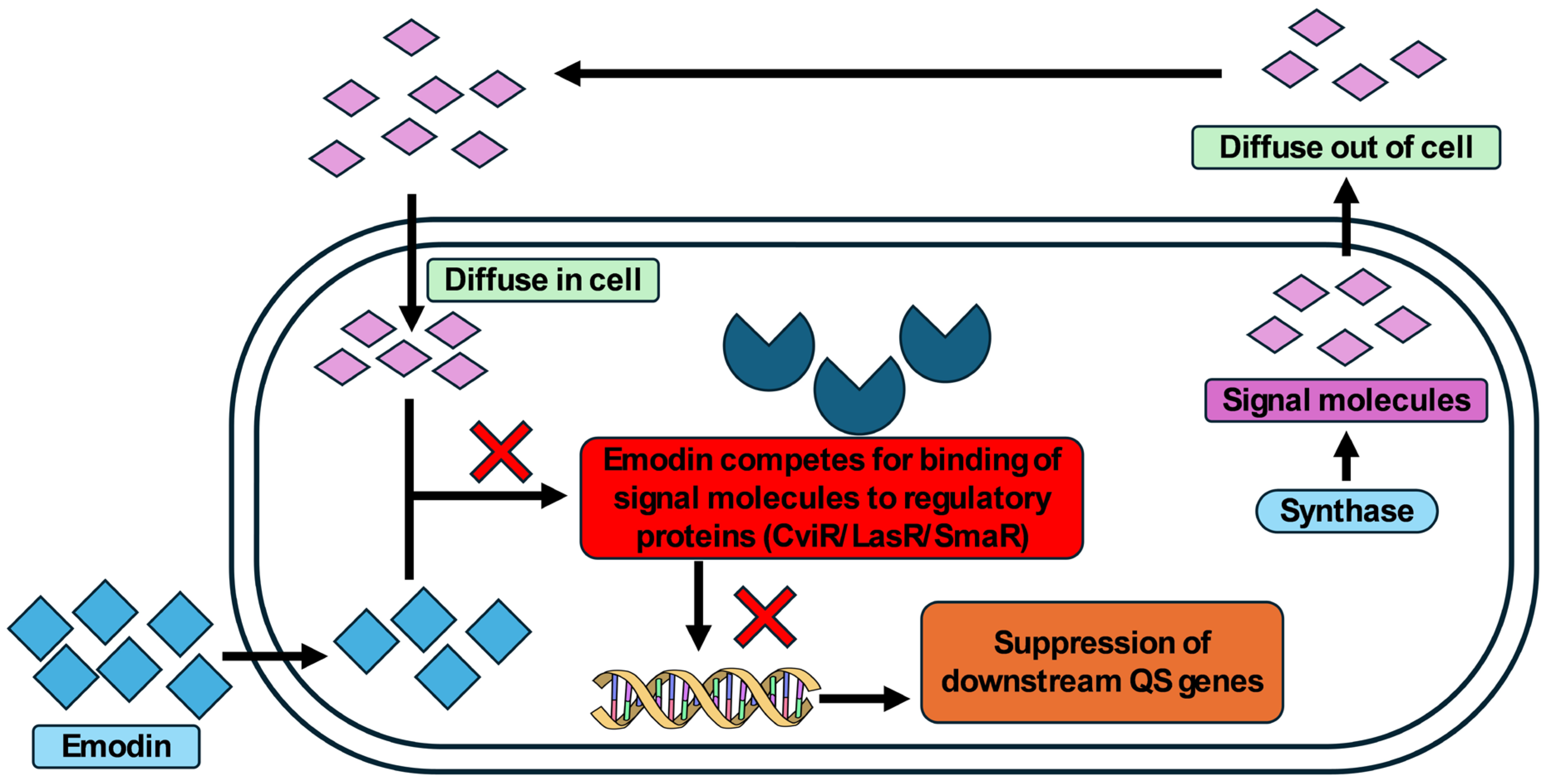
3.5. Interaction of Emodin with Regulatory Proteins of QS Using Molecular Docking
3.6. Molecular Dynamics and Simulation
4. Conclusions
5. Future Directions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, S.; Chauhan, A.; Ranjan, A.; Mathkor, D.M.; Haque, S.; Ramniwas, S.; Tuli, H.S.; Jindal, T.; Yadav, V. Emerging Challenges in Antimicrobial Resistance: Implications for Pathogenic Microorganisms, Novel Antibiotics, and Their Impact on Sustainability. Front. Microbiol. 2024, 15, 1403168. [Google Scholar] [CrossRef]
- Kariuki, S. Global Burden of Antimicrobial Resistance and Forecasts to 2050. Lancet 2024, 404, 1172–1173. [Google Scholar] [CrossRef]
- Krishnaprasad, V.H.; Nayak, V.; Kumar, S. World Health Organisation’s Bacterial Pathogen Priority List (BPPL) 2017 and BPPL 2024 to Combat Global Antimicrobial Resistance Crisis: ‘Challenges and Opportunities’. J. Antimicrob. Chemother. 2025, 80, 2061–2069. [Google Scholar] [CrossRef]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef]
- Matsumoto, T.; Darlington, O.; Miller, R.; Gordon, J.; McEwan, P.; Ohashi, T.; Taie, A.; Yuasa, A. Estimating the Economic and Clinical Value of Reducing Antimicrobial Resistance to Three Gram-Negative Pathogens in Japan. J. Health Econ. Outcomes Res. 2021, 8, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Mahizhchi, E.; Sivakumar, D.; Jayaraman, M. Antimicrobial Resistance: Techniques to Fight AMR in Bacteria—A Review. J. Pure Appl. Microbiol. 2024, 18, 16–28. [Google Scholar] [CrossRef]
- Naga, N.G.; Shaaban, M.I.; El-Metwally, M.M. An Insight on the Powerful of Bacterial Quorum Sensing Inhibition. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 2071–2081. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and Promise of Bacterial Quorum Sensing Research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The Clinical Impact of Bacterial Biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding Bacterial Biofilms: From Definition to Treatment Strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by Bacterial Human Pathogens: Clinical Relevance—Development, Composition and Regulation—Therapeutical Strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef] [PubMed]
- Manavathu, E.K.; Vazquez, J.A. The Functional Resistance of Biofilms. In Antimicrobial Drug Resistance; Springer International Publishing: Cham, Switzerland, 2017; pp. 149–162. [Google Scholar]
- Erkihun, M.; Asmare, Z.; Endalamew, K.; Getie, B.; Kiros, T.; Berhan, A. Medical Scope of Biofilm and Quorum Sensing during Biofilm Formation: Systematic Review. Bacteria 2024, 3, 118–135. [Google Scholar] [CrossRef]
- Wagner, V.E.; Gillis, R.J.; Iglewski, B.H. Transcriptome Analysis of Quorum-Sensing Regulation and Virulence Factor Expression in Pseudomonas Aeruginosa. Vaccine 2004, 22, S15–S20. [Google Scholar] [CrossRef]
- Warrier, A.; Satyamoorthy, K.; Murali, T.S. Quorum-Sensing Regulation of Virulence Factors in Bacterial Biofilm. Future Microbiol. 2021, 16, 1003–1021. [Google Scholar] [CrossRef]
- Chaieb, K.; Kouidhi, B.; Hosawi, S.B.; Baothman, O.A.S.; Zamzami, M.A.; Altayeb, H.N. Computational Screening of Natural Compounds as Putative Quorum Sensing Inhibitors Targeting Drug Resistance Bacteria: Molecular Docking and Molecular Dynamics Simulations. Comput. Biol. Med. 2022, 145, 105517. [Google Scholar] [CrossRef]
- Stompor-Gorący, M. The Health Benefits of Emodin, a Natural Anthraquinone Derived from Rhubarb—A Summary Update. Int. J. Mol. Sci. 2021, 22, 9522. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, S.; Li, X.; Liu, R. Advances in the Study of Emodin: An Update on Pharmacological Properties and Mechanistic Basis. Chin. Med. 2021, 16, 102. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, F.; Yang, W.; Yan, T.; Chen, Q. Antibiofilm Efficacy of Emodin Alone or Combined with Ampicillin against Methicillin-Resistant Staphylococcus Aureus. Sci. Rep. 2025, 15, 21904. [Google Scholar] [CrossRef]
- Ding, X.; Yin, B.; Qian, L.; Zeng, Z.; Yang, Z.; Li, H.; Lu, Y.; Zhou, S. Screening for Novel Quorum-Sensing Inhibitors to Interfere with the Formation of Pseudomonas Aeruginosa Biofilm. J. Med. Microbiol. 2011, 60, 1827–1834. [Google Scholar] [CrossRef]
- Eloff, J. A Sensitive and Quick Microplate Method to Determine the Minimal Inhibitory Concentration of Plant Extracts for Bacteria. Planta Medica 1998, 64, 711–713. [Google Scholar] [CrossRef]
- Matz, C.; Deines, P.; Boenigk, J.; Arndt, H.; Eberl, L.; Kjelleberg, S.; Jurgens, K. Impact of Violacein-Producing Bacteria on Survival and Feeding of Bacterivorous Nanoflagellates. Appl. Environ. Microbiol. 2004, 70, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.M.; Ahmad, I.; Al-Thubiani, A.S.; Abulreesh, H.H.; AlHazza, I.M.; Aqil, F. Leaf Extracts of Mangifera indica L. Inhibit Quorum Sensing—Regulated Production of Virulence Factors and Biofilm in Test Bacteria. Front. Microbiol. 2017, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Ankenbauer, R.; Sriyosachati, S.; Cox, C.D. Effects of Siderophores on the Growth of Pseudomonas aeruginosa in Human Serum and Transferrin. Infect. Immun. 1985, 49, 132–140. [Google Scholar] [CrossRef]
- Kessler, E.; Israel, M.; Landshman, N.; Chechick, A.; Blumberg, S. In Vitro Inhibition of Pseudomonas aeruginosa Elastase by Metal-Chelating Peptide Derivatives. Infect. Immun. 1982, 38, 716–723. [Google Scholar] [CrossRef]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin Influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef]
- Slater, H.; Crow, M.; Everson, L.; Salmond, G.P.C. Phosphate Availability Regulates Biosynthesis of Two Antibiotics, Prodigiosin and Carbapenem, in Serratia via Both Quorum-Sensing-Dependent and -Independent Pathways. Mol. Microbiol. 2003, 47, 303–320. [Google Scholar] [CrossRef]
- Rosenberg, M.; Gutnick, D.; Rosenberg, E. Adherence of Bacteria to Hydrocarbons: A Simple Method for Measuring Cell-Surface Hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas Aeruginosa Is a Staphylolytic Protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Pandya, P.; Agarwal, L.K.; Gupta, N.; Pal, S. Molecular Recognition Pattern of Cytotoxic Alkaloid Vinblastine with Multiple Targets. J. Mol. Graph. Model. 2014, 54, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa —A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Suryanti, E.; Wahyudi, A.T.; Akhdiya, A.; Rusmana, I. Acyl Homoserine Lactone Lactonase Bacteria Potential as Biocontrol Agent of Soft Rot Infection. Malays. Appl. Biol. 2020, 49, 131–140. [Google Scholar] [CrossRef]
- Gupta, P.; Sarkar, A.; Sandhu, P.; Daware, A.; Das, M.C.; Akhter, Y.; Bhattacharjee, S. Potentiation of Antibiotic against Pseudomonas Aeruginosa Biofilm: A Study with Plumbagin and Gentamicin. J. Appl. Microbiol. 2017, 123, 246–261. [Google Scholar] [CrossRef]
- Ran, H.; Hassett, D.J.; Lau, G.W. Human Targets of Pseudomonas Aeruginosa Pyocyanin. Proc. Natl. Acad. Sci. USA 2003, 100, 14315–14320. [Google Scholar] [CrossRef] [PubMed]
- Fothergill, J.L.; Panagea, S.; Hart, C.A.; Walshaw, M.J.; Pitt, T.L.; Winstanley, C. Widespread Pyocyanin Over-Production among Isolates of a Cystic Fibrosis Epidemic Strain. BMC Microbiol. 2007, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Kutty, S.K.; Tavallaie, R.; Ibugo, A.I.; Panchompoo, J.; Sehar, S.; Aldous, L.; Yeung, A.W.S.; Thomas, S.R.; Kumar, N.; et al. Phenazine Virulence Factor Binding to Extracellular DNA Is Important for Pseudomonas Aeruginosa Biofilm Formation. Sci. Rep. 2015, 5, 8398. [Google Scholar] [CrossRef]
- Muller, M. Pyocyanin Induces Oxidative Stress in Human Endothelial Cells and Modulates the Glutathione Re-dox Cycle. Free Radic. Biol. Med. 2002, 33, 1527–1533. [Google Scholar] [CrossRef]
- Peek, M.E.; Bhatnagar, A.; McCarty, N.A.; Zughaier, S.M. Pyoverdine, the Major Siderophore in Pseudomonas Aeruginosa, Evades NGAL Recognition. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 843509. [Google Scholar] [CrossRef]
- Das, M.C.; Sandhu, P.; Gupta, P.; Rudrapaul, P.; De, U.C.; Tribedi, P.; Akhter, Y.; Bhattacharjee, S. Attenuation of Pseudomonas Aeruginosa Biofilm Formation by Vitexin: A Combinatorial Study with Azithromycin and Gentamicin. Sci. Rep. 2016, 6, 23347. [Google Scholar] [CrossRef]
- Oh, J.; Li, X.-H.; Kim, S.-K.; Lee, J.-H. Post-Secretional Activation of Protease IV by Quorum Sensing in Pseudomonas Aeruginosa. Sci. Rep. 2017, 7, 4416. [Google Scholar] [CrossRef]
- Smith, R.S.; Iglewski, B.H.P. Aeruginosa Quorum-Sensing Systems and Virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lee, S.-H.; Byun, Y.; Park, H.-D. 6-Gingerol Reduces Pseudomonas Aeruginosa Biofilm Formation and Virulence via Quorum Sensing Inhibition. Sci. Rep. 2015, 5, 8656. [Google Scholar] [CrossRef]
- Boyd, A.; Chakrabarty, A.M. Pseudomonas Aeruginosa Biofilms: Role of the Alginate Exopolysaccharide. J. Ind. Microbiol. 1995, 15, 162–168. [Google Scholar] [CrossRef]
- Köhler, T.; Curty, L.K.; Barja, F.; van Delden, C.; Pechère, J.C. Swarming of Pseudomonas Aeruginosa Is Dependent on Cell-to-Cell Signaling and Requires Flagella and Pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [PubMed]
- O’May, C.; Tufenkji, N. The Swarming Motility of Pseudomonas Aeruginosa Is Blocked by Cranberry Proanthocyanidins and Other Tannin-Containing Materials. Appl. Environ. Microbiol. 2011, 77, 3061–3067. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Shiono, T.; Takidouchi, K.; Kato, M.; Kato, N.; Kato, J.; Ikeda, T. Inhibition of Quorum Sensing in Serratia Marcescens AS-1 by Synthetic Analogs of N-Acylhomoserine Lactone. Appl. Environ. Microbiol. 2007, 73, 6339–6344. [Google Scholar] [CrossRef]
- Goluszko, P.; Nowicki, B.; Goluszko, E.; Nowicki, S.; Kaul, A.; Pham, T. Association of Colony Variation in Serratia Marcescens with the Differential Expression of Protease and Type 1 Fimbriae. FEMS Microbiol. Lett. 1995, 133, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kida, Y.; Inoue, H.; Shimizu, T.; Kuwano, K. Serratia Marcescens Serralysin Induces Inflammatory Responses through Protease-Activated Receptor 2. Infect. Immun. 2007, 75, 164–174. [Google Scholar] [CrossRef]
- Wei, J.-R.; Lai, H.-C. N-Acylhomoserine Lactone-Dependent Cell-to-Cell Communication and Social Behavior in the Genus Serratia. Int. J. Med. Microbiol. 2006, 296, 117–124. [Google Scholar] [CrossRef]
- Salini, R.; Pandian, S.K. Interference of Quorum Sensing in Urinary Pathogen Serratia Marcescens by Anethum Graveolens. Pathog. Dis. 2015, 73, ftv038. [Google Scholar] [CrossRef]
- Srinivasan, R.; Devi, K.R.; Kannappan, A.; Pandian, S.K.; Ravi, A.V. Piper Betle and Its Bioactive Metabolite Phytol Mitigates Quorum Sensing Mediated Virulence Factors and Biofilm of Nosocomial Pathogen Serratia Marcescens In Vitro. J. Ethnopharmacol. 2016, 193, 592–603. [Google Scholar] [CrossRef]
- Kim, H.-S.; Park, H.-D. Ginger Extract Inhibits Biofilm Formation by Pseudomonas Aeruginosa PA14. PLoS ONE 2013, 8, e76106. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; De Nicola, S.; Verginelli, F.; Fiscarelli, E.; Di Bonaventura, G. Cooperative Pathogenicity in Cystic Fibrosis: Stenotrophomonas Maltophilia Modulates Pseudomonas Aeruginosa Virulence in Mixed Biofilm. Front. Microbiol. 2015, 6, 951. [Google Scholar] [CrossRef]
- Qin, N.; Tan, X.; Jiao, Y.; Liu, L.; Zhao, W.; Yang, S.; Jia, A. RNA-Seq-Based Transcriptome Analysis of Methicillin-Resistant Staphylococcus Aureus Biofilm Inhibition by Ursolic Acid and Resveratrol. Sci. Rep. 2015, 4, 5467. [Google Scholar] [CrossRef]
- Ahmed, M.; Bird, S.; Wang, F.; Palombo, E.A. In Silico Investigation of Lactone and Thiolactone Inhibitors in Bacterial Quorum Sensing Using Molecular Modeling. Int. J. Chem. 2013, 5, 9–26. [Google Scholar] [CrossRef]
- Chen, G.; Swem, L.R.; Swem, D.L.; Stauff, D.L.; O’Loughlin, C.T.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. A Strategy for Antagonizing Quorum Sensing. Mol. Cell 2011, 42, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-Aided Identification of Recognized Drugs as Pseudomonas Aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Qais, F.A.; Alomar, S.Y.; Imran, M.A.; Hashmi, M.A. In-Silico Analysis of Phytocompounds of Olea Europaea as Potential Anti-Cancer Agents to Target PKM2 Protein. Molecules 2022, 27, 5793. [Google Scholar] [CrossRef]
- Al-Jumaili, M.H.A.; Siddique, F.; Abul Qais, F.; Hashem, H.E.; Chtita, S.; Rani, A.; Uzair, M.; Almzaien, K.A. Analysis and Prediction Pathways of Natural Products and Their Cytotoxicity against HeLa Cell Line Protein Using Docking, Molecular Dynamics and ADMET. J. Biomol. Struct. Dyn. 2023, 41, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Qais, F.A.; Khan, M.S.; Ahmad, I.; Husain, F.M.; Al-kheraif, A.A.; Arshad, M.; Alam, P. Plumbagin Inhibits Quorum Sensing-Regulated Virulence and Biofilms of Gram-Negative Bacteria: In Vitro and In Silico Investigations. Biofouling 2021, 37, 724–739. [Google Scholar] [CrossRef]
- Siddiqui, S.; Ameen, F.; Jahan, I.; Nayeem, S.M.; Tabish, M. A Comprehensive Spectroscopic and Computational Investigation on the Binding of the Anti-Asthmatic Drug Triamcinolone with Serum Albumin. N. J. Chem. 2019, 43, 4137–4151. [Google Scholar] [CrossRef]
- Liao, S.-Y.; Mo, G.-Q.; Chen, J.-C.; Zheng, K.-C. Exploration of the Binding Mode between (-)-Zampanolide and Tubulin Using Docking and Molecular Dynamics Simulation. J. Mol. Model. 2014, 20, 2070. [Google Scholar] [CrossRef]





| Emodin Concentration | Prodigiosin | Protease Activity | CSH | Swarming Motility |
|---|---|---|---|---|
| 64 µM | 16.18 ± 2.80 * | 13.04 ± 3.77 * | 8.75 ± 3.50 | 4.34 ± 2.08 |
| 128 µM | 33.12 ± 4.87 * | 25.66 ± 2.66 * | 15.80 ± 2.01 * | 7.02 ± 1.53 |
| 256 µM | 41.62 ± 4.44 * | 39.27 ± 3.15 * | 33.97 ± 4.46 * | 58.19 ± 5.87 * |
| 512 µM | 55.94 ± 2.94 * | 48.80 ± 4.03 * | 41.20 ± 3.94 * | 83.27 ± 4.52 * |
| Protein | Bacteria | PDB ID | Binding Energy | Binding Constant Kb |
|---|---|---|---|---|
| CviR | C. violaceum | 3QP1 | −8.9 | 3.37 × 106 |
| LasR | P. aeruginosa | 2UV0 | −10.7 | 7.04 × 107 |
| SmaR | S. marcescens | Modelled | −6.7 | 8.20 × 104 |
| Energy | CviR-Emodin Complex | LasR-Emodin Complex | SmaR-Emodin Complex |
|---|---|---|---|
| Van der Waal energy | −35.31 ± 3.85 | −35.43 ± 2.34 | −35.16 ± 2.19 |
| Electrostatic energy | −12.56 ± 3.88 | −6.29 ± 1.37 | −11.50 ± 7.75 |
| Polar solvation energy | 30.50 ± 5.78 | 23.24 ± 2.91 | 28.20 ± 5.11 |
| SASA energy | −3.40 ± 0.03 | −3.51 ± 0.06 | −3.44 ± 0.02 |
| Binding energy | −20.78 ± 0.73 | −21.99 ± 0.91 | −21.89 ± 0.40 |
| CviR–Emodin Complex | LasR–Emodin Complex | SmaR–Emodin Complex | |||
|---|---|---|---|---|---|
| Residues | Total Energy | Residues | Total Energy | Residues | Total Energy |
| Leu57 | −1.176 ± 0.025 | Leu36 | −0.697 ± 0.030 | Ala32 | −0.500 ± 0.024 |
| Ala59 | −0.364 ± 0.012 | Phe37 | −0.334 ± 0.017 | Ala34 | −0.344 ± 0.014 |
| Leu72 | −0.768 ± 0.020 | Gly38 | −0.527 ± 0.034 | Ile46 | −0.435 ± 0.020 |
| Val75 | −0.390 ± 0.017 | Leu39 | −0.538 ± 0.022 | Trp53 | −0.318 ± 0.035 |
| Leu85 | −0.526 ± 0.036 | Leu40 | −1.132 ± 0.027 | Phe54 | −0.798 ± 0.029 |
| Tyr88 | −1.922 ± 0.053 | Tyr47 | −0.777 ± 0.057 | Tyr57 | −1.093 ± 0.037 |
| Ile99 | −1.306 ± 0.025 | Ala50 | −0.386 ± 0.020 | Val68 | −1.208 ± 0.036 |
| Leu100 | −1.219 ± 0.033 | Ile52 | −1.195 ± 0.040 | Leu69 | −0.964 ± 0.029 |
| Trp111 | −1.181 ± 0.031 | Tyr64 | −0.598 ± 0.031 | Trp81 | −1.496 ± 0.041 |
| Phe126 | −0.523 ± 0.012 | Ala70 | −0.451 ± 0.019 | Ile105 | −0.636 ± 0.026 |
| Met135 | −0.733 ± 0.029 | Val76 | −1.720 ± 0.034 | Val122 | −0.377 ± 0.025 |
| Ile153 | −0.546 ± 0.021 | Ala127 | −1.007 ± 0.037 | Met126 | −0.721 ± 0.026 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bano, F. Emodin as a Broad-Spectrum Inhibitor of QS-Regulated Pathogenicity and Biofilms: A Non-Antibiotic Strategy Against Microbial Virulence. Micro 2025, 5, 56. https://doi.org/10.3390/micro5040056
Bano F. Emodin as a Broad-Spectrum Inhibitor of QS-Regulated Pathogenicity and Biofilms: A Non-Antibiotic Strategy Against Microbial Virulence. Micro. 2025; 5(4):56. https://doi.org/10.3390/micro5040056
Chicago/Turabian StyleBano, Fareha. 2025. "Emodin as a Broad-Spectrum Inhibitor of QS-Regulated Pathogenicity and Biofilms: A Non-Antibiotic Strategy Against Microbial Virulence" Micro 5, no. 4: 56. https://doi.org/10.3390/micro5040056
APA StyleBano, F. (2025). Emodin as a Broad-Spectrum Inhibitor of QS-Regulated Pathogenicity and Biofilms: A Non-Antibiotic Strategy Against Microbial Virulence. Micro, 5(4), 56. https://doi.org/10.3390/micro5040056






