Characterization of the Microbiome and Virulence and Resistance Genes in the Howler Monkey (Alouatta seniculus) in Colombian Andean Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Locations
2.2. Sample Collection and Processing
2.3. Metagenomic Assembly
2.4. Identification of Resistance Genes and Virulence Factors
2.5. Genome Reconstruction
2.6. Resistance Genes and Virulence Factors Associated with Plasmids
3. Results
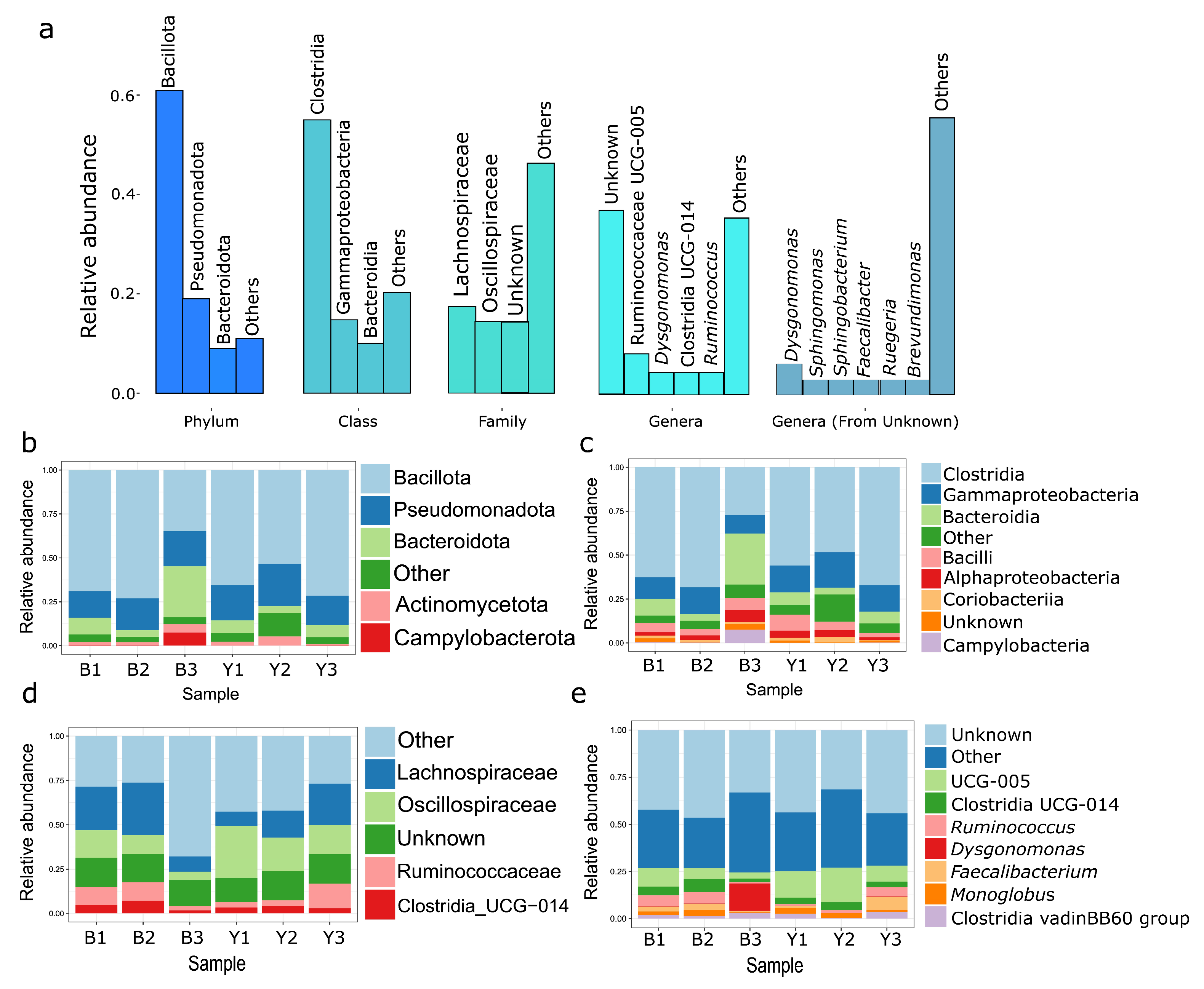
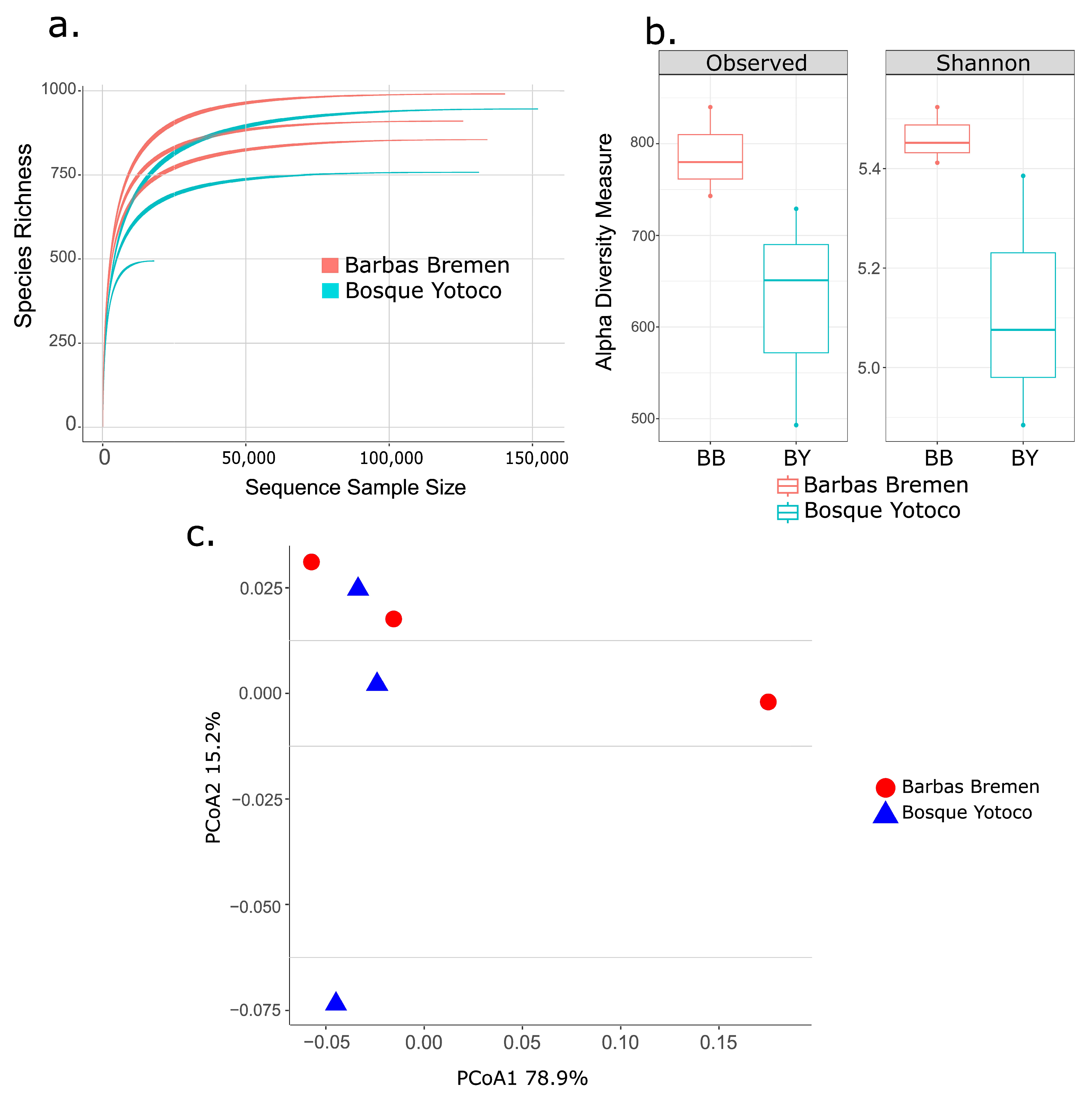
3.1. Microbiome Analysis
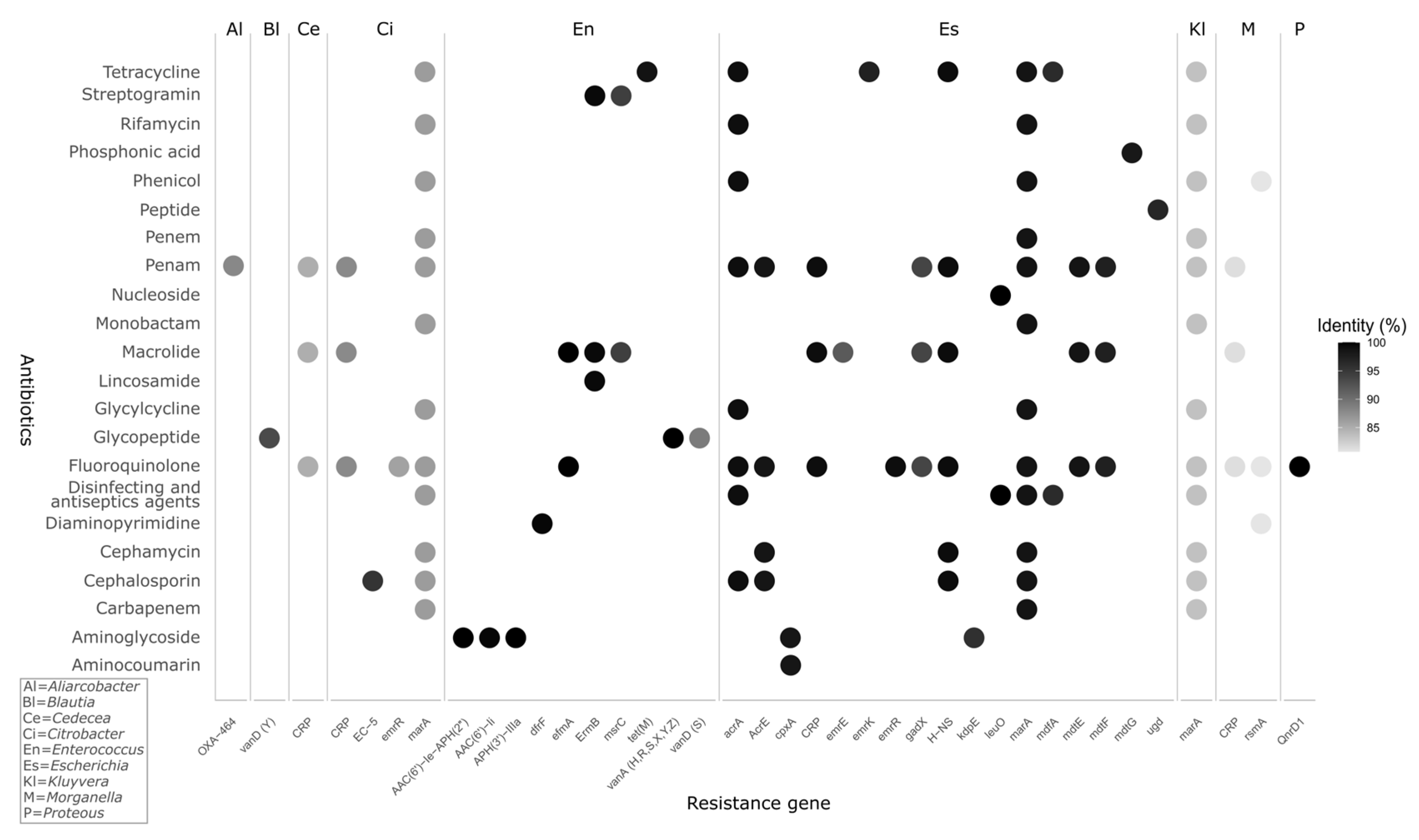
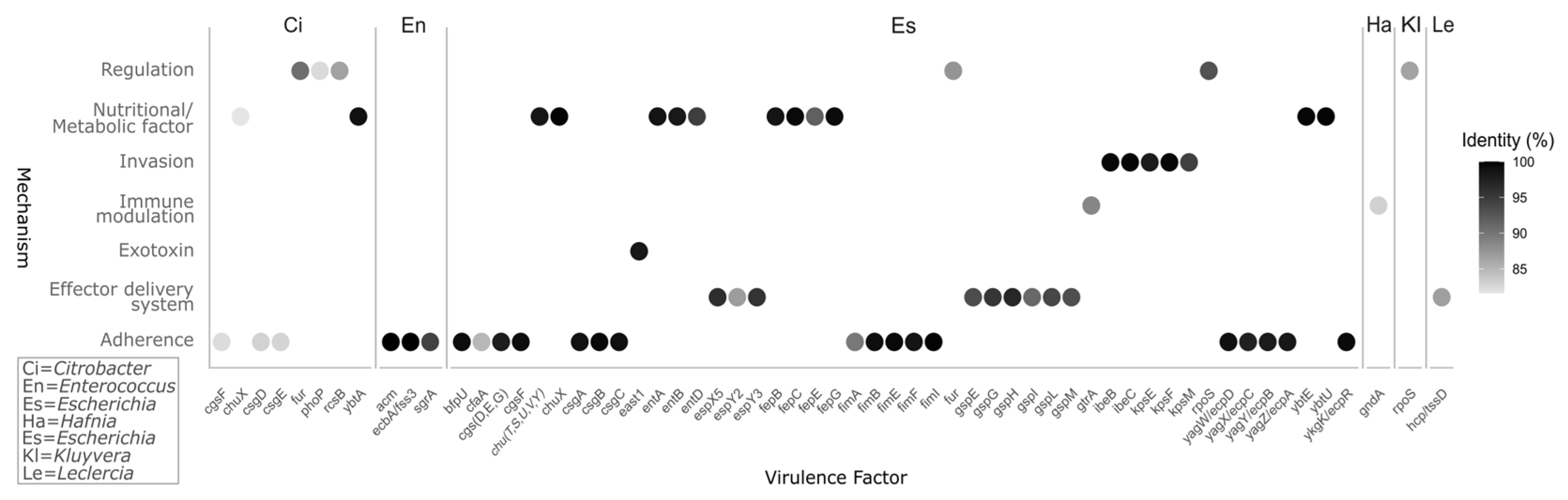
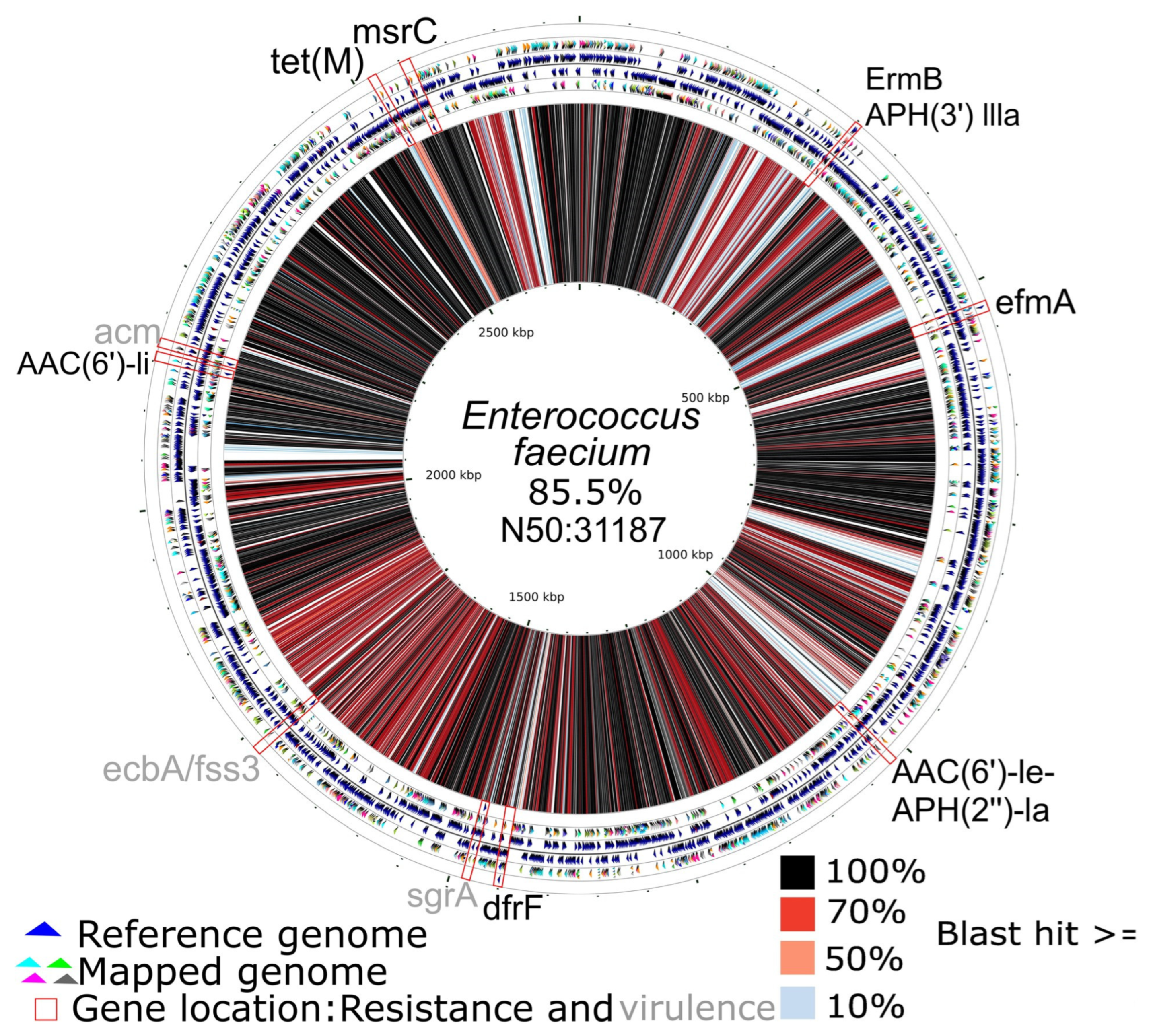
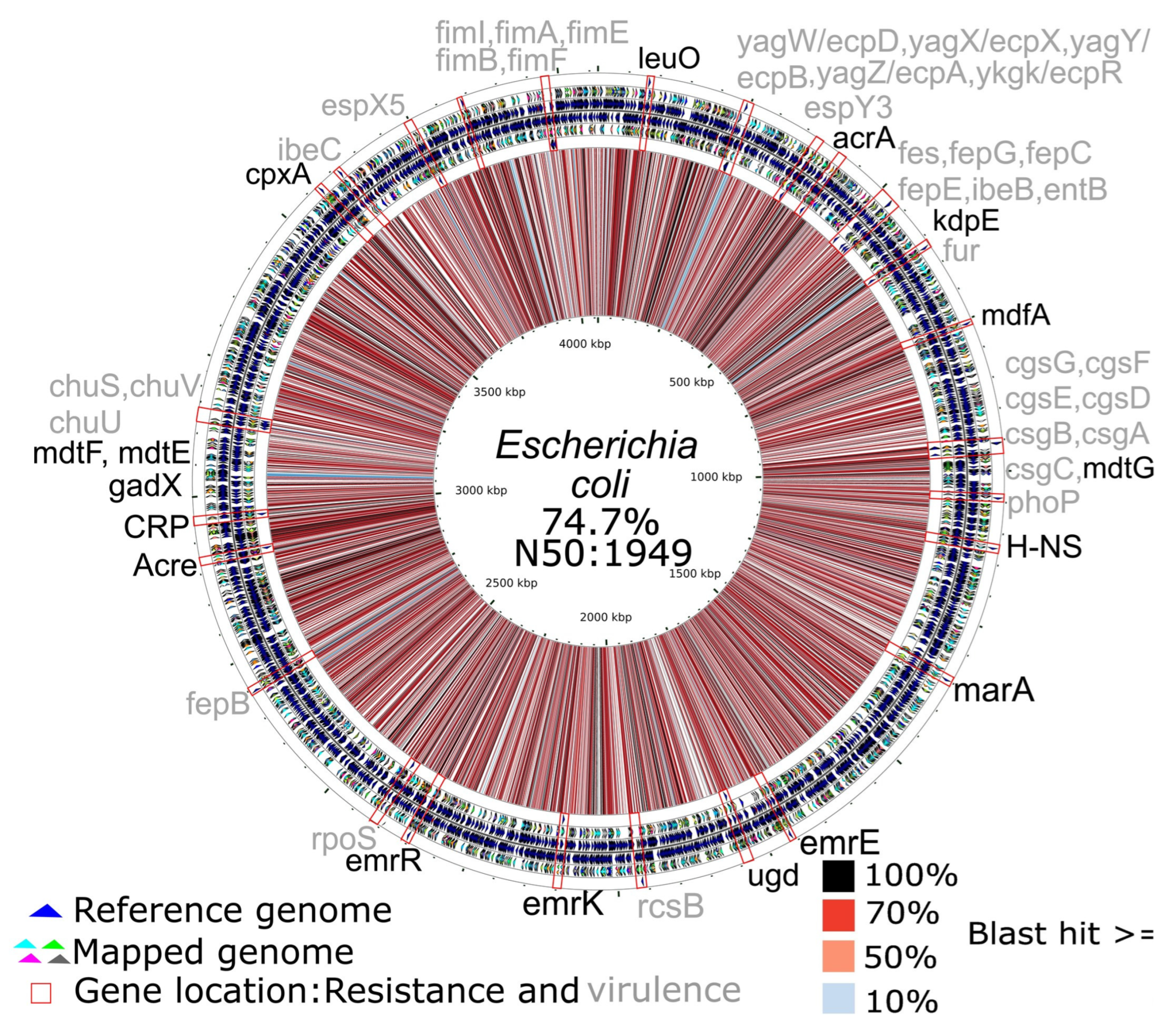
3.2. Resistance Genes and Virulence Factors
3.3. Resistance Genes and Virulence Factors Associated with Plasmids
3.4. Genome Reconstruction
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| rRNA | ribosomal RNA |
| ASV | Amplicon Sequence Variant |
| VF | Virulence Factor |
| ARG | Antibiotic Resistance Genes |
| CARD | Comprehensive Antibiotic Resistance Database |
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp. |
| NCBI | National Center for Biotechnology Information |
| WHO | World Health Organization |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic acid |
References
- Clayton, J.B.; Gomez, A.; Amato, K.; Knights, D.; Travis, D.A.; Blekhman, R.; Knight, R.; Leigh, S.; Stumpf, R.; Wolf, T.; et al. The Gut Microbiome of Nonhuman Primates: Lessons in Ecology and Evolution. Am. J. Primatol. 2018, 80, e22867. [Google Scholar] [CrossRef] [PubMed]
- Moy, M.; Diakiw, L.; Amato, K.R. Human-Influenced Diets Affect the Gut Microbiome of Wild Baboons. Sci. Rep. 2023, 13, 11886. [Google Scholar] [CrossRef]
- Manuzak, J.A.; Zevin, A.S.; Cheu, R.; Richardson, B.; Modesitt, J.; Hensley-McBain, T.; Miller, C.; Gustin, A.T.; Coronado, E.; Gott, T.; et al. Antibiotic-Induced Microbiome Perturbations Are Associated with Significant Alterations to Colonic Mucosal Immunity in Rhesus Macaques. Mucosal Immunol. 2020, 13, 471–480, Erratum in Mucosal Immunol. 2020, 13, 558. [Google Scholar] [CrossRef]
- Bolt, L.; Hadley, C.; Schreier, A. Crowded in a Fragment: High Population Density of Mantled Howler Monkeys (Alouatta palliata) in an Anthropogenically-Disturbed Costa Rican Rainforest. Primate Conserv. 2022, 36, 1–9. [Google Scholar]
- Gómez-Posada, C.; Londoño, J.M. Alouatta seniculus: Density, Home Range and Group Structure in a Bamboo Forest Fragment in the Colombian Andes. Folia Primatol. 2012, 83, 56–65. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The Gut Microbiota Appears to Compensate for Seasonal Diet Variation in the Wild Black Howler Monkey (Alouatta pigra). Microb. Ecol. 2015, 69, 434–443. [Google Scholar] [CrossRef]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Rex Gaskins, H.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat Degradation Impacts Black Howler Monkey (Alouatta pigra) Gastrointestinal Microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Bahrndorff, S.; Alemu, T.; Alemneh, T.; Lund Nielsen, J. The Microbiome of Animals: Implications for Conservation Biology. Int. J. Genom. 2016, 2016, 5304028. [Google Scholar] [CrossRef] [PubMed]
- Laborda, P.; Sanz-García, F.; Ochoa-Sánchez, L.E.; Gil-Gil, T.; Hernando-Amado, S.; Martínez, J.L. Wildlife and Antibiotic Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 873989. [Google Scholar] [CrossRef]
- Mbehang Nguema, P.P.; Onanga, R.; Ndong Atome, G.R.; Tewa, J.J.; Mabika Mabika, A.; Muandze Nzambe, J.U.; Obague Mbeang, J.C.; Bitome Essono, P.Y.; Bretagnolle, F.; Godreuil, S. High Level of Intrinsic Phenotypic Antimicrobial Resistance in Enterobacteria from Terrestrial Wildlife in Gabonese National Parks. PLoS ONE 2021, 16, e0257994. [Google Scholar] [CrossRef]
- Skarżyńska, M.; Leekitcharoenphon, P.; Hendriksen, R.S.; Aarestrup, F.M.; Wasyl, D. A Metagenomic Glimpse into the Gut of Wild and Domestic Animals: Quantification of Antimicrobial Resistance and More. PLoS ONE 2020, 15, e0242987. [Google Scholar] [CrossRef]
- Willmann, M.; Vehreschild, M.J.G.T.; Biehl, L.M.; Vogel, W.; Dörfel, D.; Hamprecht, A.; Seifert, H.; Autenrieth, I.B.; Peter, S. Distinct Impact of Antibiotics on the Gut Microbiome and Resistome: A Longitudinal Multicenter Cohort Study. BMC Biol. 2019, 17, 76. [Google Scholar] [CrossRef]
- Kang, J.; Liu, Y.; Chen, X.; Xu, F.; Wang, H.; Xiong, W.; Li, X. Metagenomic Insights into the Antibiotic Resistomes of Typical Chinese Dairy Farm Environments. Front. Microbiol. 2022, 13, 990272. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Fan, P.; Liu, T.; Yang, A.; Boughton, R.K.; Pepin, K.M.; Miller, R.S.; Jeong, K.C. Transmission of Antibiotic Resistance at the Wildlife-Livestock Interface. Commun. Biol. 2022, 5, 585. [Google Scholar] [CrossRef]
- Vezeau, N.; Kahn, L. Current Understanding and Knowledge Gaps Regarding Wildlife as Reservoirs of Antimicrobial Resistance. Am. J. Vet. Res. 2024, 85, ajvr.24.02.0040. [Google Scholar] [CrossRef]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial Resistance in Escherichia coli Isolates from Swine and Wild Small Mammals in the Proximity of Swine Farms and in Natural Environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef]
- Vásquez-Aguilar, A.A.; Toledo-Manuel, F.O.; Barbachano-Guerrero, A.; Hernández-Rodríguez, D. Detection of Antimicrobial Resistance Genes in Escherichia coli Isolated from Black Howler Monkeys (Alouatta pigra) and Domestic Animals in Fragmented Rain-Forest Areas in Tabasco, Mexico. J. Wildl. Dis. 2020, 56, 922–927. [Google Scholar] [CrossRef]
- Tegner, C.; Sunil-Chandra, N.P.; Wijesooriya, W.R.P.L.I.; Perera, B.V.; Hansson, I.; Fahlman, Å. Detection, Identification, and Antimicrobial Susceptibility of Campylobacter spp. and Salmonella spp. from Free-Ranging Nonhuman Primates in Sri Lanka. J. Wildl. Dis. 2019, 55, 879–884. [Google Scholar] [CrossRef]
- Cristóbal-Azkarate, J.; Dunn, J.C.; Day, J.M.W.; Amábile-Cuevas, C.F. Resistance to Antibiotics of Clinical Relevance in the Fecal Microbiota of Mexican Wildlife. PLoS ONE 2014, 9, e107719. [Google Scholar] [CrossRef] [PubMed]
- García-Restrepo, S.; Montilla, S.O. Taxonomy of the Primates of Colombia: Changes in the Last Twenty Years (2000–2019) and Annotations on Type Localities. Neotrop. Mammal. 2021, 28, 584. [Google Scholar]
- Leal, S.A.Z.; Defler, T.R. Sympatric Distribution of Two Species of Alouatta (A. seniculus and A. palliata: Primates) in Chocó, Colombia. Neotrop. Primates 2013, 20, 1–11. [Google Scholar] [CrossRef]
- Palma, A.C.; Vélez, A.; Gómez-Posada, C.; López, H.; Zárate, D.A.; Stevenson, P.R. Use of Space, Activity Patterns, and Foraging Behavior of Red Howler Monkeys (Alouatta seniculus) in an Andean Forest Fragment in Colombia. Am. J. Primatol. 2011, 73, 1062–1071. [Google Scholar] [CrossRef]
- Dechner, A. Searching for Alouatta palliata in Northern Colombia: Considerations for the Species Detection, Monitoring and Conservation in the Dry Forests of Bolívar, Colombia. Neotrop. Primates 2011, 18, 1–8. [Google Scholar] [CrossRef]
- Riaño Ospina, K. Aspectos Ecológicos de Diez Especies Pioneras Arbóreas en Corredores de Conexión Barbas-Bremen. Bachelor’s Thesis, Universidad del Quindío, Armenia, Colombia, 2003. [Google Scholar]
- Vargas-Salinas, F.; López-Aranda, F. Las Carreteras Pueden Restringir El Movimiento de Pequeños Mamíferos En Bosques Andinos de Colombia? Estudio de Caso En El Bosque de Yotoco, Valle Del Cauca. Caldasia 2012, 34, 409–420. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package; Bioconductor: Boston, MA, USA, 2019. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Tools for Microbiome Analysis in R. Version 2.1.28; 2017–2020. Available online: http://microbiome.github.com/microbiome (accessed on 20 October 2024).
- Oksanen, J.; Simpson, G.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.8-0; 2025. Available online: https://vegandevs.github.io/vegan/ (accessed on 20 October 2024).
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Hahsler, M.; Nagar, A. rBLAST: R Interface for the Basic Local Alignment Search Tool. Bioconductor Version: Release (3.19); R Package Version 0.99.4; 2024. Available online: https://doi.org/10.18129/B9.bioc.rBLAST (accessed on 20 October 2024).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive Analysis of Metagenomics Data for Microbiome Studies and Pathogen Identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Author Correction: Metagenome Analysis Using the Kraken Software Suite. Nat. Protoc. 2024. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing Thousands of Circular Genomes Using the CGView Comparison Tool. BMC Genom. 2012, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shang, J.; Ji, Y.; Sun, Y. PLASMe: A Tool to Identify PLASMid Contigs from Short-Read Assemblies Using Transformer. Nucleic Acids Res. 2023, 51, e83. [Google Scholar] [CrossRef]
- García-Feria, L.; Aguilar-Faisal, J.; Pastor-Nieto, R.; Serio-Silva, J. Changes in vegetation at small landscape scales and captivity alter the gut microbiota of black howler monkeys (Alouatta pigra: Atelidae) Cambios En La Vegetación a Pequeñas Escalas de Paisaje y El Cautiverio Alteran La Microbiota Intestinal de Los Monos Aulladores Negros (Alouatta pigra: Atelidae). Acta Biológica Colomb. 2023, 28, 154–164. [Google Scholar] [CrossRef]
- Bilen, M.; Fonkou, M.D.M.; Dubourg, G.; Tomei, E.; Richez, M.; Delerce, J.; Levasseur, A.; Daoud, Z.; Raoult, D.; Cadoret, F. Dysgonomonas massiliensis sp. nov., a New Species Isolated from the Human Gut and Its Taxonogenomic Description. Antonie Van Leeuwenhoek 2019, 112, 935–945. [Google Scholar] [CrossRef]
- Qin, Z.; Wang, S.; Wu, Y.; Sun, J.; Zhao, F. Seasonal Dynamics of Intestinal Microbiota in Juvenile Chinese Mitten Crab (Eriocheir sinensis) in the Yangtze Estuary. Front. Cell. Infect. Microbiol. 2024, 14, 1436547. [Google Scholar] [CrossRef]
- Izawa, K. Soil-Eating by Alouatta and Ateles. Int. J. Primatol. 1993, 14, 229–242. [Google Scholar] [CrossRef]
- Xi, L.; Wen, X.; Jia, T.; Han, J.; Qin, X.; Zhang, Y.; Wang, Z. Comparative Study of the Gut Microbiota in Three Captive Rhinopithecus Species. BMC Genom. 2023, 24, 398. [Google Scholar] [CrossRef]
- Clayton, J.B.; Al-Ghalith, G.A.; Long, H.T.; Tuan, B.V.; Cabana, F.; Huang, H.; Vangay, P.; Ward, T.; Minh, V.V.; Tam, N.A.; et al. Associations Between Nutrition, Gut Microbiome, and Health in A Novel Nonhuman Primate Model. Sci. Rep. 2018, 8, 11159. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Qu, Q.; Wang, M.; Huang, M.; Zhou, W.; Wei, F. Global Landscape of Gut Microbiome Diversity and Antibiotic Resistomes across Vertebrates. Sci. Total Environ. 2022, 838, 156178. [Google Scholar] [CrossRef] [PubMed]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial Resistance in Wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Yan, Y.; Li, H.; Fayyaz, A.; Gai, Y. Metagenomic and Network Analysis Revealed Wide Distribution of Antibiotic Resistance Genes in Monkey Gut Microbiota. Microbiol. Res. 2022, 254, 126895. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.B.; Travis, D.A.; Lonsdorf, E.V.; Lipende, I.; Elchoufi, D.; Gilagiza, B.; Collins, A.; Kamenya, S.; Tauxe, R.V.; Gillespie, T.R. Antimicrobial Resistance Creates Threat to Chimpanzee Health and Conservation in the Wild. Pathogens 2021, 10, 477. [Google Scholar] [CrossRef]
- Trościańczyk, A.; Nowakiewicz, A.; Osińska, M.; Łagowski, D.; Gnat, S.; Chudzik-Rząd, B. Comparative Characteristics of Sequence Types, Genotypes and Virulence of Multidrug-Resistant Enterococcus Faecium Isolated from Various Hosts in Eastern Poland. Spread of Clonal Complex 17 in Humans and Animals. Res. Microbiol. 2022, 173, 103925. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Barrett, J.B.; Frye, J.G.; Jackson, C.R. Antimicrobial Resistance Gene Detection and Plasmid Typing Among Multidrug Resistant Enterococci Isolated from Freshwater Environment. Microorganisms 2020, 8, 1338. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; López, M.; Guerra, A.; Petrucci-Fonseca, F.; Alcaide, E.; Zorrilla, I.; Serra, R.; Torres, C.; et al. Detection of Vancomycin-Resistant Enterococci from Faecal Samples of Iberian Wolf and Iberian Lynx, Including Enterococcus Faecium Strains of CC17 and the New Singleton ST573. Sci. Total Environ. 2011, 410–411, 266–268. [Google Scholar] [CrossRef]
- Ruiz, C.; Levy Stuart, B. Many Chromosomal Genes Modulate MarA-Mediated Multidrug Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2125–2134. [Google Scholar] [CrossRef]
- Schaffner, S.H.; Lee, A.V.; Pham, M.T.N.; Kassaye, B.B.; Li, H.; Tallada, S.; Lis, C.; Lang, M.; Liu, Y.; Ahmed, N.; et al. Extreme Acid Modulates Fitness Trade-Offs of Multidrug Efflux Pumps MdtEF-TolC and AcrAB-TolC in Escherichia coli K-12. Appl. Environ. Microbiol. 2021, 87, e00724-21. [Google Scholar] [CrossRef]
- Smith, H.E.; Blair, J.M.A. Redundancy in the Periplasmic Adaptor Proteins AcrA and AcrE Provides Resilience and an Ability to Export Substrates of Multidrug Efflux. J. Antimicrob. Chemother. 2014, 69, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Martin Robert, G.; Rosner Judah, L.; Tavio, M. Mar; Vila Jordi Constitutive SoxS Expression in a Fluoroquinolone-Resistant Strain with a Truncated SoxR Protein and Identification of a New Member of the marA-soxS-Rob Regulon, mdtG. Antimicrob. Agents Chemother. 2010, 54, 1218–1225. [Google Scholar] [CrossRef]
- Deng, Z.; Shan, Y.; Pan, Q.; Gao, X.; Yan, A. Anaerobic Expression of the gadE-mdtEF Multidrug Efflux Operon Is Primarily Regulated by the Two-Component System ArcBA through Antagonizing the H-NS Mediated Repression. Front. Microbiol. 2013, 4, 55557. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.; Pasqua, M.; Colonna, B.; Prosseda, G.; Grossi, M. Expression Profile of Multidrug Resistance Efflux Pumps During Intracellular Life of Adherent-Invasive Escherichia coli Strain LF82. Front. Microbiol. 2020, 11, 1935. [Google Scholar] [CrossRef]
- Nishino, K.; Senda, Y.; Yamaguchi, A.; Nishino, K.; Yamaguchi, A.; Nishino, K.; Yamaguchi, A. The AraC-Family Regulator GadX Enhances Multidrug Resistance in Escherichia coli by Activating Expression of mdtEF Multidrug Efflux Genes. J. Infect. Chemother. 2008, 14, 23–29. [Google Scholar] [CrossRef]
- Nishino, K.; Senda, Y.; Yamaguchi, A. CRP Regulator Modulates Multidrug Resistance of Escherichia coli by Repressing the mdtEF Multidrug Efflux Genes. J. Antibiot. 2008, 61, 120–127. [Google Scholar] [CrossRef]
- Maghembe, R.S.; Magulye, M.A.K.; Eilu, E.; Sekyanzi, S.; Makaranga, A.; Mwesigwa, S.; Katagirya, E. A Sophisticated Virulence Repertoire and Colistin Resistance of Citrobacter Freundii ST150 from a Patient with Sepsis Admitted to ICU in a Tertiary Care Hospital in Uganda, East Africa: Insight from Genomic and Molecular Docking Analyses. Infect. Genet. Evol. 2024, 120, 105591. [Google Scholar] [CrossRef]
- Behera, D.U.; Dixit, S.; Gaur, M.; Mishra, R.; Sahoo, R.K.; Sahoo, M.; Behera, B.K.; Subudhi, B.B.; Bharat, S.S.; Subudhi, E. Sequencing and Characterization of M. Morganii Strain UM869: A Comprehensive Comparative Genomic Analysis of Virulence, Antibiotic Resistance, and Functional Pathways. Genes 2023, 14, 1279. [Google Scholar] [CrossRef]
- Thompson, D.K.; Sharkady, S.M. Expanding Spectrum of Opportunistic Cedecea Infections: Current Clinical Status and Multidrug Resistance. Int. J. Infect. Dis. 2020, 100, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Kerek, Á.; Török, B.; Laczkó, L.; Kardos, G.; Bányai, K.; Somogyi, Z.; Kaszab, E.; Bali, K.; Jerzsele, Á. In Vitro Microevolution and Co-Selection Assessment of Florfenicol Impact on Escherichia coli Resistance Development. Antibiotics 2023, 12, 1728. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.S.X.; Ahmad Kamar, A.; Chong, C.W.; Yap, I.K.S.; Teh, C.S.J. Whole Genome Analysis of Multidrug-Resistant Citrobacter Freundii B9-C2 Isolated from Preterm Neonate’s Stool in the First Week. J. Glob. Antimicrob. Resist. 2020, 21, 246–251. [Google Scholar] [CrossRef]
- Shimada, T.; Bridier, A.; Briandet, R.; Ishihama, A. Novel Roles of LeuO in Transcription Regulation of E. coli Genome: Antagonistic Interplay with the Universal Silencer H-NS. Mol. Microbiol. 2011, 82, 378–397. [Google Scholar] [CrossRef] [PubMed]
- Teelucksingh, T.; Thompson, L.K.; Cox, G. The Evolutionary Conservation of Escherichia coli Drug Efflux Pumps Supports Physiological Functions. J. Bacteriol. 2020, 202, e00367-20. [Google Scholar] [CrossRef]
- Aguirre-Sánchez, J.R.; Valdez-Torres, J.B.; del Campo, N.C.; Martínez-Urtaza, J.; del Campo, N.C.; Lee, B.G.; Quiñones, B.; Chaidez-Quiroz, C. Phylogenetic Group and Virulence Profile Classification in Escherichia coli from Distinct Isolation Sources in Mexico. Infect. Genet. Evol. 2022, 106, 105380. [Google Scholar] [CrossRef]
- Ye, D.; Nguyen, P.T.; Bourgault, S.; Couture, M. The Heme Binding Protein ChuX Is a Regulator of Heme Degradation by the ChuS Protein in Escherichia coli O157:H7. J. Inorg. Biochem. 2024, 256, 112575. [Google Scholar] [CrossRef]
- Yuan, C.; Yin, Z.; Wang, J.; Qian, C.; Wei, Y.; Zhang, S.; Jiang, L.; Liu, B. Comparative Genomic Analysis of Citrobacter and Key Genes Essential for the Pathogenicity of Citrobacter Koseri. Front. Microbiol. 2019, 10, 2774. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Swasthi, H.M.; Basalla, J.L.; Dudley, C.E.; Vecchiarelli, A.G.; Chapman, M.R. Cell Surface-Localized CsgF Condensate Is a Gatekeeper in Bacterial Curli Subunit Secretion. Nat. Commun. 2023, 14, 2392. [Google Scholar] [CrossRef]
- Hu, L. Prevalence of Curli Genes among Cronobacter Species and Their Roles in Biofilm Formation and Cell-Cell Aggregation. Int. J. Food Microbiol. 2018, 265, 65–73. [Google Scholar] [CrossRef]
- Aguirre-Sánchez, J.R.; Quiñones, B.; Ortiz-Muñoz, J.A.; Prieto-Alvarado, R.; Vega-López, I.F.; Martínez-Urtaza, J.; Lee, B.G.; Chaidez, C. Comparative Genomic Analyses of Virulence and Antimicrobial Resistance in Citrobacter Werkmanii, an Emerging Opportunistic Pathogen. Microorganisms 2023, 11, 2114. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, R.L. Lag Phase-Associated Iron Accumulation Is Likely a Microbial Counter-Strategy to Host Iron Sequestration: Role of the Ferric Uptake Regulator (Fur). J. Theor. Biol. 2014, 359, 72–79. [Google Scholar] [CrossRef]
- Botta, A.; Barra, N.G.; Lam, N.H.; Chow, S.; Pantopoulos, K.; Schertzer, J.D.; Sweeney, G. Iron Reshapes the Gut Microbiome and Host Metabolism. J. Lipid Atheroscler. 2021, 10, 160–183. [Google Scholar] [CrossRef] [PubMed]
- Schellhorn, H.E. Function, Evolution, and Composition of the RpoS Regulon in Escherichia coli. Front. Microbiol. 2020, 11, 560099. [Google Scholar] [CrossRef]
- Chiang, S.M.; Schellhorn, H.E. Evolution of the RpoS Regulon: Origin of RpoS and the Conservation of RpoS-Dependent Regulation in Bacteria. J. Mol. Evol. 2010, 70, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, J.D. EAST1 Toxin: An Enigmatic Molecule Associated with Sporadic Episodes of Diarrhea in Humans and Animals. J. Microbiol. 2019, 57, 541–549. [Google Scholar] [CrossRef]
- Silva, L.E.; Souza, T.B.; Silva, N.P.; Scaletsky, I.C. Detection and Genetic Analysis of the Enteroaggregative Escherichia coli Heat-Stable Enterotoxin (EAST1) Gene in Clinical Isolates of Enteropathogenic Escherichia coli (EPEC) Strains. BMC Microbiol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Agius, J.E.; Phalen, D.N.; Rose, K.; Eden, J.-S. Genomic Insights Into the Pathogenicity of a Novel Biofilm-Forming Enterococcus sp. Bacteria (Enterococcus lacertideformus) Identified in Reptiles. Front. Microbiol. 2021, 12, 635208. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Top, J.; McNally, A.; Puranen, S.; Pesonen, M.; Pensar, J.; Marttinen, P.; Braat, J.C.; Rogers, M.R.C.; van Schaik, W.; et al. Plasmids Shaped the Recent Emergence of the Major Nosocomial Pathogen Enterococcus Faecium. mBio 2020, 11, e03284-19. [Google Scholar] [CrossRef]
- Simjee, S.; White, D.G.; McDermott, P.F.; Wagner, D.D.; Zervos, M.J.; Donabedian, S.M.; English, L.L.; Hayes, J.R.; Walker, R.D. Characterization of Tn1546 in Vancomycin-Resistant Enterococcus Faecium Isolated from Canine Urinary Tract Infections: Evidence of Gene Exchange between Human and Animal Enterococci. J. Clin. Microbiol. 2002, 40, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Suzuki, M.; Kobayashi, S.; Hirahara, Y.; Kurushima, J.; Hirakawa, H.; Nomura, T.; Tanimoto, K.; Tomita, H. Enterococcal Linear Plasmids Adapt to Enterococcus Faecium and Spread within Multidrug-Resistant Clades. Antimicrob. Agents Chemother. 2023, 67, e01619-22. [Google Scholar] [CrossRef] [PubMed]
- Woegerbauer, M.; Zeinzinger, J.; Springer, B.; Hufnagl, P.; Indra, A.; Korschineck, I.; Hofrichter, J.; Kopacka, I.; Fuchs, R.; Steinwider, J.; et al. Prevalence of the Aminoglycoside Phosphotransferase Genes Aph(3′)-IIIa and Aph(3′)-IIa in Escherichia coli, Enterococcus faecalis, Enterococcus faecium, Pseudomonas aeruginosa, Salmonella enterica subsp. enterica and Staphylococcus aureus Isolates in Austria. J. Med. Microbiol. 2014, 63, 210–217. [Google Scholar] [PubMed]
- Huys, G.; D’Haene, K.; Collard, J.-M.; Swings, J. Prevalence and Molecular Characterization of Tetracycline Resistance in Enterococcus Isolates from Food. Appl. Environ. Microbiol. 2004, 70, 1555–1562. [Google Scholar] [CrossRef]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. Front. Microbiol. 2020, 11, 256. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florez, A.; Patiño-Montoya, A.; Florez-Ríos, H.; Piedrahita, M.; Marmolejo, J.P.A.; Roncancio-Duque, N.; López-Alvarez, D.; Castillo, A. Characterization of the Microbiome and Virulence and Resistance Genes in the Howler Monkey (Alouatta seniculus) in Colombian Andean Forests. Appl. Microbiol. 2025, 5, 129. https://doi.org/10.3390/applmicrobiol5040129
Florez A, Patiño-Montoya A, Florez-Ríos H, Piedrahita M, Marmolejo JPA, Roncancio-Duque N, López-Alvarez D, Castillo A. Characterization of the Microbiome and Virulence and Resistance Genes in the Howler Monkey (Alouatta seniculus) in Colombian Andean Forests. Applied Microbiology. 2025; 5(4):129. https://doi.org/10.3390/applmicrobiol5040129
Chicago/Turabian StyleFlorez, Anyelo, Angie Patiño-Montoya, Hernan Florez-Ríos, Madelaine Piedrahita, Juan Pablo Arias Marmolejo, Néstor Roncancio-Duque, Diana López-Alvarez, and Andrés Castillo. 2025. "Characterization of the Microbiome and Virulence and Resistance Genes in the Howler Monkey (Alouatta seniculus) in Colombian Andean Forests" Applied Microbiology 5, no. 4: 129. https://doi.org/10.3390/applmicrobiol5040129
APA StyleFlorez, A., Patiño-Montoya, A., Florez-Ríos, H., Piedrahita, M., Marmolejo, J. P. A., Roncancio-Duque, N., López-Alvarez, D., & Castillo, A. (2025). Characterization of the Microbiome and Virulence and Resistance Genes in the Howler Monkey (Alouatta seniculus) in Colombian Andean Forests. Applied Microbiology, 5(4), 129. https://doi.org/10.3390/applmicrobiol5040129






