Assessment of Stress Tolerance of Enterococcus faecium and Enterococcus durans Strains by Flow Cytometry Using NADS Protocol and Traditional Culture Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Stress Treatments on Exponential Phase Cells
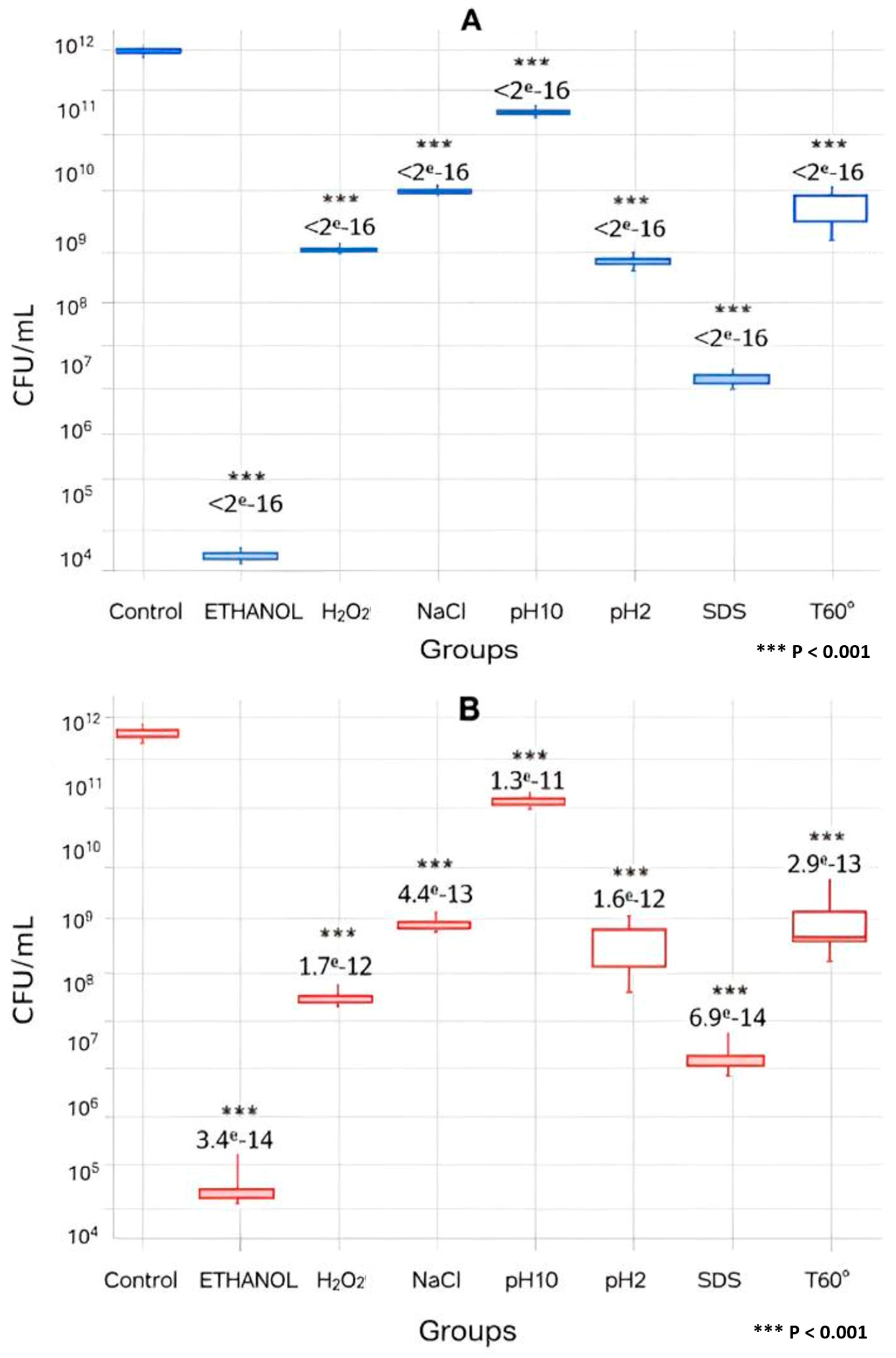
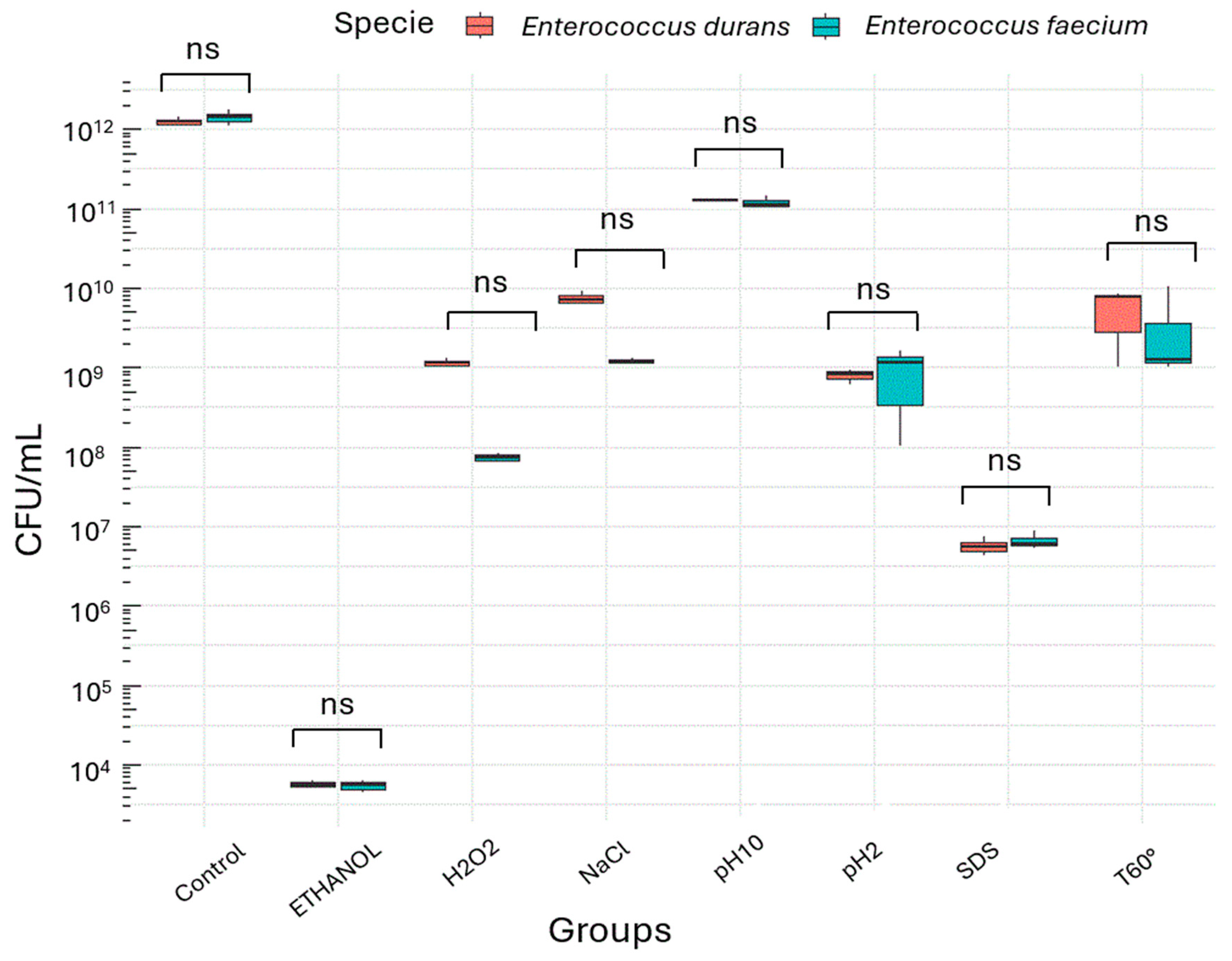
2.3. Enumeration of the Total Viable Count and Survival Percentage by CFU
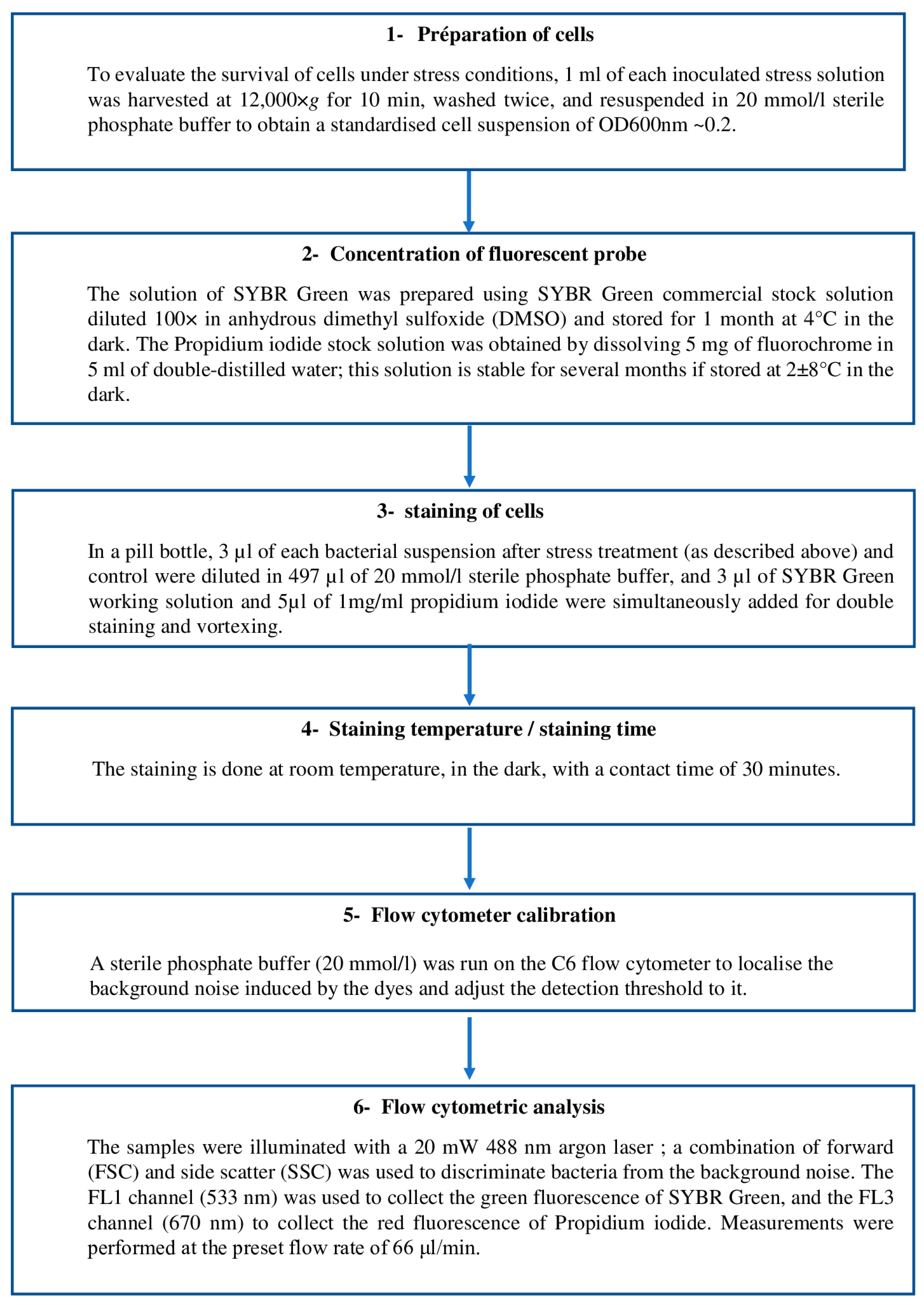
2.4. Enumeration of the Total Viable Count by FCM
2.4.1. Instrument
2.4.2. Fluorescent Probes
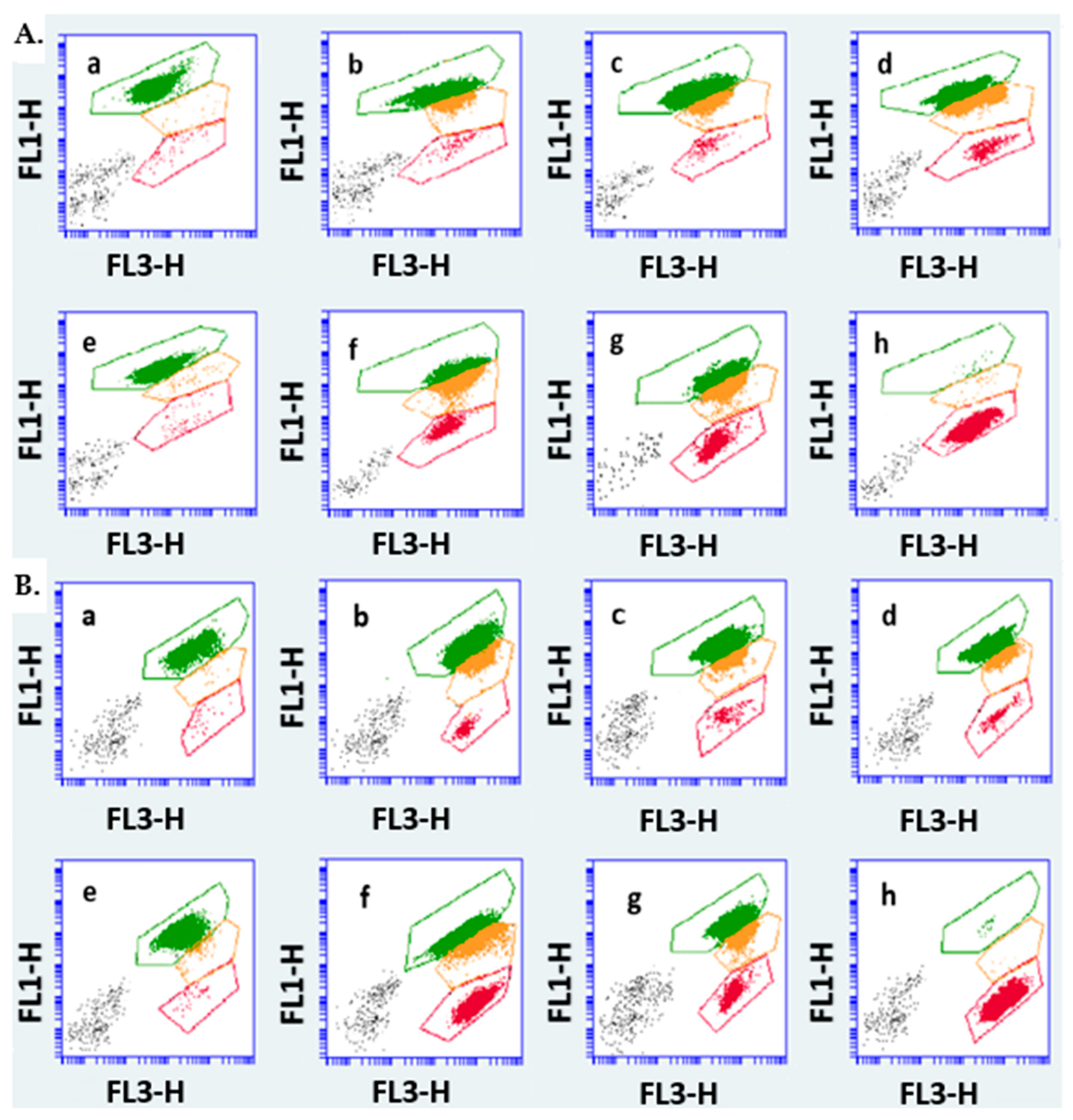
2.4.3. Staining Methodology and Flow Cytometric Assessment
2.5. Statistical Analysis
3. Results
3.1. Evaluation of Viability During the Exponential Phase by CFU
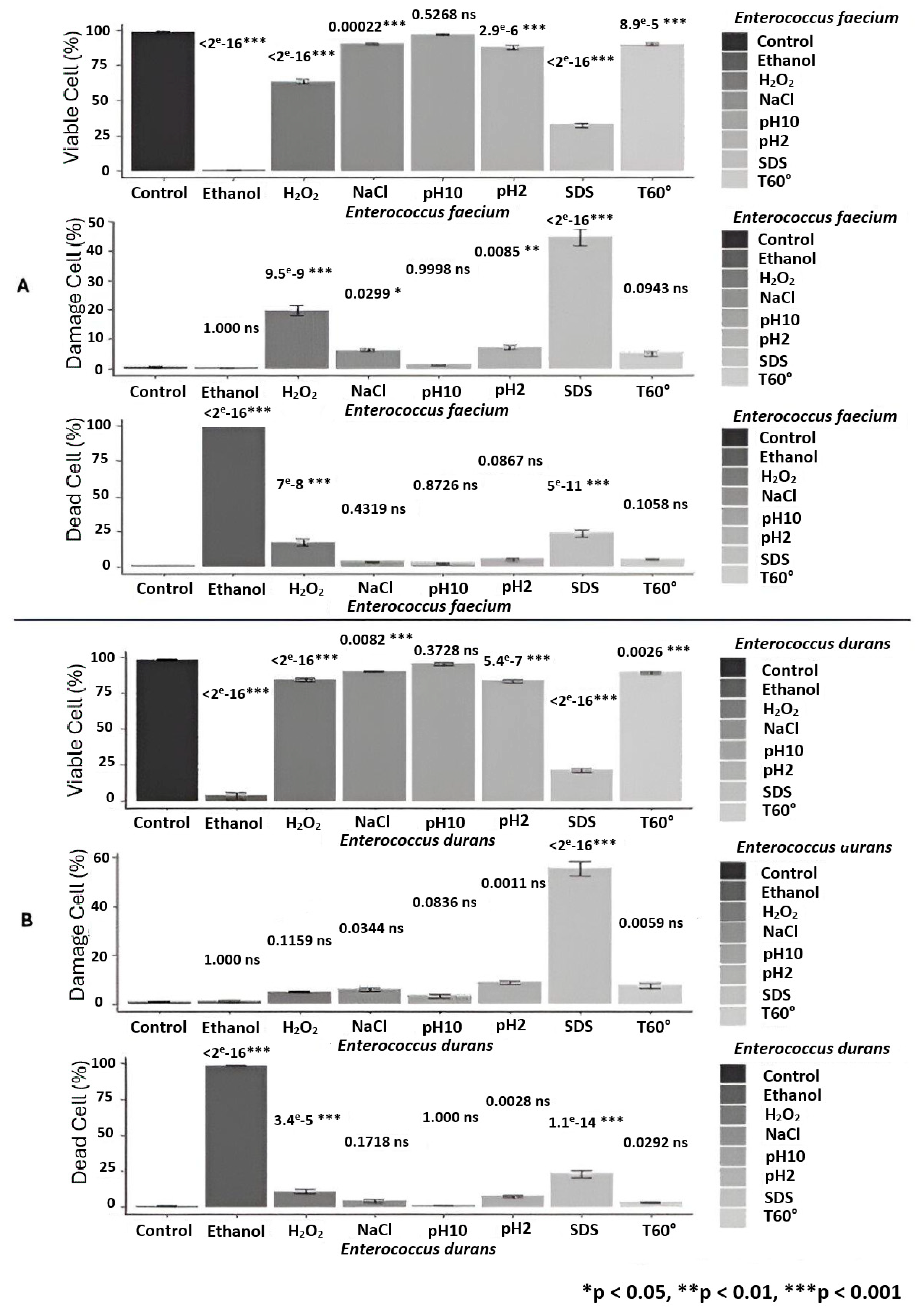
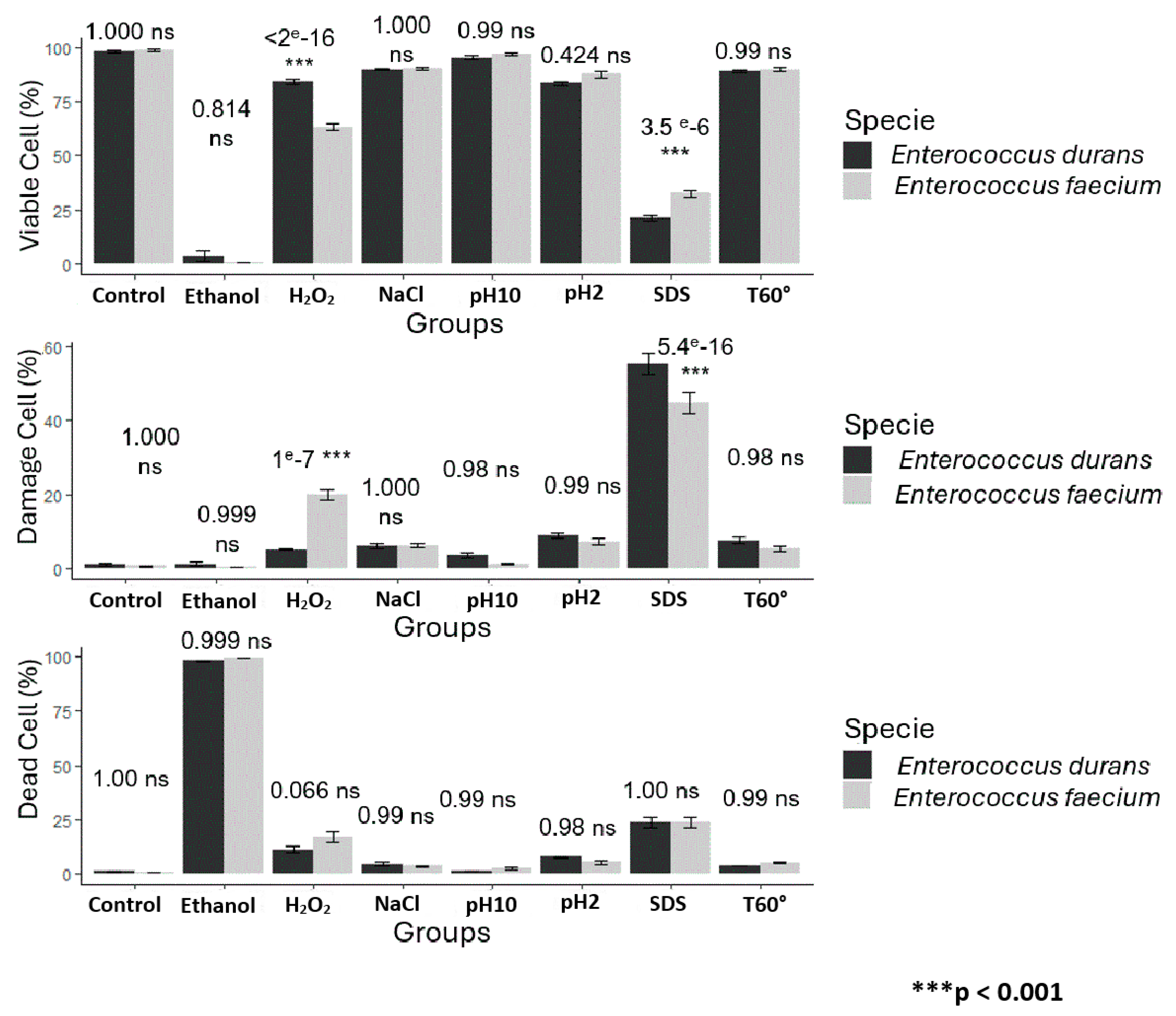
3.2. Assessment of Cell Viability and Membrane Integrity by FCM
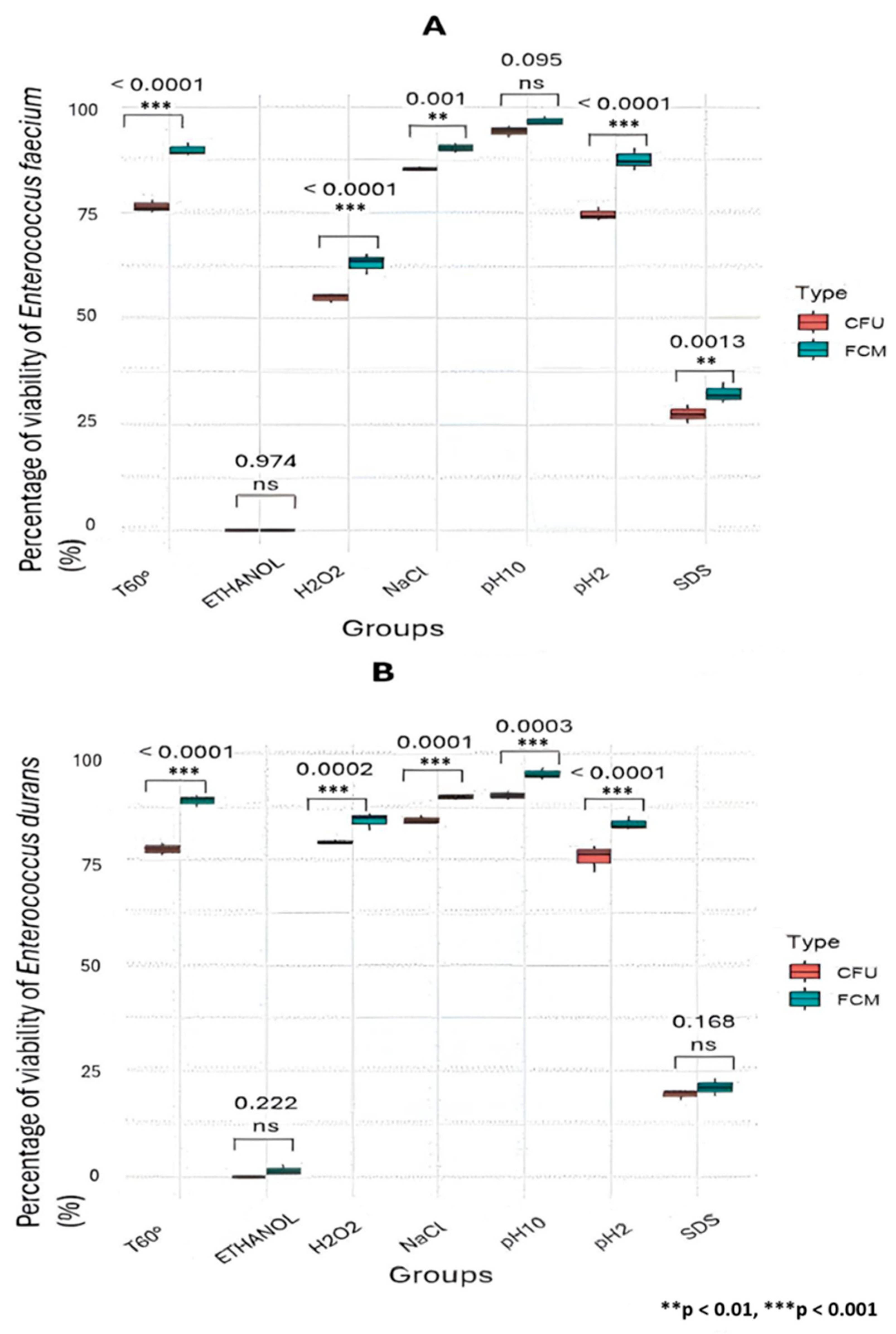
3.3. Validation of the Viability Method by Flow Cytometry Compared to the Classical Plate Count Method
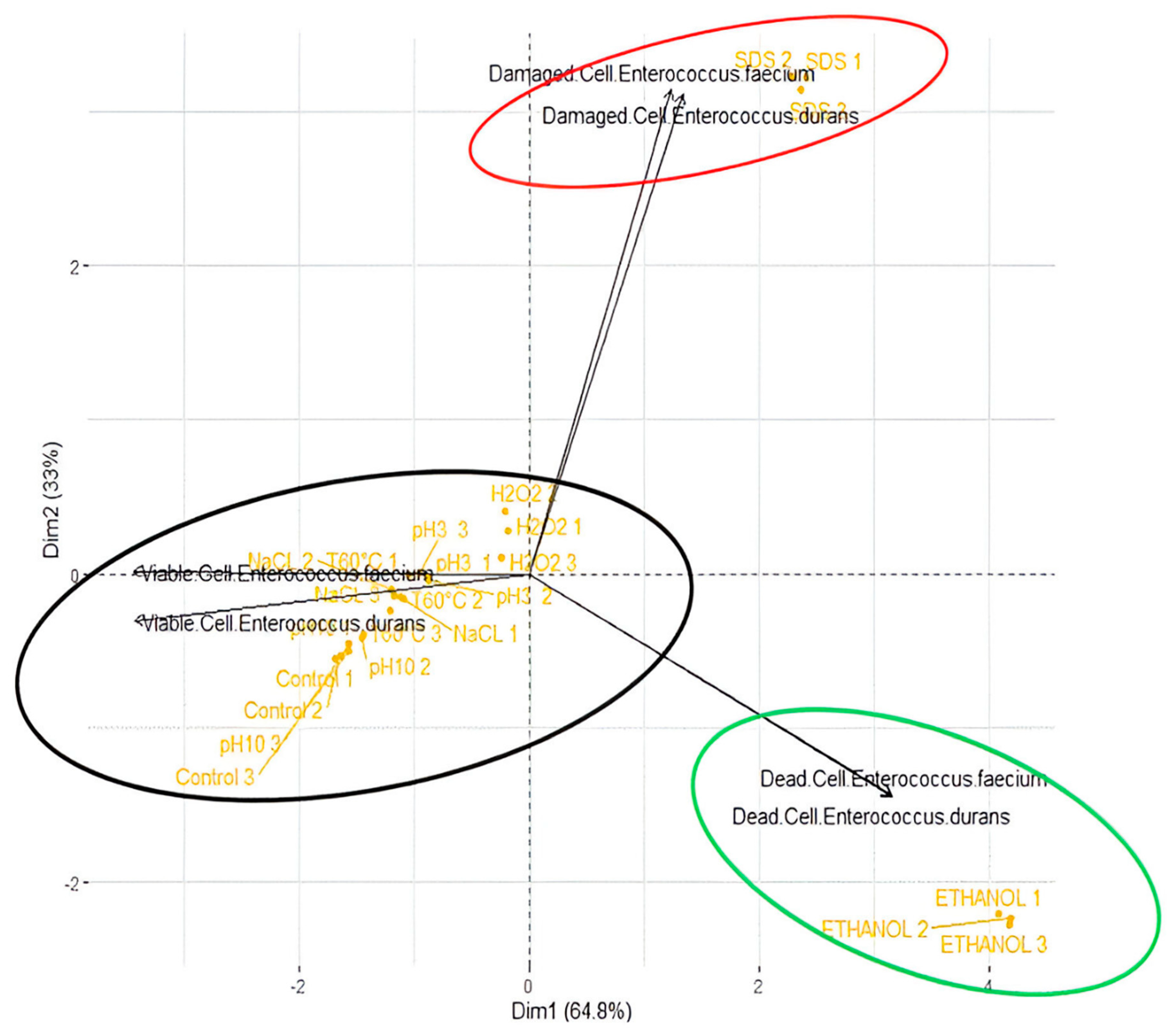
3.4. The Principal Component Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NADS | nucleic-acid double staining |
| CFU | colony-forming units |
| FCM | Flow cytometry |
| SY | SYBR green |
| PI | Propidium iodide |
| VBNC | viable non-cultivable cells |
References
- Papadimitriou, K.; Pot, B.; Tsakalidou, E. How microbes adapt to a diversity of food niches. Curr. Opin. Food Sci. 2015, 2, 29–35. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress Physiology of Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Salek, F.; Mirzaei, H.; Khandaghi, J.; Javadi, A.; Nami, Y. Apoptosis induction in cancer cell lines and anti-inflammatory and anti-pathogenic properties of proteinaceous metabolites secreted from potential probiotic Enterococcus faecalis KUMS-T48. Sci. Rep. 2023, 13, 7813. [Google Scholar] [CrossRef]
- Zaghloul, E.H.; Abuohashish, H.M.; El Sharkawy, A.S.; Abbas, E.M.; Ahmed, M.M.; Al-Rejaie, S.S. Probiotic Potential of the Marine Isolate Enterococcus faecium EA9 and In Vivo Evaluation of Its Antisepsis Action in Rats. Mar. Drugs 2023, 21, 45. [Google Scholar] [CrossRef]
- Casey, M.G.; Häni, J.P.; Gruskovnjak, J.; Schaeren, W.; Wechsler, D. Characterisation of the non-starter lactic acid bacteria (NSLAB) of Gruyère PDO cheese. Le Lait 2006, 86, 407–414. [Google Scholar] [CrossRef]
- Shi, Y.; Zhai, M.; Li, J.; Li, B. Evaluation of safety and probiotic properties of a strain of Enterococcus faecium isolated from chicken bile. J. Food Sci. Technol. 2020, 57, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Dinçer, E.; Kıvanç, M. In vitro evaluation of probiotic potential of Enterococcus faecium strains isolated from Turkish pastırma. Arch. Microbiol. 2021, 203, 2831–2841. [Google Scholar] [CrossRef]
- Abril, A.G.; Quintela-Baluja, M.; Villa, T.G.; Calo-Mata, P.; Barros-Velázquez, J.; Carrera, M. Proteomic Characterization of Virulence Factors and Related Proteins in Enterococcus Strains from Dairy and Fermented Food Products. Int. J. Mol. Sci. 2022, 23, 10971. [Google Scholar] [CrossRef]
- Hassanzadazar, H.; Ehsani, A.; Mardani, K. Antibacterial activity of Enterococcus faecium derived from Koopeh cheese against Listeria monocytogenes in probiotic ultra-filtrated cheese Prevalence, serotypes and antimicrobial resistance of isolated salmonella from Eggs marketed in Zanjan city View project Monitoring of edible oils quality in resteurants and fast food centers using peroxide and Acid value View project. Vet. Res. Forum 2014, 5, 169–175. [Google Scholar] [PubMed]
- Martino, G.P.; Quintana, I.M.; Espariz, M.; Blancato, V.S.; Magni, C. Aroma compounds generation in citrate metabolism of Enterococcus faecium: Genetic characterization of type I citrate gene cluster. Int. J. Food Microbiol. 2016, 218, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Zareie, Z.; Moayedi, A.; Garavand, F.; Tabar-Heydar, K.; Khomeiri, M.; Maghsoudlou, Y. Probiotic Properties, Safety Assessment, and Aroma-Generating Attributes of Some Lactic Acid Bacteria Isolated from Iranian Traditional Cheese. Fermentation 2023, 9, 338. [Google Scholar] [CrossRef]
- Gotova, I.; Dimitrov, Z. Dimitrov Enterococcus faecium strain used as an adjunct culture in a starter for kashkaval cheese plays important role to proteolytic processes and release of bioactive peptides during ripening. J. BioSci. Biotechnol. 2015, 119–123. Available online: https://api.semanticscholar.org/CorpusID:6293506.
- Akpinar, A.; Saygili, D.; Yerlikaya, O. Production of set-type yoghurt using Enterococcus faecium and Enterococcus durans strains with probiotic potential as starter adjuncts. Int. J. Dairy Technol. 2020, 73, 726–736. [Google Scholar] [CrossRef]
- AlKalbani, N.S.; Turner, M.S.; Ayyash, M.M. Isolation, identification, and potential probiotic characterization of isolated lactic acid bacteria and in vitro investigation of the cytotoxicity, antioxidant, and antidiabetic activities in fermented sausage. Microb. Cell Fact. 2019, 18, 188. [Google Scholar] [CrossRef] [PubMed]
- Ghazisaeedi, F.; Meens, J.; Hansche, B.; Maurischat, S.; Schwerk, P.; Goethe, R.; Wieler, L.H.; Fulde, M.; Tedin, K.A. Virulence factor as a therapeutic: The probiotic Enterococcus faecium SF68 arginine deiminase inhibits innate immune signaling pathways. Gut Microbes 2022, 14, 2106105. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns-An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Yerlikaya, O.; Akbulut, N. In vitro characterisation of probiotic properties of Enterococcus faecium and Enterococcus durans strains isolated from raw milk and traditional dairy products. Int. J. Dairy Technol. 2019, 73, 98–107. [Google Scholar] [CrossRef]
- Ferchichi, M.; Sebei, K.; Boukerb, A.M.; Karray-Bouraoui, N.; Chevalier, S.; Feuilloley, M.G.J.; Connil, N.; Zommiti, M. Enterococcus spp.: Is It a Bad Choice for a Good Use—A Conundrum to Solve? Microorganisms 2021, 9, 2222. [Google Scholar] [CrossRef] [PubMed]
- Krishna, K.V.; Koujalagi, K.; Surya, R.U.; Namratha, M.P.; Malaviya, A. Enterococcus species and their probiotic potential: Current status and future prospects. J. Appl. Biol. Biotechnol. 2023, 11, 36–44. [Google Scholar] [CrossRef]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.G. Flow Cytometry and Food Microbiology: Challenges, opportunities and progress to date. Técnicas Lab. 2016, 417, 722–728. [Google Scholar]
- Sielatycka, K.; Poniewierska-Baran, A.; Nurek, K.; Torbé, A.; Ratajczak, M.Z. Novel View on Umbilical Cord Blood and Maternal Peripheral Blood-an Evidence for an Increase in the Number of Circulating Stem Cells on Both Sides of the Fetal-Maternal Circulation Barrier. Stem Cell Rev. Rep. 2017, 13, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.G. Flow cytometry as a potential method of measuring bacterial viability in probiotic products: A review. Trends Food Sci. Technol. 2018, 78, 1–10. [Google Scholar] [CrossRef]
- Wen, G.; Cao, R.; Wan, Q.; Tan, L.; Xu, X.; Wang, J.; Huang, T. Development of fungal spore staining methods for flow cytometric quantification and their application in chlorine-based disinfection. Chemosphere 2020, 243, 125453. [Google Scholar] [CrossRef]
- Doherty, S.B.; Wang, L.; Ross, R.P.; Stanton, C.; Fitzgerald, G.F.; Brodkorb, A. Use of viability staining in combination with flow cytometry for rapid viability assessment of Lactobacillus rhamnosus GG in complex protein matrices. J. Microbiol. Methods 2010, 82, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Sielatycka, K.; Juzwa, W.; Śliwa-Dominiak, J.; Kaczmarczyk, M.; Łoniewski, I.; Marlicz, W. Multiparameter flow cytometric enumeration of probiotic-containing commercial powders. Innov. Food Sci. Emerg. Technol. 2021, 68, 102598. [Google Scholar] [CrossRef]
- Tracey, H.; Coates, N.; Hulme, E.; John, D.; Michael, D.R.; Plummer, S.F. Insights into the enumeration of mixtures of probiotic bacteria by flow cytometry. BMC Microbiol. 2023, 23, 48. [Google Scholar] [CrossRef]
- Barbesti, S.; Citterio, S.; Labra, M.; Baroni, M.D.; Neri, M.G.; Sgorbati, S. Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry 2000, 40, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Grégori, G.; Citterio, S.; Ghiani, A.; Labra, M.; Sgorbati, S.; Brown, S.; Denis, M. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl. Environ. Microbiol. 2001, 67, 4662–4670. [Google Scholar] [CrossRef]
- Falcioni, T.; Papa, S.; Gasol, J.M. Evaluating the flow-cytometric nucleic acid double-staining protocol in realistic situations of planktonic bacterial death. Appl. Environ. Microbiol. 2008, 74, 1767–1779. [Google Scholar] [CrossRef] [PubMed]
- Grégori, G.; Denis, M.; Sgorbati, S.; Citterio, S. Resolution of Viable and Membrane-Compromised Free Bacteria in Aquatic Environments by Flow Cytometry. Curr. Protoc. Cytom. 2018, 85, e42. [Google Scholar] [CrossRef] [PubMed]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analyses. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef] [PubMed]
- Terzaghi, B.E.; Sandine, W.E. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 1975, 29, 807–813. [Google Scholar] [CrossRef]
- Parente, E.; Ciocia, F.; Ricciardi, A.; Zotta, T.; Felis, G.E.; Torriani, S. Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: A multivariate screening study. Int. J. Food Microbiol. 2010, 144, 270–279. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Kimbara, K. Rapid assessment of the physiological status of the polychlorinated biphenyl degrader Comamonas testosteroni TK102 by flow cytometry. Appl. Environ. Microbiol. 2002, 68, 2031–2035. [Google Scholar] [CrossRef]
- Van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Serrazanetti, D.I.; Guerzoni, M.E.; Corsetti, A.; Vogel, R. Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol. 2009, 26, 700–711. [Google Scholar] [CrossRef]
- Martínez, S.; López, M.; Bernardo, A. Thermal inactivation of Enterococcus faecium: Effect of growth temperature and physiological state of microbial cells. Lett. Appl. Microbiol. 2003, 37, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.M.F.; Silva, L.F.; Casarotti, S.N.; Nascimento, L.C.S.; Penna, A.L.B. Enterococcus faecium and Enterococcus durans isolated from cheese: Survival in the presence of medications under simulated gastrointestinal conditions and adhesion properties. J. Dairy Sci. 2017, 100, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Casella, T.; Gomes, E.S.; Nogueira, M.C.L.; De Dea Lindner, J.; Penna, A.L.B. Diversity of Lactic Acid Bacteria Isolated from Brazilian Water Buffalo Mozzarella Cheese. J. Food Sci. 2015, 80, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Kramer, V.C.; Nickerson, K.W.; Hamlett, N.V.; O’Hara, C. Prevalence of extreme detergent resistance among the Enterobacteriaceae. Can. J. Microbiol. 1984, 30, 711–713. [Google Scholar] [CrossRef]
- Jung, J.M.; Savin, G.; Pouzot, M.; Schmitt, C.; Mezzenga, R. Structure of heat-induced beta-lactoglobulin ag gregates and their complexes with sodium-dodecyl sulfate. Biomacromolecules 2008, 9, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Zegarra, F.C.; Homouz, D.; Eliaz, Y.; Gasic, A.G.; Cheung, M.S. Impact of hydrodynamic interactions on protein folding rates depends on temperature. Phys. Rev. E 2018, 97, 032402. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Pratsinis, H.; Nebe-von-Caron, G.; Kletsas, D.; Tsakalidou, E. Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 2007, 73, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, A.; Rochat, T.; Gratadoux, J.J.; Le Loir, Y.; Oliveira, S.C.; Langella, P.; Azevedo, V. Oxidative stress in Lactococcus lactis. Genet. Mol. Res. 2003, 2, 348–359. [Google Scholar]
- Graça da Silveira, M.; Vitória San Romão, M.; Loureiro-Dias, M.C.; Rombouts, F.M.; Abee, T. Flow cytometric assessment of membrane integrity of ethanol-stressed Oenococcus oeni cells. Appl. Environ. Microbiol. 2002, 68, 6087–6093. [Google Scholar] [CrossRef] [PubMed]
- Leão, C.; Van Uden, N. Effects of ethanol and other alkanols on passive proton influx in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1984, 774, 43–48. [Google Scholar] [CrossRef]
- Osman, Y.A.; Ingram, L.O. Mechanism of ethanol inhibition of fermentation in Zymomonas mobilis CP4. J. Bacteriol. 1985, 164, 173–180. [Google Scholar] [CrossRef]
- Wendel, U. Assessing Viability and Stress Tolerance of Probiotics—A Review. Front. Microbiol. 2022, 12, 81846. [Google Scholar] [CrossRef]
- Giraffa, G.; Neviani, E. DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 2001, 67, 19–34. [Google Scholar] [CrossRef]
- Davis, C. Enumeration of probiotic strains: Review of culture-dependent and alternative techniques to quantify viable bacteria. J. Microbiol. Methods 2014, 103, 9–17. [Google Scholar] [CrossRef]
- Hansen, S.J.Z.; Tang, P.; Kiefer, A.; Galles, K.; Wong, C.; Morovic, W. Droplet Digital PCR Is an Improved Alternative Method for High-Quality Enumeration of Viable Probiotic Strains. Front. Microbiol. 2020, 10, 3025. [Google Scholar] [CrossRef] [PubMed]
| Stress | Solutions |
|---|---|
| Osmotic stress | 3 mol/L NaCl solution, 1 h, 30 °C. |
| Heat stress | 10 mmol/L phosphate buffer pH7 (PB7), 30 min, 60 °C. |
| Acid stress | 0.1 mol/L Citrate buffer pH 2.0, 1 h, 30 °C. |
| Alkaline stress | 0.2 mol/L glycine-NaOH buffer pH 11, 1 h, 30 °C. |
| Detergent stress | 0.05% (w/v), sodium dodecyl sulfate solution (SDS) 30 min, 30 °C |
| Oxidative stress | 0.1% v/v H2O2 solution, 30 min, 30 °C. |
| Alcoholic stress | 20% ethanol solution, 30 min, 30 °C. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Aouimeur, H.; Boublenza, F.; Gerald, G.; Barani, A.; Makhlouf, Y. Assessment of Stress Tolerance of Enterococcus faecium and Enterococcus durans Strains by Flow Cytometry Using NADS Protocol and Traditional Culture Methods. Appl. Microbiol. 2026, 6, 24. https://doi.org/10.3390/applmicrobiol6020024
Aouimeur H, Boublenza F, Gerald G, Barani A, Makhlouf Y. Assessment of Stress Tolerance of Enterococcus faecium and Enterococcus durans Strains by Flow Cytometry Using NADS Protocol and Traditional Culture Methods. Applied Microbiology. 2026; 6(2):24. https://doi.org/10.3390/applmicrobiol6020024
Chicago/Turabian StyleAouimeur, Hayet, Faiza Boublenza, Grégori Gerald, Aude Barani, and Yasmina Makhlouf. 2026. "Assessment of Stress Tolerance of Enterococcus faecium and Enterococcus durans Strains by Flow Cytometry Using NADS Protocol and Traditional Culture Methods" Applied Microbiology 6, no. 2: 24. https://doi.org/10.3390/applmicrobiol6020024
APA StyleAouimeur, H., Boublenza, F., Gerald, G., Barani, A., & Makhlouf, Y. (2026). Assessment of Stress Tolerance of Enterococcus faecium and Enterococcus durans Strains by Flow Cytometry Using NADS Protocol and Traditional Culture Methods. Applied Microbiology, 6(2), 24. https://doi.org/10.3390/applmicrobiol6020024






