Biodegradation Potential and Taxonomic Composition of Hydrocarbon-Degrading Bacterial Consortia in Diesel-Contaminated Agricultural Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Development of Hydrocarbon-Degrading Bacterial Consortia
2.3. Microcosm-Scale Bioremediation Assays
2.3.1. Evaluation of Biodegradation Potential
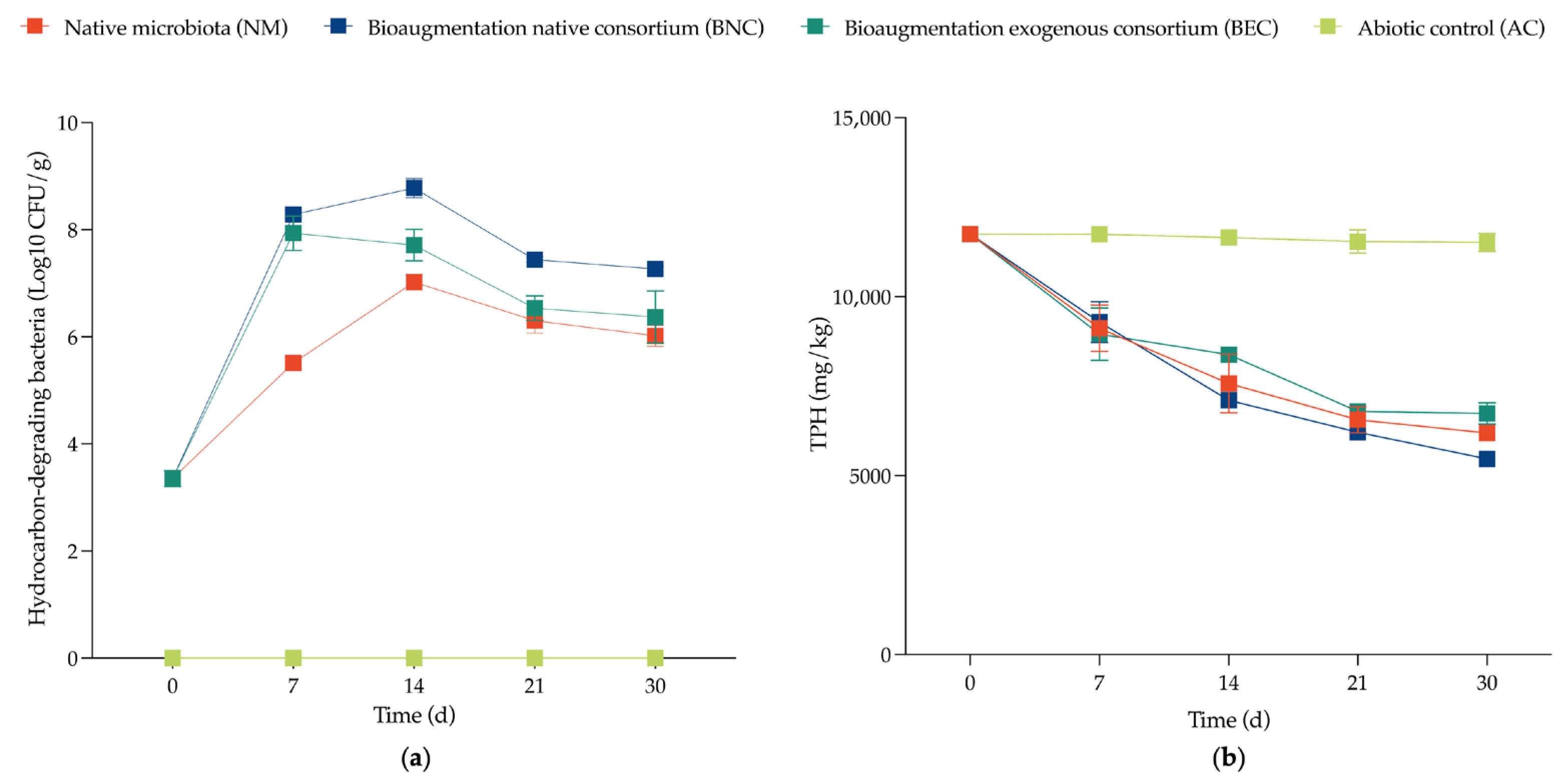
Total Petroleum Hydrocarbons (TPH)
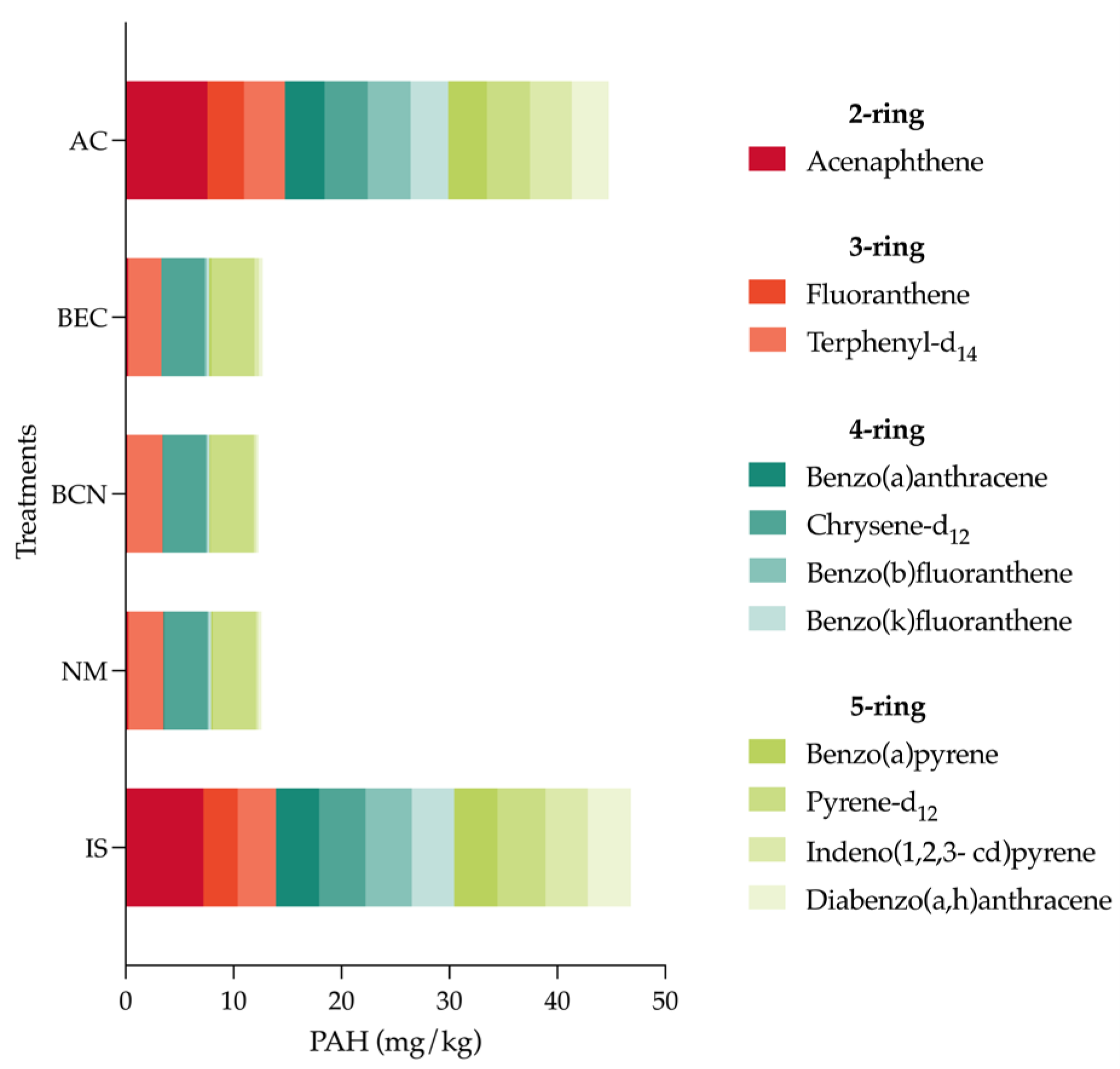
Polycyclic Aromatic Hydrocarbons (PAHs)
2.4. Analysis of Bacterial Community Composition
2.4.1. Dynamics of Cultivable Biodegrading Bacterial Populations
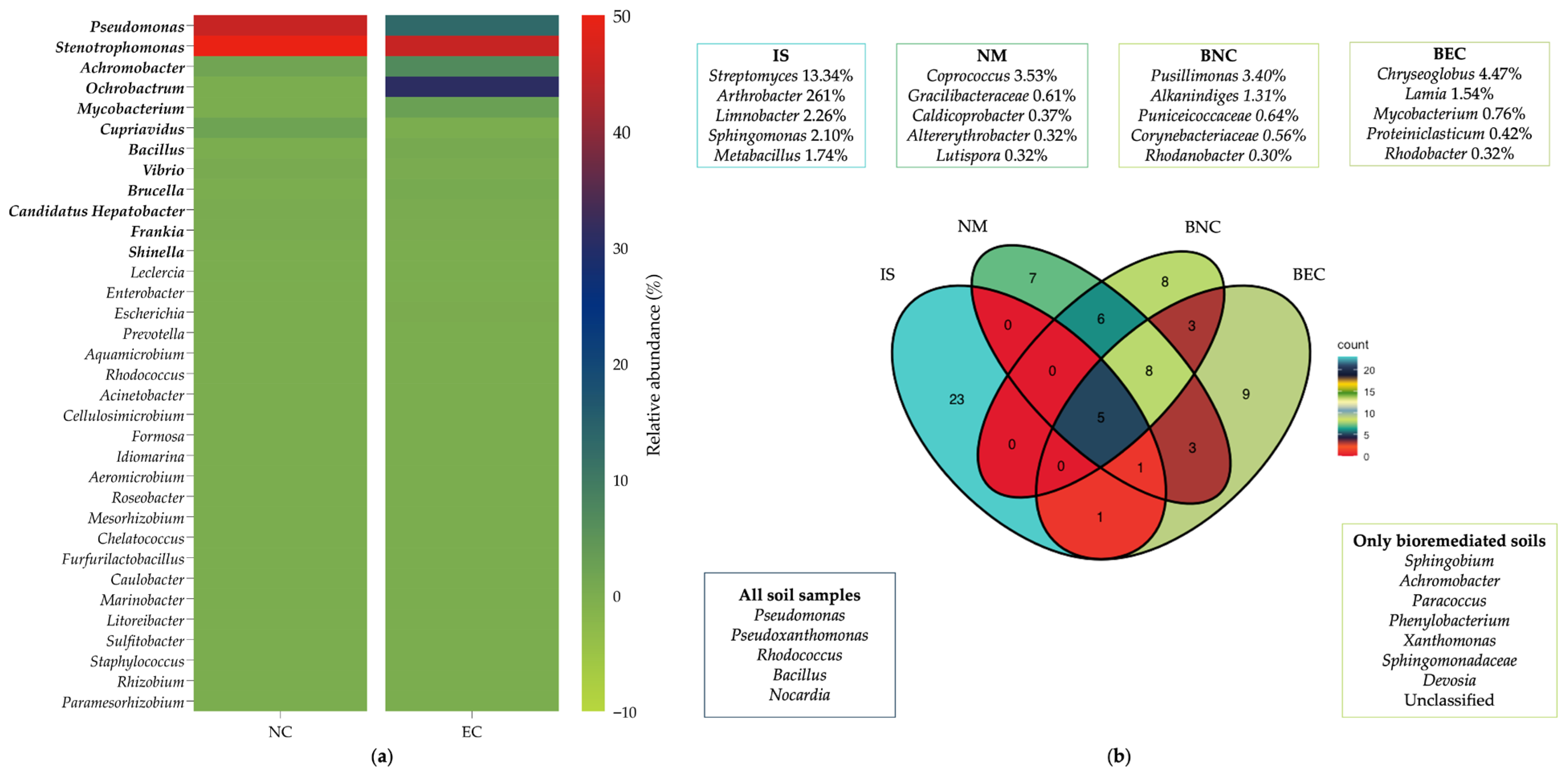
2.4.2. Bacterial Community Taxonomic Profile
2.5. Statistical Analysis
3. Results
3.1. Samples
3.2. Microcosm-Scale Bioremediation Assays
Evaluation of Biodegradation Potential
3.3. Analysis of Bacterial Community Composition
3.3.1. Dynamics of Cultivable Biodegrading Bacterial Populations
3.3.2. Bacterial Community Taxonomic Profile
4. Discussion
4.1. Microcosm-Scale Bioremediation Assays
Evaluation of Biodegradation Potential
4.2. Analysis of Bacterial Community Composition
4.2.1. Dynamics of Cultivable Biodegrading Bacterial Populations
4.2.2. Bacterial Community Taxonomic Profile
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TPH | Total Petroleum Hydrocarbons |
| PAHs | Polycyclic Aromatic Hydrocarbons |
| NC | Native Consortium |
| EC | Exogenous Consortium |
| NM | Native Microbiota |
| BNC | Bioaugmentation with Native Consortium |
| BEC | Bioaugmentation with Exogenous Consortium |
| AC | Abiotic Control |
| CFU | Colony Forming Units |
| DNA | Deoxyribonucleic Acid |
| rRNA | Ribosomal Ribonucleic Acid |
| PCR | Polymerase Chain Reaction |
| ANOVA | Analysis of Variance |
| OTU | Operational Taxonomic Unit |
References
- Stepanova, A.Y.; Gladkov, E.A.; Osipova, E.S.; Gladkova, O.V.; Tereshonok, D.V. Bioremediation of soil from petroleum contamination. Processes 2020, 10, 1224. [Google Scholar] [CrossRef]
- Tripathi, V.; Kumar, V.; Kaur, I.; Kumar, P.; Manickam, N. Unlocking bioremediation potential for site restoration: A comprehensive approach for crude oil degradation in agricultural soil and phytotoxicity assessment. J. Environ. Manag. 2024, 355, 120508. [Google Scholar] [CrossRef]
- Arroyo, S.; Rosano-Ortega, G.; Martínez-Gallegos, S.; Pérez-Armendariz, B.; Vega-Lebrún, C.A. Reduction of hydrocarbons in contaminated soil through paired sorption and advanced oxidation processes. Soil Secur. 2021, 4, 100013. [Google Scholar] [CrossRef]
- Flores-Martinez, I. Oil Theft in Mexico: A Hidden Threat to Public Health; SSRN: Rochester, NY, USA, 2024. [Google Scholar]
- Ambaye, T.G.; Chebbi, A.; Formicola, F.; Prasad, S.; Gomez, F.H.; Franzetti, A.; Vaccari, M. Remediation of soil polluted with petroleum hydrocarbons and its reuse for agriculture: Recent progress, challenges, and perspectives. Chemosphere 2022, 293, 133572. [Google Scholar] [CrossRef]
- Mohanta, S.; Pradhan, B.; Behera, I.D. Impact and remediation of petroleum hydrocarbon pollutants on agricultural land: A review. Geomicrobiol. J. 2023, 41, 345–359. [Google Scholar] [CrossRef]
- Kebede, G.; Tafese, T.; Abda, E.M.; Kamaraj, M.; Assefa, F. Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: Mechanisms and impacts. J. Chem. 2021, 2021, 9823362. [Google Scholar] [CrossRef]
- Chakravarty, P.; Chowdhury, D.; Deka, H. Ecological risk assessment of priority PAHs pollutants in crude oil contaminated soil and its impacts on soil biological properties. J. Hazard. Mater. 2022, 437, 129325. [Google Scholar] [CrossRef]
- Kisić, I.; Hrenović, J.; Zgorelec, Z.; Durn, G.; Brkić, V.; Delač, D. Bioremediation of agriculture soil contaminated by organic pollutants. Energies 2022, 15, 1561. [Google Scholar] [CrossRef]
- Ji, J.; Jiang, M.; Zhang, Y.; Hou, J.; Sun, S. Polycyclic aromatic hydrocarbons contamination in edible oils: A Review. Food Rev. Int. 2022, 39, 6977–7003. [Google Scholar] [CrossRef]
- Elijah, A.A. A review of the petroleum hydrocarbons contamination of soil, water and air and the available remediation techniques, taking into consideration the sustainable development goals. Earthline J. Chem. Sci. 2022, 7, 97–113. [Google Scholar] [CrossRef]
- Davis, S.J.; Alexander, K.; Moreno-Cruz, J.; Hong, C.; Shaner, M.; Caldeira, K.; McKay, I. Food without agriculture. Nat. Sustain. 2023, 7, 90–95. [Google Scholar] [CrossRef]
- Wei, Y.J.; Zhang, Y.J.; Zhu, X.D.; Gu, H.M.; Zhu, Z.Q.; Liu, S.H.; Sun, X.Y.; Jiang, X.L. Effects of diesel hydrocarbon components on cetane number and engine combustion and emission characteristics. Appl. Sci. 2022, 12, 3549. [Google Scholar] [CrossRef]
- Yang, Z.; Shah, K.; Pilon-McCullough, C.; Faragher, R.; Azmi, P.; Hollebone, B.; Fieldhouse, B.; Yang, C.; Dey, D.; Lambert, P.; et al. Characterization of renewable diesel, petroleum diesel and renewable diesel/biodiesel/petroleum diesel blends. Renew. Energy 2024, 224, 120151. [Google Scholar] [CrossRef]
- Curiel-Alegre, S.; Velasco-Arroyo, B.; Rumbo, C.; Khan, A.H.A.; Tamayo-Ramos, J.A.; Rad, C.; Gallego, J.L.R.; Barros, R. Evaluation of biostimulation, bioaugmentation, and organic amendments application on the bioremediation of recalcitrant hydrocarbons of soil. Chemosphere 2022, 307, 135638. [Google Scholar] [CrossRef]
- Haber, L.T.; Pecquet, A.M.; Vincent, M.J.; White, L.M. The long goodbye: Finally moving on from the relative potency approach to a mixtures approach for polycyclic aromatic hydrocarbons (PAHs). Int. J. Environ. Res. Public Health 2022, 19, 9490. [Google Scholar] [CrossRef]
- Barbosa, F.; Rocha, B.A.; Souza, M.C.O.; Bocato, M.Z.; Azevedo, L.F.; Adeyemi, J.A.; Santana, A.; Campiglia, A.D. Polycyclic aromatic hydrocarbons (PAHs): Updated aspects of their determination, kinetics in the human body, and toxicity. J. Toxicol. Environ. Health Part B 2023, 26, 28–65. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Dong, Q.; Li, F.; Wang, T.; Qiu, X.; Zhu, T. A systematic review of polycyclic aromatic hydrocarbon derivatives: Occurrences, levels, biotransformation, exposure biomarkers, and toxicity. Environ. Sci. Technol. 2023, 57, 15314–15335. [Google Scholar] [CrossRef]
- Hrdina, A.I.H.; Kohale, I.N.; Kaushal, S.; Kelly, J.; Selin, N.E.; Engelward, B.P.; Kroll, J.H. The parallel transformations of polycyclic aromatic hydrocarbons in the body and in the atmosphere. Environ. Health Perspect. 2022, 130, 025004. [Google Scholar] [CrossRef]
- Maciejczyk, M.; Tyrpień, K.; Janoszka, B.; Gierat, B.; Muzyka, R. Mutagenic and carcinogenic polycyclic aromatic hydrocarbons (PAHs) in food—Occurrence, human health effects, and assessment methods of exposure. Environ. Med. 2023, 26, 8–15. [Google Scholar] [CrossRef]
- Vijayanand, M.; Ramakrishnan, A.; Subramanian, R.; Issac, P.K.; Nasr, M.; Khoo, K.S.; Rajagopal, R.; Greff, B.; Wan Azelee, N.I.; Jeon, B.H.; et al. Polyaromatic hydrocarbons (PAHs) in the water environment: A review on toxicity, microbial biodegradation, systematic biological advancements, and environmental fate. Environ. Res. 2023, 227, 115716. [Google Scholar] [CrossRef]
- Mekonnen, B.A.; Aragaw, T.A.; Genet, M.B. Bioremediation of petroleum hydrocarbon contaminated soil: A review on principles, degradation mechanisms, and advancements. Front. Environ. Sci. 2024, 12, 1354422. [Google Scholar] [CrossRef]
- Aparicio, J.D.; Raimondo, E.E.; Saez, J.M.; Costa-Gutierrez, S.B.; Álvarez, A.; Benimeli, C.S.; Polti, M.A. The current approach to soil remediation: A review of physicochemical and biological technologies, and the potential of their strategic combination. J. Environ. Chem. Eng. 2022, 10, 107141. [Google Scholar] [CrossRef]
- Baskaran, D.; Byun, H.S. Current trend of polycyclic aromatic hydrocarbon bioremediation: Mechanism, artificial mixed microbial strategy, machine learning, ground application, cost and policy implications. J. Chem. Eng. 2024, 498, 155334. [Google Scholar] [CrossRef]
- Yaashikaa, P.R.; Kumar, P.S. Bioremediation of hazardous pollutants from agricultural soils: A sustainable approach for waste management towards urban sustainability. Environ. Pollut. 2022, 312, 120031. [Google Scholar] [CrossRef]
- Esa, F.N.; Nik Him, N.R. Bioaugmentation as bioremediation approach for contaminated soil: A review. J. Teknol. 2024, 86, 89–102. [Google Scholar] [CrossRef]
- Karishma, S.; Saravanan, A.; Deivayanai, V.C.; Ajithkumar, U.; Yaashikaa, P.R.; Vickram, A.S. Emerging strategies for enhancing microbial degradation of petroleum hydrocarbons: Prospects and challenges. Bioresour. Technol. Rep. 2024, 26, 101866. [Google Scholar] [CrossRef]
- Saleem, H.; Farooq, H.; Mazhar, R.; Shakil, S.; Fazal, S. A review on bioremediation of heavy metals and hydrocarbons through plant growth-promoting bacteria and composting. J. Bioresour. Manag. 2024, 11, 168–185. Available online: https://corescholar.libraries.wright.edu/jbm/vol11/iss1/15/ (accessed on 7 September 2025).
- Chicca, I.; Becarelli, S.; Gregorio, S.D. Microbial involvement in the bioremediation of total petroleum hydrocarbon polluted soils: Challenges and perspectives. Environments 2022, 9, 52. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Q.; Gong, Z. Microbial remediation of petroleum-contaminated soil focused on the mechanism and microbial response: A review. Environ. Sci. Pollut. Res. 2024, 31, 33325–33346. [Google Scholar] [CrossRef]
- Muter, O. Current trends in bioaugmentation tools for bioremediation: A critical review of advances and knowledge gaps. Microorganisms 2023, 11, 710. [Google Scholar] [CrossRef]
- Mishra, P.; Kiran, N.S.; Ferreira, L.F.R.; Yadav, K.K.; Mulla, S.I. New insights into the bioremediation of petroleum contaminants: A systematic review. Chemosphere 2023, 326, 138391. [Google Scholar] [CrossRef]
- Rojas-Vargas, J.; Adaya, L.; Silva-Jiménez, H.; Licea-Navarro, A.F.; Sanchez-Flores, A.; Gracia, A.; Pardo-López, L. Oil-degrading bacterial consortium from Gulf of Mexico designed by a factorial method, reveals stable population dynamics. Front. Mar. Sci. 2022, 9, 962071. [Google Scholar] [CrossRef]
- Wu, H.; Du, X.; Zheng, J.; Song, Q.; Li, X.; Yan, Y.; Tong, K.; Chen, H.; Li, J. Top-Down Consortia for Crude Oil Degradation: Structure and Function Dynamic, Co-Occurrence Network and Bacterial Isolation; SSRN: Rochester, NY, USA, 2024. [Google Scholar] [CrossRef]
- Chen, Y.S.; Huang, Y.H.; Huixiong, L.; Zhao, H.M.; Xiang, L.; Li, H.; Mo, C.H.; Li, Y.W.; Cai, Q.Y. Simultaneous biodegradation of polycyclic aromatic hydrocarbons and phthalates by bacterial consortium and its bioremediation for complex polluted soil and sewage sludge. Bioresour. Technol. 2024, 408, 131161. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, H. Microbial consortia are needed to degrade soil pollutants. Microorganisms 2022, 10, 261. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, X.; Geng, P.; Zeng, Y.; Hu, F.; Sun, P.; Zhuang, G.; Ma, A. Advancing biodegradation of petroleum contaminants by indigenous microbial consortia through assembly strategy innovations. J. Chem. Eng. 2023, 475, 146142. [Google Scholar] [CrossRef]
- Stancu, M.M. Investigating the potential of native soil bacteria for diesel biodegradation. Microorganisms 2025, 13, 564. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Idris, O.A.; Erasmus, M. Metagenomic approaches for optimising hydrocarbon pollution rhizoremediation. Int. J. Environ. Res. 2024, 19, 46. [Google Scholar] [CrossRef]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front. Bioeng. Biotechnol. 2021, 9, 632059. [Google Scholar] [CrossRef]
- Sengupta, K.; Pal, S. A review on microbial diversity and genetic markers involved in methanogenic degradation of hydrocarbons: Futuristic prospects of biofuel recovery from contaminated regions. Environ. Sci. Pollut. Res. 2021, 28, 40288–40307. [Google Scholar] [CrossRef]
- Chunyan, X.; Qaria, M.A.; Qi, X.; Daochen, Z. The role of microorganisms in petroleum degradation: Current development and prospects. Sci. Total Environ. 2023, 865, 161112. [Google Scholar] [CrossRef] [PubMed]
- Diario Oficial de la Federación. Norma Oficial Mexicana NOM-021-SEMARNAT-2000. Especificaciones de Fertilidad, Salinidad y Clasificación de Suelos. Estudios, Muestreo y Análisis. Available online: https://dof.gob.mx/nota_detalle.php?codigo=717582&fecha=31/12/2002#gsc.tab=0 (accessed on 7 September 2025).
- Garrido-Sanz, D.; Manzano, J.; Martín, M.; Redondo-Nieto, M.; Rivilla, R. Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front. Microbiol. 2018, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Mnif, S.; Chebbi, A.; Mhiri, N.; Sayadi, S.; Chamkha, M. Biodegradation of phenanthrene by a bacterial consortium enriched from Sercina oilfield. Process Saf. Environ. Prot. 2017, 107, 44–53. [Google Scholar] [CrossRef]
- Poorsoleiman, M.S.; Hosseini, S.A.; Etminan, A.; Abtahi, H.; Koolivand, A. Bioremediation of Petroleum Hydrocarbons by using a two-step inoculation composting process scaled-up from a mineral-based medium: Effect of biostimulation of an indigenous bacterial strain. Waste Biomass Valori. 2020, 12, 2089–2096. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Method 9071 B. n-Hexane Extractable Material (HEM) for Sludge, Sediment, and Solid Samples. Available online: https://www.epa.gov/sites/default/files/2015-12/documents/9071b.pdf (accessed on 7 September 2025).
- Bolotnik, T.A.; Plyushchenko, I.V.; Smolenkov, A.D.; Pirogov, A.V.; Popik, M.V.; Shpigun, O.A. Identification of spillages of semi-volatile hydrocarbon fuels in soils by as chromatography–mass spectrometry. J. Anal. Chem. 2018, 73, 570–575. [Google Scholar] [CrossRef]
- Ukpaka, C.P.; Ugiri, A.C. Biodegradation kinetics of petroleum hydrocarbon in soil environment using Mangnifera indica seed biomass: A mathematical approach. Chem. Int. 2022, 8, 77–88. [Google Scholar] [CrossRef]
- Wokem, V.C.; Madufuro, C. Application of cowdung and sawdust as biostimulants for enhanced bioremediation of diesel contaminated soil. J. Appl. Sci. Envir. Manag. 2020, 24, 49. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 7, e1. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Jing, G.; Sun, Z.; Wang, H.; Gong, Y.; Huang, S.; Ning, K.; Xu, J.; Su, X. Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 2017, 7, 40371. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Available online: https://ggplot2.tidyverse.org (accessed on 7 September 2025).
- Gao, C.H.; Chen, C.; Akyol, T.; Dușa, A.; Yu, G.; Cao, B.; Cai, P. ggVennDiagram: Intuitive Venn diagram software extended. iMeta 2024, 3, e177. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Kumar, V.; Chaure, K.; Pandey, P. Environmental restoration of polyaromatic hydrocarbon-contaminated soil through sustainable rhizoremediation: Insights into bioeconomy and high-throughput systematic analysis. Environ. Sci. Adv. 2025, 4, 842–883. [Google Scholar] [CrossRef]
- Eze, M.O.; Thiel, V.; Hose, G.C.; George, S.C.; Daniel, R. Enhancing rhizoremediation of petroleum hydrocarbons through bioaugmentation with a plant growth-promoting bacterial consortium. Chemosphere 2022, 289, 133143. [Google Scholar] [CrossRef]
- Curiel-Alegre, S.; Fuente-Vivas, D.; Ali, A.H.; García-Tojal, J.; Velasco-Arroyo, B.; Rumbo, C.; Soja, G.; Rad, C.; Barros, R. Unveiling the capacity of bioaugmentation application, in comparison with biochar and rhamnolipid for TPHs degradation in aged hydrocarbons polluted soil. Environ. Res. 2024, 252, 118880. [Google Scholar] [CrossRef]
- Abena, M.T.B.; Li, T.; Shah, M.N.; Zhong, W. Biodegradation of total petroleum hydrocarbons (TPH) in highly contaminated soils by natural attenuation and bioaugmentation. Chemosphere 2019, 234, 864–874. [Google Scholar] [CrossRef]
- Doustaky, M.; Ebrahimi, S.; Movahedi Naeini, A.R.; Olamaee, M. Monitoring TPH biodegradation in soil around ray oil refinery by natural attenuation, biostimulation and bioaugmentation treatments. DLSR 2022, 1, 57–72. [Google Scholar] [CrossRef]
- Jia, W.; Cheng, L.; Tan, Q.; Liu, Y.; Dou, J.; Yang, K.; Yang, Q.; Wang, S.; Li, J.; Niu, G.; et al. Response of the soil microbial community to petroleum hydrocarbon stress shows a threshold effect: Research on aged realistic contaminated fields. Front. Microbiol. 2023, 14, 1188229. [Google Scholar] [CrossRef]
- Lipatov, A.; Belyanova, E.; Petunina, I. Prediction the biodegradation rate of soil contaminated with different oil concentrations. Results Nonlinear Anal. 2024, 7, 24–34. Available online: https://nonlinear-analysis.com/index.php/pub/article/view/252 (accessed on 4 November 2025).
- Liu, L.; Chen, Y.; Shen, J.; Pan, Y.; Lin, W. Metabolic versatility of soil microbial communities below the rocks of the hyperarid Dalangtan Playa. Appl. Environ. Microbiol. 2023, 89, e01072-23. [Google Scholar] [CrossRef]
- Gupta, N.; Hoque, R.R.; Balachandran, S. In bioremediation strategies of chrysene: A carcinogenic polycyclic aromatic hydrocarbons. Rajesh Publ. 2024, 1, 230–236. [Google Scholar]
- Vaidya, S.; Devpura, N.; Jain, K.; Madamwar, D. Degradation of chrysene by enriched bacterial consortium. Front. Microbiol. 2018, 9, 1333. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Wu, Y.; Fan, Q.; Li, P.; Liang, J.; Liu, Y.H.; Ma, R.; Li, R.; Shi, L. Influence mechanism of organic matter and low-molecular-weight organic acids on the interaction between minerals and PAHs. Sci. Total Environ. 2022, 862, 160872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Luo, Y.; Zhu, Y.; Zhang, H.; Wang, X.; Li, W.; Li, P.; Han, J. Insights into the mechanisms underlying the biodegradation of phenanthrene in biochar-amended soil: From bioavailability to soil microbial communities. Biochar 2023, 5, 14. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yan, K.; Lou, J.; Ding, J.; Snyder, S.A.; Wu, L.; Xu, J. Metabolic interactions in a bacterial co-culture accelerate phenanthrene degradation. J. Hazard. Mater. 2020, 403, 123825. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Qu, C.; Yu, T.; Du, M. Bioremediation of clay with high oil content and biological response after restoration. Sci. Rep. 2021, 11, 9725. [Google Scholar] [CrossRef]
- Fei-Baffoe, B.; Ebenezer, E.Y.A.; Annan, E.L.; Sulemana, A.; Sackey, L.N.A.; Miezah, K.; Bentil, J.; Nang, D.B.; Kazapoe, R.W. Synergistic use of cattle bile, compost and fertilizer amendments in enhancing the bioremediation of hydrocarbon-contaminated soils. Clean. Circ. Bioecon. 2024, 9, 100116. [Google Scholar] [CrossRef]
- Sogbanmu, T.O.; Doherty, V.F.; Buraimoh, O.M.; Arokoyu, O.F. Influence of a dispersant on the types and growth of microbial hydrocarbon degraders in a crude oil-contaminated medium. J. Health Pollut. 2017, 7, 62–70. [Google Scholar] [CrossRef]
- Diario Oficial de la Federación. Norma Oficial Mexicana NOM-138-SEMARNAT/SSA1-2012. Límites Máximos Permisibles de Hidrocarburos en Suelos y Lineamientos para el Muestreo en la Caracterización y Especificaciones para la Remediación. Available online: https://www.dof.gob.mx/nota_detalle_popup.php?codigo=5313544 (accessed on 7 September 2025).
- Galitskaya, P.; Biktasheva, L.; Kuryntseva, P.; Selivanovskaya, S. Response of soil bacterial communities to high petroleum content in the absence of remediation procedures. Environ. Sci. Pollut. Res. 2020, 28, 9610–9627. [Google Scholar] [CrossRef]
- Belykh, E.; Maystrenko, T.; Velegzhaninov, I.; Tavleeva, M.; Rasova, E.; Rybak, A. Taxonomic diversity and functional traits of soil bacterial communities under radioactive contamination: A review. Microorganisms 2024, 12, 733. [Google Scholar] [CrossRef]
- Rahmeh, R.; Akbar, A.; Kumar, V.; Al-Mansour, H.; Kishk, M.; Ahmed, N.; Al-Shamali, M.; Boota, A.; Al-Ballam, Z.; Shajan, A.; et al. Insights into bacterial community involved in bioremediation of aged oil-contaminated soil in arid environment. Evol. Bioinform. Online 2021, 17, 11769343211016887. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Chen, H.; Sun, M.; Wen, J. Comparison of bacterial community structure and function under different petroleum hydrocarbon degradation conditions. Bioprocess Biosyst. Eng. 2020, 43, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L.; Yin, X.; Zhang, T. Polycyclic aromatic hydrocarbon (PAH) biodegradation capacity revealed by a genome-function relationship approach. Environ. Microbiome 2023, 18, 39. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Xin, Y.; Li, C.; Huang, T. Metagenomics-metabolomics analysis of microbial function and metabolism in petroleum-contaminated soil. Braz. J. Microbiol. 2023, 54, 935–947. [Google Scholar] [CrossRef]
- Huang, L.; Ye, J.; Jiang, K.; Wang, Y.; Li, Y. Oil contamination drives the transformation of soil microbial communities: Co-occurrence pattern, metabolic enzymes and culturable hydrocarbon-degrading bacteria. Ecotoxicol. Environ. Saf. 2021, 225, 112740. [Google Scholar] [CrossRef]
- Nong, J.; Peng, P.; Pan, J.; Shen, T.; Xi, Q. Effect of bioaugmentation and biostimulation on hydrocarbon degradation and bacterial community composition in different petroleum-contaminated soil layers. Water Air Soil Pollut. 2023, 234, 189. [Google Scholar] [CrossRef]
- Larsbrink, L.; McKee, L.S. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar] [CrossRef]
- Zhan, C.Y. Microbial decomposition and soil health: Mechanisms and ecological implications. Mol. Soil Biol. 2024, 15, 59–70. [Google Scholar] [CrossRef]
- Wu, B.; Xiu, J.; Yu, L.; Huang, L.; Yi, L.; Ma, Y. Degradation of crude oil in a co-culture system of Bacillus subtilis and Pseudomonas aeruginosa. Front. Microbiol. 2023, 14, 1132831. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Lin, S.; Zhu, X.; Zhang, Z.; Shen, B.; Zhou, S. Fulvic acid enhancing pyrene biodegradation by immobilized Stenotrophomonas maltophilia: Effect and mechanism. Bioresour. Technol. 2024, 403, 130857. [Google Scholar] [CrossRef]
- Song, Y.J.; Zhao, N.L.; Dai, D.R.; Bao, R. Prospects of Pseudomonas in microbial fuel, bioremediation, and sustainability. ChemSusChem 2024, 18, e202401324. [Google Scholar] [CrossRef] [PubMed]
- Vandera, E.; Koukkou, A.I. Bacterial community response to hydrocarbon contamination in soils and marine sediments: A critical review of case studies. In Microbial Ecotoxicology, 1st ed.; Cravo-Laureau, C., Cagnon, C., Lauga, B., Duran, R., Eds.; Springer: Cham, Switzerland, 2017; Volume 1, pp. 185–226. [Google Scholar] [CrossRef]
- Medić, A.B.; Karadžić, I.M. Pseudomonas in environmental bioremediation of hydrocarbons and phenolic compounds- key catabolic degradation enzymes and new analytical platforms for comprehensive investigation. World J. Microbiol. Biotechnol. 2022, 38, 165. [Google Scholar] [CrossRef] [PubMed]
- Yesankar, P.J.; Patil, A.; Kapley, A.; Qureshi, A. Catalytic resilience of multicomponent aromatic ring-hydroxylating dioxygenases in Pseudomonas for degradation of polycyclic aromatic hydrocarbons. World J. Microbiol. Biotechnol 2023, 39, 166. [Google Scholar] [CrossRef] [PubMed]
- Freitas, J.F.; de Lima-Silva, D.F.; Castro, J.N.F.; Agnez-Lima, L.F. Genomic and phenotypic characterization of novel Ochrobactrum species isolated from Brazilian oil reservoirs: Genomic diversity and bioremediation potential. Process Biochem. 2024, 149, 74–84. [Google Scholar] [CrossRef]
- Shi, K.; Yang, Y.; Qiao, Y.; Jiang, Q.; Cheng, D.; Xue, J. Oil degradation ability difference and microbial community successions by Ochrobactrum and Shewanella in different oil-polluted seawater. J. Environ. Chem. Eng. 2022, 10, 108392. [Google Scholar] [CrossRef]
- Jing, J.; Wang, T.; Guo, X.; Huang, P.; Li, C.; Qu, Y. Continuous exogenous bioaugmented remediation of petroleum-contaminated soil: Ecological effects, microbial communities, and mechanisms. J. Environ. Manag. 2025, 393, 127007. [Google Scholar] [CrossRef]
- Haque, S.; Srivastava, N.; Pal, D.B.; Alkhanani, M.F.; Almalki, A.H.; Areeshi, M.Y.; Naiudu, R.; Gupta, V.K. Functional microbiome strategies for the bioremediation of petroleum-hydrocarbon and heavy metal contaminated soils: A review. Sci. Total Environ. 2022, 833, 155222. [Google Scholar] [CrossRef]
- Alori, E.T.; Gabasawa, A.I.; Elenwo, C.E.; Agbeyegbe, O.O. Bioremediation techniques as affected by limiting factors in soil environment. Front. Soil Sci. 2022, 2, 937186. [Google Scholar] [CrossRef]
- Huang, J.; Ai, G.; Liu, N.; Huang, Y. Environmental adaptability and organic pollutant degradation capacity of a novel Rhodococcus species derived from soil in the uninhabited area of the Qinghai-Tibet Plateau. Microorganisms 2022, 10, 1935. [Google Scholar] [CrossRef]
- Zhu, N.; Sun, S.; Guo, X.; Luo, W.; Zhuang, Y.; Lei, T.; Leng, F.; Chen, J.; Wang, Y. Integration of physiology, genomics and microbiomics analyses reveal the biodegradation mechanism of petroleum hydrocarbons by Medicago sativa L. and growth-promoting bacterium Rhodococcus erythropolis KB1. Bioresour. Technol. 2024, 415, 131659. [Google Scholar] [CrossRef]
- Saeed, M.; Ilyas, N.; Bibi, F.; Jayachandran, K.; Dattamudi, S.; Elgorban, A.M. Biodegradation of PAHs by Bacillus marsiflavi, genome analysis and its plant growth promoting potential. Environ. Pollut. 2021, 292, 118343. [Google Scholar] [CrossRef] [PubMed]
- Nonthakaew, N.; Sharkey, L.K.R.; Pidot, S.J. The genus Nocardia as a source of new antimicrobials. Npj Antimicrob. Resist. 2025, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, F.; Lee, S.J.; Lahrach, Z.; St-Arnaud, M.; Hijri, M. Draft Genome of Nocardia canadensis sp. nov. Isolated from petroleum-hydrocarbon-contaminated Soil. Microorganisms 2023, 11, 2972. [Google Scholar] [CrossRef] [PubMed]
- Afordoanyi, D.M.; Akosah, Y.A.; Shnakhova, L.; Saparmyradov, K.; Gilles, R.; Validov, S. biotechnological key genes of the rhodococcus erythropolis mgmm8 genome: Genes for bioremediation, antibiotics, plant protection, and growth stimulation. Microorganisms 2023, 12, 88. [Google Scholar] [CrossRef]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for plant growth promotion and stress resilience: What have we learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef]
- Cao, Z.; Yan, W.; Ding, M.; Yuan, Y. Construction of microbial consortia for microbial degradation of complex compounds. Front. Bioeng. Biotechnol. 2022, 10, 1051233. [Google Scholar] [CrossRef]
- Liu, Z.S.; Wang, K.H.; Han, Q.; Jiang, C.Y.; Liu, S.J.; Li, D.F. Sphingobium sp. SJ10-10 encodes a not-yet-reported chromate reductase and the classical Rieske dioxygenases to simultaneously degrade PAH and reduce chromate. J. Hazard. Mater. 2024, 475, 134889. [Google Scholar] [CrossRef]
- Usman, S.; Yakasai, H.M.; Gimba, M.Y.; Shehu, D.; Jagaba, A.H. Anthracene degradation by Achromobacter xylosoxidans strain BUK_BTEG6 isolated from petrochemical contaminated soil. Case Stud. Chem. Environ. Eng. 2023, 8, 100418. [Google Scholar] [CrossRef]
- Chen, W.; Teng, Y.; Ren, W.; Luo, Y.; Yu, Y. A highly effective polycyclic aromatic hydrocarbon-degrading bacterium, Paracoccus sp. HPD-2, shows opposite remediation potential in two soil types. Pedosphere 2022, 32, 673–685. [Google Scholar] [CrossRef]
- Rodríguez-Uribe, M.L.; Peña-Cabriales, J.J.; del Carmen Rivera-Cruz, M.; Délano-Frier, J.P. Native bacteria isolated from weathered petroleum oil-contaminated soils in Tabasco, Mexico, accelerate the degradation petroleum hydrocarbons in saline soil microcosms. Environ. Technol. Innov. 2021, 23, 101781. [Google Scholar] [CrossRef]
- Osadebe, A.U.; Okoye, O.K. Biodegradation of complex hydrocarbon compounds by Xanthomonas campestris isolated from polluted soil: Influence of monoammonium phosphate as a source of inorganic nutrients. Soil Environ. 2023, 42, 165–176. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, S.; Chen, G.; Sun, S.; Liu, Y.; Chen, H.; Meng, L.; Han, Z.; Zheng, D. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by a novel species of the genus Devosia isolated from the deep-sea region of the Kermadec Trench. Front. Microbiol. 2025, 16, 1584496. [Google Scholar] [CrossRef]
- Yang, S.; Wen, X.; Shi, Y.; Liebner, S.; Jin, H.; Perfumo, A. Hydrocarbon degraders establish at the costs of microbial richness, abundance and keystone taxa after crude oil contamination in permafrost environments. Sci. Rep. 2016, 6, 37473. [Google Scholar] [CrossRef]


| Bioremediation Treatment | Code | Description * |
|---|---|---|
| Native microbiota | NM | Soil |
| Bioaugmentation with native consortium | BNC | Soil + Native consortium ** |
| Bioaugmentation with exogenous consortium | BEC | Soil + Exogenous consortium ** |
| Abiotic control | AC | Soil + HgCl2 2% |
| Treatment | Linear Equation | R-Squared | Biodegradation Rate (mg/kg per Day) |
|---|---|---|---|
| Native microbiota (NM) | y = −181.8x + 10857 | 0.8829 | 181.8 |
| Bioaugmentation with native consortium (BNC) | y = −208.5x + 10967 | 0.9133 | 208.5 |
| Bioaugmentation with exogenous consortium (BEC) | y = −161.9x + 10854 | 0.8564 | 161.9 |
| Abiotic control (AC) | y = −8.975x+ 11771 | 0.9263 | 8.975 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valencia-Luna, G.A.; Lozada-Campos, D.; Pardo-López, L.; Millán-López, K.S.; Loera, O.; Tapia-Hernández, A.; Pérez-Armendáriz, B. Biodegradation Potential and Taxonomic Composition of Hydrocarbon-Degrading Bacterial Consortia in Diesel-Contaminated Agricultural Soils. Appl. Microbiol. 2025, 5, 126. https://doi.org/10.3390/applmicrobiol5040126
Valencia-Luna GA, Lozada-Campos D, Pardo-López L, Millán-López KS, Loera O, Tapia-Hernández A, Pérez-Armendáriz B. Biodegradation Potential and Taxonomic Composition of Hydrocarbon-Degrading Bacterial Consortia in Diesel-Contaminated Agricultural Soils. Applied Microbiology. 2025; 5(4):126. https://doi.org/10.3390/applmicrobiol5040126
Chicago/Turabian StyleValencia-Luna, Gloria Anaí, Damián Lozada-Campos, Liliana Pardo-López, Karla Sofía Millán-López, Octavio Loera, Armando Tapia-Hernández, and Beatriz Pérez-Armendáriz. 2025. "Biodegradation Potential and Taxonomic Composition of Hydrocarbon-Degrading Bacterial Consortia in Diesel-Contaminated Agricultural Soils" Applied Microbiology 5, no. 4: 126. https://doi.org/10.3390/applmicrobiol5040126
APA StyleValencia-Luna, G. A., Lozada-Campos, D., Pardo-López, L., Millán-López, K. S., Loera, O., Tapia-Hernández, A., & Pérez-Armendáriz, B. (2025). Biodegradation Potential and Taxonomic Composition of Hydrocarbon-Degrading Bacterial Consortia in Diesel-Contaminated Agricultural Soils. Applied Microbiology, 5(4), 126. https://doi.org/10.3390/applmicrobiol5040126









