Ruminal Planktonic, Weakly, and Tightly Feed-Adhered Bacterial Community as Affected by Two Trichoderma reesei Enzyme Preparations Fed to Lactating Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. DNA Extraction
2.3. Bioinformatics
2.4. Statistical Analysis
3. Results
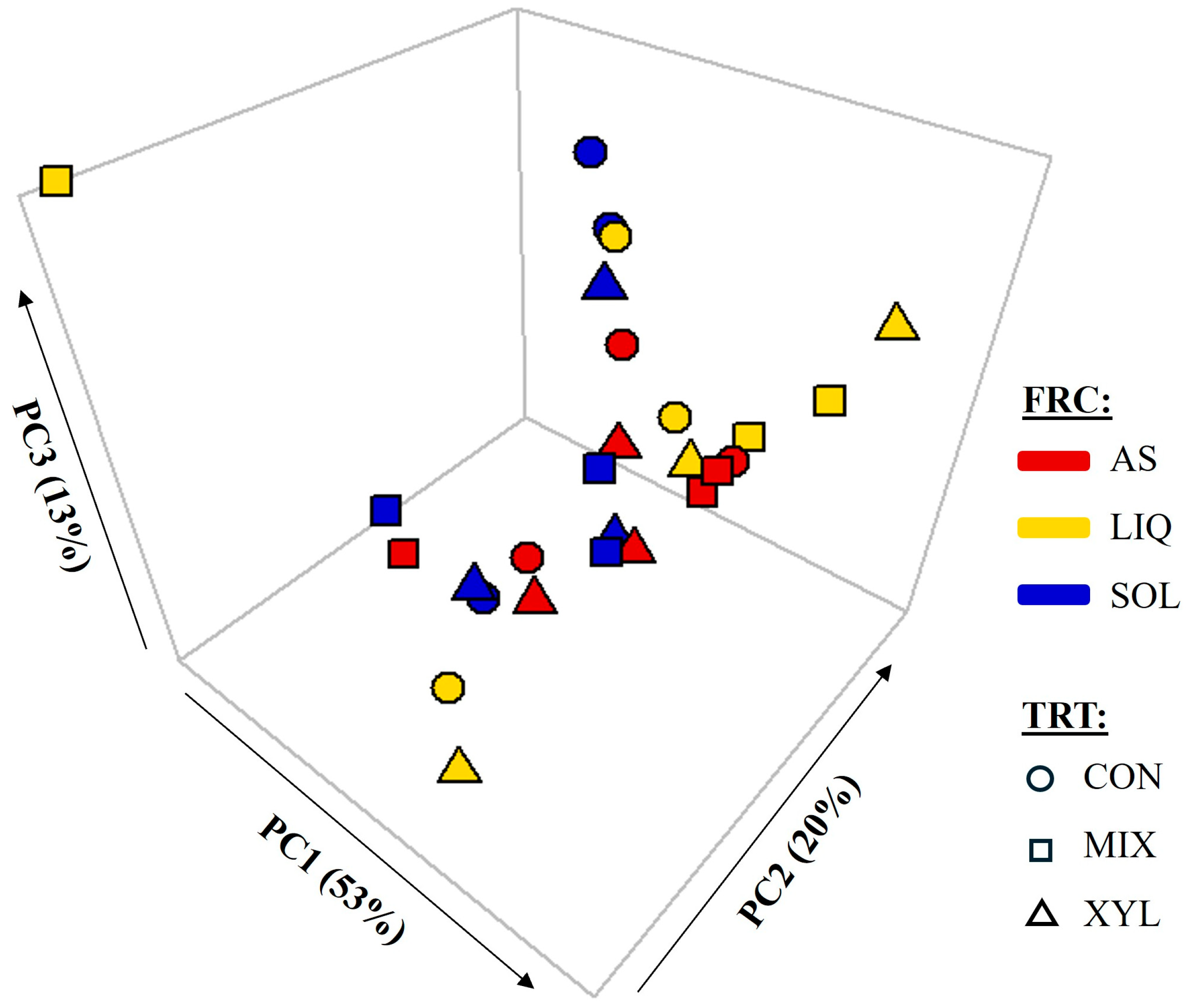
3.1. Alpha and Beta Diversity
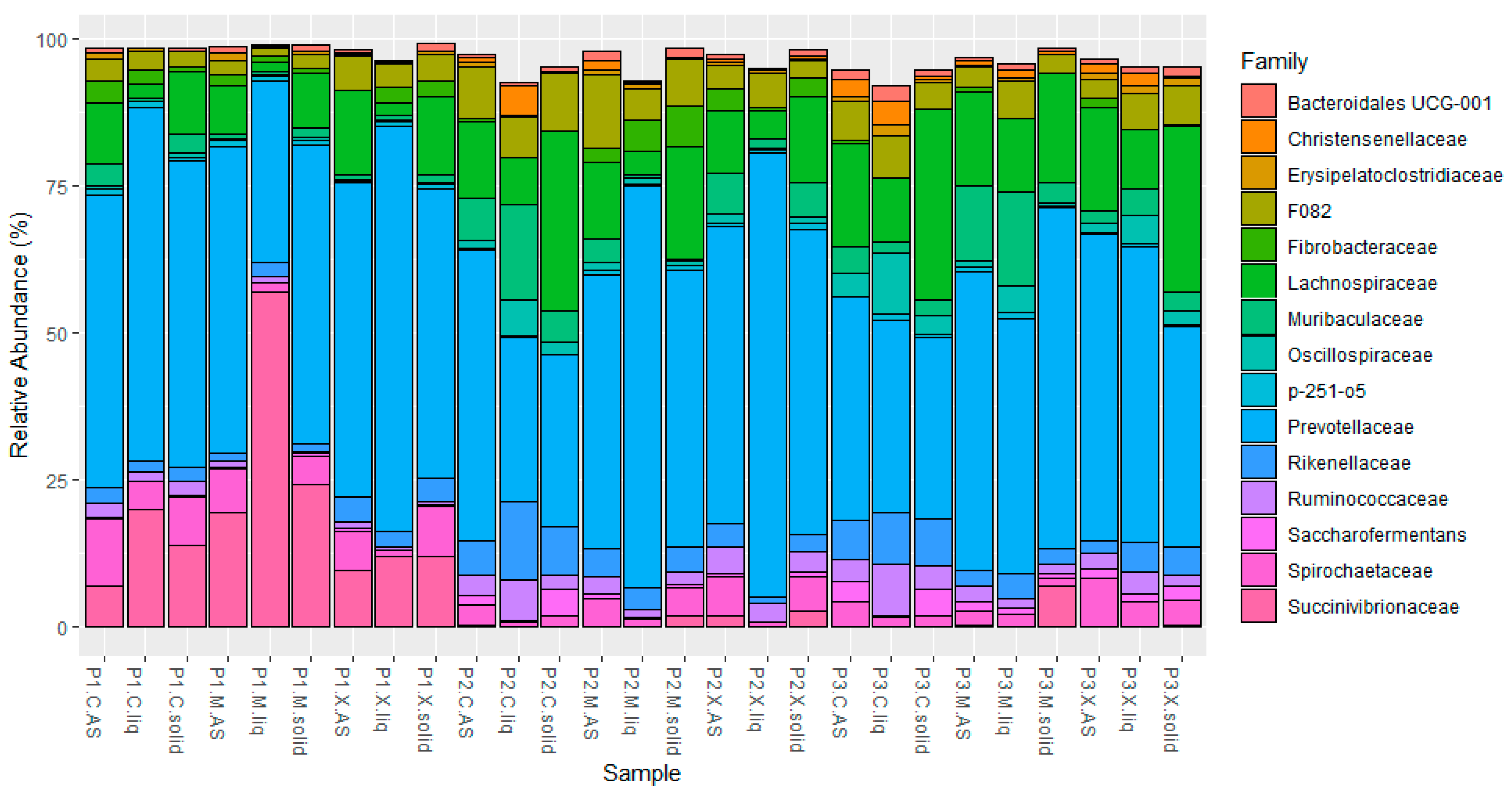
3.2. Relative Abundance
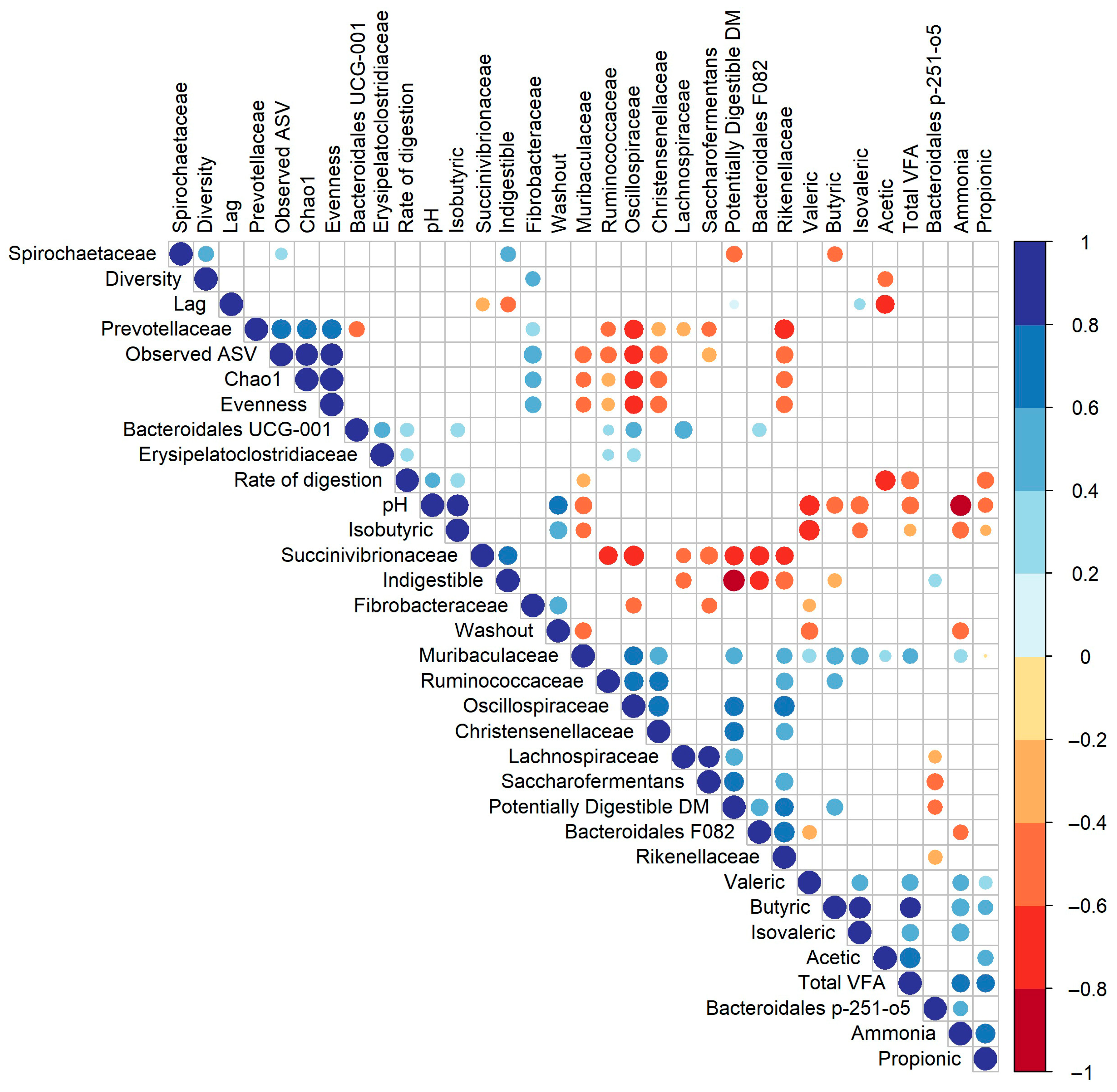
3.3. Correlations
4. Discussion
4.1. Spatial Distribution of Rumen Bacterial Communities
4.2. Supplementation of Fibrolytic Enzymes
4.3. Relationships of Bacterial Relative Abundance with Nutritional Measures
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASV | Amplicon sequence variant |
| AS | Loosely particle-adhered bacterial fraction |
| CON | Untreated control group |
| DM | Dry matter |
| EFE | Exogenous fibrolytic enzymes |
| FRC | Bacterial rumen content fraction |
| LIQ | Planktonic or free-floating bacterial fraction |
| MIX | Cellulase/xylanase enzyme treatment |
| NDF | Neutral detergent fiber |
| PD | Phylogenetic diversity |
| RA | Relative abundance |
| SOL | Tightly particle-adhered bacterial fraction |
| TRT | Treatment |
| XYL | High-xylanase enzyme treatment |
| VFA | Volatile fatty acids |
References
- Nagaraja, T.G. Microbiology of the rumen. In Rumenology; Millen, D.D., De Beni Arrigoni, M., Lauritano Pacheco, R.D., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 39–62. [Google Scholar]
- Moran, J. (Ed.) How the rumen works. In Tropical Dairy Farming: Feeding Management for Small Holder Dairy Farmers in the Humid Tropics; Landlinks Press: Melbourne, Australia, 2005; pp. 41–49. [Google Scholar]
- Weimer, P.J. Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms. Microorganisms 2022, 10, 2345. [Google Scholar] [CrossRef] [PubMed]
- Golder, H.M.; Rehberger, J.; Smith, A.H.; Block, E.; Lean, I.J. Ruminal bacterial communities differ in early-lactation dairy cows with differing risk of ruminal acidosis. Front. Microbiomes 2023, 2, 1212255. [Google Scholar] [CrossRef]
- Mouriño, F.; Akkarawongsa, R.; Weimer, P.J. Initial pH as a Determinant of Cellulose Digestion Rate by Mixed Ruminal Microorganisms In Vitro1. J. Dairy Sci. 2001, 84, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.A.; Penner, G.B.; Chaucheyras-Durand, F.; Guan, L.L. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J. Dairy Sci. 2016, 99, 4955–4966. [Google Scholar] [CrossRef]
- Adesogan, A.T.; Arriola, K.G.; Jiang, Y.; Oyebade, A.; Paula, E.M.; Pech-Cervantes, A.A.; Romero, J.J.; Ferraretto, L.F.; Vyas, D. Symposium review: Technologies for improving fiber utilization. J. Dairy Sci. 2019, 102, 5726–5755. [Google Scholar] [CrossRef] [PubMed]
- Krueger, N.A.; Adesogan, A.T.; Staples, C.R.; Krueger, W.K.; Dean, D.B.; Littell, R.C. The potential to increase digestibility of tropical grasses with a fungal, ferulic acid esterase enzyme preparation. Anim. Feed. Sci. Technol. 2008, 145, 95–108. [Google Scholar] [CrossRef]
- Tirado-González, D.N.; Miranda-Romero, L.A.; Ruíz-Flores, A.; Medina-Cuéllar, S.E.; Ramírez-Valverde, R.; Tirado-Estrada, G. Meta-analysis: Effects of exogenous fibrolytic enzymes in ruminant diets. J. Appl. Anim. Res. 2018, 46, 771–783. [Google Scholar] [CrossRef]
- Beauchemin, K.; Holtshausen, L. Developments in enzyme usage in ruminants. Enzym. Farm Anim. Nutr. 2010, 2, 206–230. [Google Scholar] [CrossRef]
- Arriola, K.G.; Kim, S.C.; Staples, C.R.; Adesogan, A.T. Effect of fibrolytic enzyme application to low- and high-concentrate diets on the performance of lactating dairy cattle. J. Dairy Sci. 2011, 94, 832–841. [Google Scholar] [CrossRef]
- Romero, J.J.; Zarate, M.A.; Arriola, K.G.; Gonzalez, C.F.; Silva-Sanchez, C.; Staples, C.R.; Adesogan, A.T. Screening exogenous fibrolytic enzyme preparations for improved in vitro digestibility of bermudagrass haylage. J. Dairy Sci. 2015, 98, 2555–2567. [Google Scholar] [CrossRef]
- Romero, J.J.; Zarate, M.A.; Adesogan, A.T. Effect of the dose of exogenous fibrolytic enzyme preparations on preingestive fiber hydrolysis, ruminal fermentation, and in vitro digestibility of bermudagrass haylage. J. Dairy Sci. 2015, 98, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Romero, J.J.; Ma, Z.X.; Gonzalez, C.F.; Adesogan, A.T. Effect of adding cofactors to exogenous fibrolytic enzymes on preingestive hydrolysis, in vitro digestibility, and fermentation of bermudagrass haylage. J. Dairy Sci. 2015, 98, 4659–4672. [Google Scholar] [CrossRef]
- Romero, J.J.; Macias, E.G.; Ma, Z.X.; Martins, R.M.; Staples, C.R.; Beauchemin, K.A.; Adesogan, A.T. Improving the performance of dairy cattle with a xylanase-rich exogenous enzyme preparation. J. Dairy Sci. 2016, 99, 3486–3496. [Google Scholar] [CrossRef]
- Wang, Y.; Ramirez-Bribiesca, J.E.; Yanke, L.J.; Tsang, A.; McAllister, T.A. Effect of Exogenous Fibrolytic Enzyme Application on the Microbial Attachment and Digestion of Barley Straw In vitro. Asian-Australas. J. Anim. Sci. 2012, 25, 66–74. [Google Scholar] [CrossRef][Green Version]
- Larue, R.; Yu, Z.; Parisi, V.A.; Egan, A.R.; Morrison, M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 2005, 7, 530–543. [Google Scholar] [CrossRef]
- Mertens, D.R. Dietary fiber components: Relationship to the rate and extent of ruminal digestion. Fed. Proc. 1977, 36, 187–192. [Google Scholar] [PubMed]
- Noel, R.J.; Hambleton, L.G. Collaborative study of a semiautomated method for the determination of crude protein in animal feeds. J. Assoc. Off. Anal. Chem. 1976, 59, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons Ltd.: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Simpson, G.; Solymos, P.; Stevens, M.H.; Szoecs, E.; Wagner, H. Vegan: Community Ecology Package, Version 2.5-7. Comprehensive R Archive Network. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 27 August 2025).
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix, Version 0.84. Comprehensive R Archive Network. 2017. Available online: https://cran.r-project.org/web/packages/corrplot/index.html (accessed on 27 August 2025).
- Montgomery, D.C. Randomized Blocks, Latin Squares, and Related Designs. In Design and Analysis of Experiments, 8th ed.; Montgomery, D.C., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 139–181. [Google Scholar]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Abecia, L.; Angarita, E.; Aravena, P.; Nora Arenas, G.; Ariza, C.; et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Yang, W.Z.; Beauchemin, K.A.; Rode, L.M. Effect of dietary factors on distribution and chemical composition of liquid- or solid-associated bacterial populations in the rumen of dairy cows. J. Anim. Sci. 2001, 79, 2736–2746. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- AlZahal, O.; Li, F.; Guan, L.L.; Walker, N.D.; McBride, B.W. Factors influencing ruminal bacterial community diversity and composition and microbial fibrolytic enzyme abundance in lactating dairy cows with a focus on the role of active dry yeast. J. Dairy Sci. 2017, 100, 4377–4393. [Google Scholar] [CrossRef]
- Deusch, S.; Camarinha-Silva, A.; Conrad, J.; Beifuss, U.; Rodehutscord, M.; Seifert, J. A Structural and Functional Elucidation of the Rumen Microbiome Influenced by Various Diets and Microenvironments. Front. Microbiol. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Palmonari, A.; Federiconi, A.; Formigoni, A. Animal board invited review: The effect of diet on rumen microbial composition in dairy cows. Animal 2024, 18, 101319. [Google Scholar] [CrossRef]
- Pinnell, L.J.; Reyes, A.A.; Wolfe, C.A.; Weinroth, M.D.; Metcalf, J.L.; Delmore, R.J.; Belk, K.E.; Morley, P.S.; Engle, T.E. Bacteroidetes and Firmicutes Drive Differing Microbial Diversity and Community Composition Among Micro-Environments in the Bovine Rumen. Front. Vet. Sci. 2022, 9, 897996. [Google Scholar] [CrossRef]
- Kong, Y.; Teather, R.; Forster, R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol. Ecol. 2010, 74, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Wirth, R.; Kádár, G.; Kakuk, B.; Maróti, G.; Bagi, Z.; Szilágyi, Á.; Rákhely, G.; Horváth, J.; Kovács, K.L. The Planktonic Core Microbiome and Core Functions in the Cattle Rumen by Next Generation Sequencing. Front. Microbiol. 2018, 9, 2285. [Google Scholar] [CrossRef]
- Purushe, J.; Fouts, D.E.; Morrison, M.; White, B.A.; Mackie, R.I.; Coutinho, P.M.; Henrissat, B.; Nelson, K.E.; The North American Consortium for Rumen Bacteria. Comparative Genome Analysis of Prevotella ruminicola and Prevotella bryantii: Insights into Their Environmental Niche. Microb. Ecol. 2010, 60, 721–729. [Google Scholar] [CrossRef]
- Ren, Q.; Si, H.; Yan, X.; Liu, C.; Ding, L.; Long, R.; Li, Z.; Qiu, Q. Bacterial communities in the solid, liquid, dorsal, and ventral epithelium fractions of yak (Bos grunniens) rumen. MicrobiologyOpen 2020, 9, e963. [Google Scholar] [CrossRef]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef]
- Michalet-Doreau, B.; Fernandez, I.; Peyron, C.; Millet, L.; Fonty, G. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents. Reprod. Nutr. Dev. 2001, 41, 187–194. [Google Scholar] [CrossRef]
- Takizawa, S.; Abe, K.; Fukuda, Y.; Feng, M.; Baba, Y.; Tada, C.; Nakai, Y. Recovery of the fibrolytic microorganisms from rumen fluid by flocculation for simultaneous treatment of lignocellulosic biomass and volatile fatty acid production. J. Clean. Prod. 2020, 257, 120626. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Colombatto, D.; Morgavi, D.P.; Yang, W.Z.; Rode, L.M. Mode of action of exogenous cell wall degrading enzymes for ruminants. Can. J. Anim. Sci. 2004, 84, 13–22. [Google Scholar] [CrossRef]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of Probiotics/Prebiotics on Cattle Health and Productivity. Microbes Environ. 2015, 30, 126–132. [Google Scholar] [CrossRef]
- Westland, A.; Martin, R.; White, R.; Martin, J.H. Mannan oligosaccharide prepartum supplementation: Effects on dairy cow colostrum quality and quantity. Animal 2017, 11, 1779–1782. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, K.A.; Yang, W.Z.; Rode, L.M. Effects of grain source and enzyme additive on site and extent of nutrient digestion in dairy cows. J. Dairy Sci. 1999, 82, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Zhou, M.; Holtshausen, L.; Alexander, T.W.; McAllister, T.A.; Guan, L.L.; Oba, M.; Beauchemin, K.A. A fibrolytic enzyme additive for lactating Holstein cow diets: Ruminal fermentation, rumen microbial populations, and enteric methane emissions. J. Dairy Sci. 2012, 95, 1419–1427. [Google Scholar] [CrossRef]
- McCann, J.C.; Wickersham, T.A.; Loor, J.J. High-throughput Methods Redefine the Rumen Microbiome and Its Relationship with Nutrition and Metabolism. Bioinform. Biol. Insights 2014, 2014, 109–125. [Google Scholar] [CrossRef]
- Paz, H.A.; Anderson, C.L.; Muller, M.J.; Kononoff, P.J.; Fernando, S.C. Rumen Bacterial Community Composition in Holstein and Jersey Cows Is Different under Same Dietary Condition and Is Not Affected by Sampling Method. Front. Microbiol. 2016, 7, 1206. [Google Scholar] [CrossRef]
- Liu, Z.K.; Li, Y.; Zhao, C.C.; Liu, Z.J.; Wang, L.M.; Li, X.Y.; Pellikaan, W.F.; Yao, J.H.; Cao, Y.C. Effects of a combination of fibrolytic and amylolytic enzymes on ruminal enzyme activities, bacterial diversity, blood profile and milk production in dairy cows. Animal 2022, 16, 100595. [Google Scholar] [CrossRef]
- Silva, K.G.S.; Favero, I.G.; Nardi, K.T.; Kondratovich, L.B.; Hoffmann, C.A.; Hinds, J.K.; Hall, N.; Henry, D.D.; Sarturi, J.O. Effects of exogenous fibrolytic enzymes on beef cattle fed growing diets: Ruminal microbiome. J. Anim. Sci. 2020, 98, 425. [Google Scholar] [CrossRef]
- Li, M.; Zi, X.; Yang, H.; Ji, F.; Tang, J.; Lv, R.; Zhou, H. Effects of king grass and sugarcane top in the absence or presence of exogenous enzymes on the growth performance and rumen microbiota diversity of goats. Trop. Anim. Health Prod. 2021, 53, 106. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, J.M.; Callaway, T.R.; Kieran, T.J.; Glenn, T.C.; McCann, J.C.; Stewart, R.L. Analysis of the Rumen Microbiota of Beef Calves Supplemented During the Suckling Phase. Front. Microbiol. 2019, 10, 1131. [Google Scholar] [CrossRef]
- Wu, X.; Huang, S.; Huang, J.; Peng, P.; Liu, Y.; Han, B.; Sun, D. Identification of the Potential Role of the Rumen Microbiome in Milk Protein and Fat Synthesis in Dairy Cows Using Metagenomic Sequencing. Animals 2021, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Guan, L.L.; Liu, J.X. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef]
- Lopes, D.R.G.; de Souza Duarte, M.; La Reau, A.J.; Chaves, I.Z.; de Oliveira Mendes, T.A.; Detmann, E.; Bento, C.B.P.; Mercadante, M.E.Z.; Bonilha, S.F.M.; Suen, G.; et al. Assessing the relationship between the rumen microbiota and feed efficiency in Nellore steers. J. Anim. Sci. Biotechnol. 2021, 12, 79. [Google Scholar] [CrossRef]
- Rubino, F.; Carberry, C.; Waters, S.M.; Kenny, D.; McCabe, M.S.; Creevey, C.J. Divergent functional isoforms drive niche specialisation for nutrient acquisition and use in rumen microbiome. ISME J. 2017, 11, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef]
- Cunha, C.S.; Veloso, C.M.; Marcondes, M.I.; Mantovani, H.C.; Tomich, T.R.; Pereira, L.G.R.; Ferreira, M.F.L.; Dill-McFarland, K.A.; Suen, G. Assessing the impact of rumen microbial communities on methane emissions and production traits in Holstein cows in a tropical climate. Syst. Appl. Microbiol. 2017, 40, 492–499. [Google Scholar] [CrossRef]
- Indugu, N.; Vecchiarelli, B.; Baker, L.D.; Ferguson, J.D.; Vanamala, J.K.P.; Pitta, D.W. Comparison of rumen bacterial communities in dairy herds of different production. BMC Microbiol. 2017, 17, 190. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, M.L.; Cersosimo, L.M.; Wright, A.D.; Kraft, J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 2016, 92, fiw059. [Google Scholar] [CrossRef] [PubMed]

| EFE 1 | Endoglucanase | Exoglucanase | Xylanase | Protein |
|---|---|---|---|---|
| XYL | 2714 | 1.21 | 26,926 | 92.07 |
| MIX | 2087 | 1.65 | 8791 | 79.65 |
| Treatment | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|
| CON | MIX | XYL | Mean | SEM | TRT | FRC | TRT × FRC | |
| Observed ASVs | 46.2 | 0.026 | 0.629 | 0.565 | ||||
| SOL | 685 | 800 | 760 | 748 | ||||
| AS | 730 | 757 | 822 | 770 | ||||
| LIQ | 656 | 729 | 819 | 735 | ||||
| Mean | 690 b | 762 ab | 800 a | |||||
| Chao1 | 49.8 | 0.042 | 0.531 | 0.302 | ||||
| SOL | 773 | 864 | 805 | 814 | ||||
| AS | 789 | 799 | 879 | 822 | ||||
| LIQ | 687 | 773 | 884 | 782 | ||||
| Mean | 750 b | 812 ab | 856 a | |||||
| Simpson’s Evenness | 0.02 | 0.651 | 0.965 | 0.851 | ||||
| SOL | 0.574 | 0.557 | 0.577 | 0.569 | ||||
| AS | 0.570 | 0.566 | 0.582 | 0.573 | ||||
| LIQ | 0.553 | 0.478 | 0.584 | 0.574 | ||||
| Mean | 0.566 | 0.569 | 0.581 | |||||
| Phylogenetic Diversity | 1.61 | 0.380 | 0.705 | 0.948 | ||||
| SOL | 36.2 | 33.6 | 36.3 | 35.4 | ||||
| AS | 36.1 | 35.7 | 37.4 | 36.4 | ||||
| LIQ | 35.9 | 35.0 | 35.9 | 35.6 | ||||
| Mean | 36.1 | 34.8 | 36.5 | |||||
| Treatment | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|
| CON | MIX | XYL | Mean | SEM | TRT | FRC | TRT × FRC | |
| Bacteroidota | 3.25 | 0.006 | <0.001 | 0.182 | ||||
| SOL | 54.3 | 63.0 | 60.6 | 59.3 C | ||||
| AS | 63.9 | 67.0 | 64.7 | 65.2 B | ||||
| LIQ | 62.7 | 76.2 | 77.8 | 72.2 A | ||||
| Mean | 60.3 b | 68.7 a | 67.7 a | |||||
| Firmicutes | 3.53 | <0.001 | 0.003 | 0.334 | ||||
| SOL | 36.4 | 19.6 | 26.0 | 27.3 A | ||||
| AS | 25.4 | 19.5 | 22.1 | 22.3 B | ||||
| LIQ | 26.2 | 12.6 | 14.2 | 17.6 B | ||||
| Mean | 29.3 a | 17.2 b | 20.7 b | |||||
| Proteobacteria | 1.78 | 0.078 | 0.208 | 0.281 | ||||
| SOL | 4.7 | 11.1 | 5.1 | 7.0 | ||||
| AS | 2.5 | 6.7 | 4.0 | 4.4 | ||||
| LIQ | 6.9 | 5.3 | 4.2 | 5.4 | ||||
| Mean | 4.7 ab | 7.7 a | 4.4 b | |||||
| Spirochaetota | 1.18 | 0.251 | 0.003 | 0.814 | ||||
| SOL | 4.0 | 3.7 | 6.1 | 4.6 A | ||||
| AS | 6.4 | 4.9 | 7.1 | 6.2 A | ||||
| LIQ | 2.4 | 1.7 | 2.0 | 2.0 B | ||||
| Mean | 4.3 | 3.4 | 5.1 | |||||
| Fibrobacterota | 1.02 | 0.339 | 0.919 | 0.821 | ||||
| SOL | 0.3 | 2.5 | 2.0 | 1.6 | ||||
| AS | 1.6 | 1.7 | 1.7 | 1.7 | ||||
| LIQ | 0.8 | 2.1 | 1.2 | 1.4 | ||||
| Mean | 0.9 | 2.1 | 1.6 | |||||
| Treatment | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|
| CON | MIX | XYL | Mean | SEM | TRT | FRC | TRT × FRC | |
| Prevotellaceae | 6.29 | 0.032 | 0.456 | 0.269 | ||||
| SOL | 37.4 | 51.9 | 46.1 | 45.1 | ||||
| AS | 45.6 | 49.7 | 51.9 | 49.1 | ||||
| LIQ | 40.1 | 47.5 | 64.7 | 50.8 | ||||
| Mean | 41.0 b | 49.7 ab | 54.2 a | |||||
| Lachnospiraceae | 2.26 | 0.146 | <0.001 | 0.359 | ||||
| SOL | 24.5 | 15.8 | 18.6 | 19.6 A | ||||
| AS | 13.6 | 12.3 | 14.1 | 13.3 B | ||||
| LIQ | 7.08 | 6.10 | 5.63 | 6.27 C | ||||
| Mean | 15.1 | 11.4 | 12.8 | |||||
| Succinivibrionaceae | 1.79 | 0.092 | 0.207 | 0.285 | ||||
| SOL | 4.66 | 11.1 | 5.04 | 6.92 | ||||
| AS | 2.47 | 6.59 | 3.89 | 4.32 | ||||
| LIQ | 6.70 | 5.02 | 4.09 | 5.27 | ||||
| Mean | 4.61 ab | 7.55 a | 4.34 b | |||||
| Bacteroidales F082 | 1.38 | 0.503 | 0.809 | 0.842 | ||||
| SOL | 5.62 | 4.52 | 4.83 | 5.00 | ||||
| AS | 6.43 | 6.12 | 4.33 | 5.63 | ||||
| LIQ | 5.80 | 4.36 | 5.32 | 5.16 | ||||
| Mean | 5.95 | 5.00 | 4.83 | |||||
| Spirochaetaceae | 1.18 | 0.251 | 0.003 | 0.814 | ||||
| SOL | 4.00 | 3.67 | 6.14 | 4.60 A | ||||
| AS | 6.42 | 4.92 | 7.13 | 6.15 A | ||||
| LIQ | 2.40 | 1.66 | 2.03 | 2.03 B | ||||
| Mean | 4.27 | 3.42 | 5.10 | |||||
| Rikenellaceae | 1.26 | 0.003 | 0.556 | 0.534 | ||||
| SOL | 6.09 | 2.73 | 3.90 | 4.24 | ||||
| AS | 5.07 | 2.95 | 3.48 | 3.83 | ||||
| LIQ | 8.00 | 3.37 | 3.00 | 4.79 | ||||
| Mean | 6.38 a | 3.02 b | 3.46 b | |||||
| Muribaculaceae | 2.67 | 0.625 | 0.637 | 0.853 | ||||
| SOL | 3.74 | 1.76 | 3.49 | 3.00 | ||||
| AS | 5.12 | 5.86 | 3.32 | 4.77 | ||||
| LIQ | 6.14 | 5.59 | 2.26 | 4.66 | ||||
| Mean | 5.00 | 4.41 | 3.02 | |||||
| Ruminococcaceae | 0.69 | 0.003 | 0.172 | 0.179 | ||||
| SOL | 2.90 | 1.36 | 1.95 | 2.07 | ||||
| AS | 3.31 | 2.20 | 2.68 | 2.73 | ||||
| LIQ | 5.73 | 1.44 | 2.40 | 3.19 | ||||
| Mean | 4.00 a | 1.67 b | 2.34 b | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Killerby, M.A.; Romero, J.J.; Ma, Z.; Adesogan, A.T. Ruminal Planktonic, Weakly, and Tightly Feed-Adhered Bacterial Community as Affected by Two Trichoderma reesei Enzyme Preparations Fed to Lactating Cattle. Appl. Microbiol. 2025, 5, 93. https://doi.org/10.3390/applmicrobiol5030093
Killerby MA, Romero JJ, Ma Z, Adesogan AT. Ruminal Planktonic, Weakly, and Tightly Feed-Adhered Bacterial Community as Affected by Two Trichoderma reesei Enzyme Preparations Fed to Lactating Cattle. Applied Microbiology. 2025; 5(3):93. https://doi.org/10.3390/applmicrobiol5030093
Chicago/Turabian StyleKillerby, Marjorie A., Juan J. Romero, Zhengxin Ma, and Adegbola T. Adesogan. 2025. "Ruminal Planktonic, Weakly, and Tightly Feed-Adhered Bacterial Community as Affected by Two Trichoderma reesei Enzyme Preparations Fed to Lactating Cattle" Applied Microbiology 5, no. 3: 93. https://doi.org/10.3390/applmicrobiol5030093
APA StyleKillerby, M. A., Romero, J. J., Ma, Z., & Adesogan, A. T. (2025). Ruminal Planktonic, Weakly, and Tightly Feed-Adhered Bacterial Community as Affected by Two Trichoderma reesei Enzyme Preparations Fed to Lactating Cattle. Applied Microbiology, 5(3), 93. https://doi.org/10.3390/applmicrobiol5030093






